Abstract
Follicle center lymphoma (FCL) is an indolent low-grade B-cell non-Hodgkin's lymphoma (NHL) that frequently transforms to aggressive diffuse large B-cell lymphoma (DLBCL). Histologic transformation of FCL is commonly associated with accumulation of secondary genetic alterations. The BCL-6 gene is altered by chromosomal rearrangements and mutations clustering in its 5′ noncoding regulatory region in up to 70% of primary DLBCL, but in a significantly smaller subset of FCL. Previous studies have shown that both chromosomal rearrangements and mutations could deregulateBCL-6 expression. To evaluate the association between progressive accumulation of BCL-6 regulatory region mutations and the histologic transformation of FCL, we analyzed by extensive cloning and sequencing paired biopsy specimens obtained at the time of FCL diagnosis and transformation (6 patients) or FCL relapse (3 patients). In an additional patient, biopsy specimens obtained at the time of diagnosis, FCL relapse, and subsequent transformation to DLBCL were evaluated. The presence of identical mutations in the paired diagnosis and posttransformation DLBCL specimens confirmed the common clonal origin of both the pretransformation and the posttransformation lymphomas. No new mutations in the 5′ noncoding regulatory region of the BCL-6 gene were detected in any of the specimens evaluated at the time of FCL relapse. In contrast, 5 of the 7 transformed specimens contained new mutations not found in the paired original biopsy specimens obtained at the time of FCL diagnosis or relapse. The number of these new mutations ranged from 1 to 6 per specimen. Some of the new mutations tended to cluster in certain areas of the 5′ noncoding regulatory region of the BCL-6 gene. Our results show that transformation of FCL to DLBCL is associated with accumulation of new mutations in the 5′ noncoding regulatory region of the BCL-6 gene, that by deregulation of theBCL-6 gene expression may play a role in lymphoma transformation.
Follicle center lymphoma (FCL) accounts for approximately 40% of all non-Hodgkin's lymphomas (NHLs) occurring in adults.1 It is generally characterized by a relatively indolent clinical course and long survival. Nonetheless, it is well recognized that FCL may transform to a more aggressive lymphoma in 25% to 60% of patients.2,3 This transformation is associated with an increasing proportion of large cells, rapidly progressive clinical course, and short survival. Identification of the molecular mechanisms associated with morphologic transformation and clinical progression of FCL are critical issues in understanding the pathogenesis of these lymphomas. Several secondary genetic abnormalities, including nonrandom chromosomal changes,4-6c-myc gene rearrangement,7,8 p53 tumor suppressor gene mutations,9,10 somatic mutations of the translocatedBCL-2 gene,11 and p16 and p15 inactivation by deletions, mutations, and hypermethylation12 13 were associated with histologic transformation of FCL. However, the marked heterogeneity of these secondary aberrations that are only observed in a subset of transformed lymphomas suggests that other molecular mechanisms must be implicated in FCL transformation.
The gene most commonly implicated in the pathogenesis of diffuse large B-cell lymphomas (DLBCL) is the BCL-6 proto-oncogene, located at chromosome 3q27 that encodes a POZ/Zinc finger sequence-specific transcription repressor.14-18 Clonal BCL-6 gene rearrangements are observed in 30% to 40% of DLBCLs but in only 6% to 10% of FCLs.16,18,19 These rearrangements cluster within a highly conserved 4.0-kilobase (kb) regulatory region spanning the promoter, the first noncoding exon and the 5′ region of the first intron—the major breakpoint region (MBR) and result inBCL-6 expression driven by a heterologous promoter from the partner chromosomes.20 In addition, small deletions and somatic point mutations occurring in the BCL-6 regulatory region that overlap with the subdomain of MBR are reported in 70% of DLBCLs but in only 45% of FCLs.21-23 The occurrence of mutations is independent of translocation-generated rearrangements and it was demonstrated that some mutations can significantly deregulateBCL-6 expression.22 24
To gain further insight into the secondary molecular alterations associated with transformation to higher-grade NHLs, we have analyzed the nucleotide sequence of the 5′ regulatory region of theBCL-6 gene in sequential biopsy specimens from patients with FCL who underwent morphologic transformation to DLBCL.
Materials and methods
Tumor specimens
Sequential biopsy specimens from 10 patients with FCL were selected for this study, based on availability of frozen viable single cell suspensions for molecular analyses. Overall, 11 biopsy specimens obtained at the time of FCL diagnosis (2 biopsy specimens from case IL111), 4 biopsy specimens obtained at the time of FCL relapse, and 7 biopsy specimens obtained at the time of morphologic transformation to DLBCL were evaluated (Table 1). All lymphoma specimens were classified according to the Revised European-American Lymphoma Classification1 and were routinely immunophenotyped by flow cytometry for expression of Ig heavy and light chains and B- and T-cell markers. The histology of the first lymph node (9 patients) and spleen (2 patients) at the time of diagnosis was FCL, provisional cytologic grade I (10 specimens), and diffuse predominantly small cell (DPSC) lymphoma (1 specimen). The lymph node histology at the time of NHL relapse was classified as FCL, provisional cytologic grade I in 3 specimens and provisional cytologic grade 2 in 1 specimen. Lymph node histology at the time of transformation was classified as DLBCL in all 7 evaluated specimens. Specimen IL105A was evaluated and reported in our previous study.25
Summary of BCL-6 5′ regulatory region mutations in 10 cases of paired FCL and subsequent FCL or DLBCL specimens
| UPN . | Sample . | Date of biopsy . | Histology . | No. of confirmed mutations . | Type of mutation* . |
|---|---|---|---|---|---|
| IL105 | A | 2.1987 | FCL-1 | 3 | 186 (T > C)† |
| 586 (C > A) | |||||
| 671 (T > A)† | |||||
| B | 8.1990 | DLBCL | 3 | 350 (T > G) | |
| 586 (C > A) | |||||
| 671 (T > A)† | |||||
| IL114 | A | 4.1983 | FCL-DPSC | 0 | |
| B | 4.1985 | FCL-2 | 0 | ||
| C | 9.1992 | DLBCL | 6 | 182 (T > G)† | |
| 266 (T > C) | |||||
| 475 (T > C) | |||||
| 507 (C > T) | |||||
| 510 (C > A) | |||||
| 600 (C > A) | |||||
| IL115 | A | 3.1998 | FCL-1 | 1 | 110 (T > C) |
| B | 6.1999 | DLBCL | 3 | ▵▵T44† | |
| 110 (T > C) | |||||
| 188 (G > C) | |||||
| IL116 | A | 6.1987 | FCL-2 | 3 | 122 (G > A) |
| 480 (T > G)† | |||||
| 584 (C > T)† | |||||
| B | 2.1990 | DLBCL | 5 | 88 (G > C) | |
| 122 (G > A) | |||||
| 480 (T > G) | |||||
| 584 (C > T) | |||||
| 618 (G > A) | |||||
| IL117 | A | 7.1986 | FCL-1 | 0 | |
| B | 8.1991 | DLBCL | 0 | ||
| IL119 | A | 6.1985 | FCL-1 | 1 | 494 (T > A)† |
| B | 4.1993 | DLBCL | 0 | ||
| IL120 | A | 2.1989 | FCL-1 | 3 | ΔΔT44† |
| 329 (C > G)† | |||||
| 348 (C > T) | |||||
| B | 2.1992 | DLBCL | 5 | 104 (A > C) | |
| 180 (T > A) | |||||
| 348 (C > T) | |||||
| 507 (C > T)† | |||||
| 750 (C > G)† | |||||
| IL111 | A | 7.1988 | FCL-1 | 0 | |
| B | 9.1988 | FCL-1 | 0 | ||
| C | 1.1995 | FCL-1 | 0 | ||
| IL112 | A | 8.1987 | FCL-1 | 1 | 502 (G > A) |
| B | 4.1994 | FCL-1 | 1 | 502 (G > A) | |
| IL118 | A | 5.1990 | FCL-1 | 0 | |
| B | 4.1994 | FCL-1 | 0 |
| UPN . | Sample . | Date of biopsy . | Histology . | No. of confirmed mutations . | Type of mutation* . |
|---|---|---|---|---|---|
| IL105 | A | 2.1987 | FCL-1 | 3 | 186 (T > C)† |
| 586 (C > A) | |||||
| 671 (T > A)† | |||||
| B | 8.1990 | DLBCL | 3 | 350 (T > G) | |
| 586 (C > A) | |||||
| 671 (T > A)† | |||||
| IL114 | A | 4.1983 | FCL-DPSC | 0 | |
| B | 4.1985 | FCL-2 | 0 | ||
| C | 9.1992 | DLBCL | 6 | 182 (T > G)† | |
| 266 (T > C) | |||||
| 475 (T > C) | |||||
| 507 (C > T) | |||||
| 510 (C > A) | |||||
| 600 (C > A) | |||||
| IL115 | A | 3.1998 | FCL-1 | 1 | 110 (T > C) |
| B | 6.1999 | DLBCL | 3 | ▵▵T44† | |
| 110 (T > C) | |||||
| 188 (G > C) | |||||
| IL116 | A | 6.1987 | FCL-2 | 3 | 122 (G > A) |
| 480 (T > G)† | |||||
| 584 (C > T)† | |||||
| B | 2.1990 | DLBCL | 5 | 88 (G > C) | |
| 122 (G > A) | |||||
| 480 (T > G) | |||||
| 584 (C > T) | |||||
| 618 (G > A) | |||||
| IL117 | A | 7.1986 | FCL-1 | 0 | |
| B | 8.1991 | DLBCL | 0 | ||
| IL119 | A | 6.1985 | FCL-1 | 1 | 494 (T > A)† |
| B | 4.1993 | DLBCL | 0 | ||
| IL120 | A | 2.1989 | FCL-1 | 3 | ΔΔT44† |
| 329 (C > G)† | |||||
| 348 (C > T) | |||||
| B | 2.1992 | DLBCL | 5 | 104 (A > C) | |
| 180 (T > A) | |||||
| 348 (C > T) | |||||
| 507 (C > T)† | |||||
| 750 (C > G)† | |||||
| IL111 | A | 7.1988 | FCL-1 | 0 | |
| B | 9.1988 | FCL-1 | 0 | ||
| C | 1.1995 | FCL-1 | 0 | ||
| IL112 | A | 8.1987 | FCL-1 | 1 | 502 (G > A) |
| B | 4.1994 | FCL-1 | 1 | 502 (G > A) | |
| IL118 | A | 5.1990 | FCL-1 | 0 | |
| B | 4.1994 | FCL-1 | 0 |
FCL, follicle center lymphoma; DLBCL, diffuse large B-cell lymphoma; DPSC, diffuse predominantly small cell.
Mutations in bold represent new mutations observed in transformed DLBCL specimens but not in the initial FCL specimens.
Ongoing mutations present in some but not all molecular clones representing 1 allele.
DNA synthesis and polymerase chain reaction
High-molecular-weight DNA was extracted from 5.0 × 106 cells using a commercially available kit as described by the manufacturer (QIAamp Tissue Kit; Qiagen, Valencia, CA). The first intron region of the BCL-6 gene that has previously been shown to undergo extensive mutation in B cells was amplified by polymerase chain reaction (PCR) using 5′-CCGCTGCTCATGATCATTATTT and 5′-TAGACACGATACTTCATCTCAT primers, as was previously reported.25
For each PCR, a control with no added template was used to check for contamination. PCR products were analyzed by 2% agarose gel electrophoresis and staining with ethidium bromide. Bands of appropriate size were excised from the gels and purified by adsorption to a silica matrix (QIAquick columns, Qiagen).
Cloning and sequencing of polymerase chain reaction products
The purified PCR amplicons were ligated into a TA-PCR cloning vector (Invitrogen, Carlsbad, CA) and were used for transformation of competent Escherichia coli (1 Shot INV αF′; Invitrogen) according to the manufacturer instructions. Twelve to 24 white colonies were picked per sample and used in a second round of PCR. All the PCR amplicons (range: 8-21) were sequenced on a 373 automatic DNA sequencer (Applied Biosystems, Foster City, CA) using the ABI Prism Big Dye Terminator Kit (Perkin Elmer, Foster City, CA) as recommended by the manufacturer. The same primers used for the PCR were used for forward and backward sequencing. The Taq DNA polymerase error frequency in our laboratory is 0.09%, which amounts to 0.71 mutations perBCL-6 clone.
Sequence analysis was performed using the MacVector program (Oxford Molecular Group, Campbell, CA). Sequences were aligned with theBCL-6 hypermutation region germ line sequence previously established in our laboratory (GenBank accession number AF 191831).25 The first nucleotide of the amplifiedBCL-6 gene region, corresponding to the first nucleotide of the sense primer, was arbitrarily defined as position +1. Tumor mutation status was determined after excluding previously established polymorphisms.21 25 Only mutations defined as confirmed mutations, observed more than once in the BCL-6 gene clones from biopsy specimens from the same patient, were included in the analysis. Unconfirmed mutations, defined as mutation observed in only 1 of the BCL-6 gene clones from biopsy specimens from the same patient, were disregarded in our determination of the tumor mutation status, because their rate was similar to the Taq DNA polymerase error frequency in our laboratory, thus suggesting that at least some of them could be Taq DNA polymerase errors and not real mutations.
The BCL-6 alleles were easily identified by different mutation clustering on the alleles and by the presence of polymorphisms.
Results
BCL-6 gene cloning and sequencing were performed in sequential biopsy specimens from the time of FCL diagnosis and relapse in 4 patients and from the time of FCL diagnosis and transformation to DLBCL in 7 patients (1 of these patients had 3 biopsy specimens obtained at the time of FCL diagnosis, relapse, and transformation).
BCL-6 gene mutations in the paired diagnosis-relapse biopsies
Biopsy specimens from 4 patients with FCL were evaluated at the time of diagnosis (5 specimens) and at the time of FCL relapse (4 specimens) without evidence of morphologic transformation to a higher-grade NHL (Table 1). At the time of diagnosis, only 1 of the patients had confirmed mutation in the 5′ regulatory region of theBCL-6 gene, whereas in the remaining 3 patients, theBCL-6 gene was unmutated. No new mutations were observed in any of the specimens tested at the time of FCL relapse.
BCL-6 mutations in the paired diagnosis-transformation biopsies
Biopsy specimens from 7 patients with FCL were evaluated at the time of FCL diagnosis and at the time of transformation to DLBCL. Evaluation of immunoglobulin gene in sequential pretransformation and posttransformation biopsy specimens showed clonal relation between the tumors in all the cases, except case IL119, in which it was not performed (data not shown). At the time of diagnosis, confirmed mutations in the 5′ regulatory region of the BCL-6 gene were found in biopsy specimens from 5 of the 7 evaluated patients. Their number ranged from 1 to 3 per specimen, with an incidence ranging from 1.3 × 10−3 to 3.8 × 10−3 per base pair (bp). Some of the mutations were observed in all the molecular clones of the same allele, whereas others were present in some but not all clones of the same allele (Table 1 and Figure 1), confirming the presence of ongoing mutation in the BCL-6 gene of FCL that we have previously reported.25
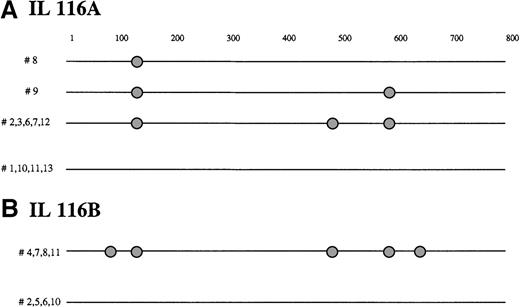
BCL-6 mutations in FCL transformation.
Schematic representation of the BCL-6 5′ noncoding region mutations in molecular clones derived from FCL diagnosis biopsy specimen (A) and transformation to DLBCL biopsy specimen (B) from representative IL116 case, demonstrating intraclonal diversification at diagnosis and acquisition of new mutations in DLBCL biopsy specimens. Each sequence is represented as a horizontal line. The first nucleotide of the amplified BCL-6 gene region, corresponding to the first nucleotide of the sense primer, is arbitrarily defined as position +1. In each sequence, mutations are indicated as circles. The nature of each mutation is specified in Table1.
BCL-6 mutations in FCL transformation.
Schematic representation of the BCL-6 5′ noncoding region mutations in molecular clones derived from FCL diagnosis biopsy specimen (A) and transformation to DLBCL biopsy specimen (B) from representative IL116 case, demonstrating intraclonal diversification at diagnosis and acquisition of new mutations in DLBCL biopsy specimens. Each sequence is represented as a horizontal line. The first nucleotide of the amplified BCL-6 gene region, corresponding to the first nucleotide of the sense primer, is arbitrarily defined as position +1. In each sequence, mutations are indicated as circles. The nature of each mutation is specified in Table1.
At the time of transformation to DLBCL, confirmed mutations in the 5′ regulatory region of the BCL-6 gene were found in the biopsy specimens from 5 patients. In 1 of these patients (IL114, Table1 and Figure 2), 2 previous biopsy specimens, obtained at the time of FCL diagnosis and relapse, were unmutated, whereas the biopsy specimen obtained at the time of transformation to DLBCL contained a BCL-6 gene with 6 confirmed mutations. In the additional 4 patients, the specimens obtained at the time of transformation contained BCL-6 gene mutations, most of which were identical to mutations observed in the paired FCL specimens, confirming their common clonal origin. In addition, new mutations not observed in the FCL diagnosis specimens were found (Figure 1). The number of these new mutations ranged from 1 to 4 per specimen (Table1). Most of the new mutations were present in all the tested molecular clones, whereas some were ongoing. Most probably, only the mutations observed in all or the majority (because of admixture of remaining follicular cells) of the molecular clones may play a role in lymphoma transformation. In the patients IL105, IL119, and IL120, some of the ongoing mutations, observed in the minority of the molecular clones evaluated at the time of FCL diagnosis, were not found in any of molecular clones evaluated at the time of transformation. Finally, in the case IL117, the paired diagnosis-transformation specimens were both unmutated.
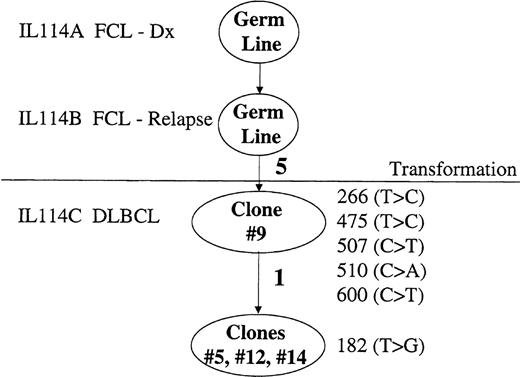
Mutation events of BCL-6 in FCL transformation.
Mutation events in the 5′ noncoding regulatory region of the BCL-6 gene derived from biopsy specimens at the time of FCL diagnosis (IL114A), FCL relapse (IL114B), and transformation to DLBCL (IL114C) in NHL patient IL116. Clonal relationship of the molecular clone sequences (ovals) is shown. Number of mutations is indicated along the lines in bold. The mutations are listed by location as is specified in the “Materials and methods.”
Mutation events of BCL-6 in FCL transformation.
Mutation events in the 5′ noncoding regulatory region of the BCL-6 gene derived from biopsy specimens at the time of FCL diagnosis (IL114A), FCL relapse (IL114B), and transformation to DLBCL (IL114C) in NHL patient IL116. Clonal relationship of the molecular clone sequences (ovals) is shown. Number of mutations is indicated along the lines in bold. The mutations are listed by location as is specified in the “Materials and methods.”
To evaluate the potential significance of these new confirmed mutations observed in the transformed but not in the FCL diagnosis specimens, we examined the distribution of these mutations. These new mutations appeared to cluster in certain areas of the 5′ regulatory region of the BCL-6 gene: identical mutation 507 (C → T) in 2 patients and mutations at positions 180, 182, and 188. We searched for mutations at these positions in the previously described compilation of all the published BCL-6 mutations in NHL and Hodgkin's lymphoma for which nucleotide change and exact mutation positions have been reported.25 Mutations at positions 507, 180, 182, and 188 have not been previously reported. However, mutations located in proximity to these positions (especially positions 186 and 190) have been reported repeatedly. To evaluate the potential influence of these mutations on BCL-6 regulation, we searched for possible transcription factor-binding sites in and around these mutation positions (TFSEARCH on the Internet). Mutations 180 (T → A) and 182 (T → G) occurred in the potential binding site of the GATA 2 transcription factor. It is possible that additional potential binding sites of presently unknown transcription factors may be affected by some of these mutations.
Discussion
Following recent reports that showed BCL-6 gene alterations in NHL, and especially in DLBCL, by either chromosomal rearrangements16,18 or somatic mutations in its 5′ noncoding regulatory region21,22,25 and previous demonstration that some of these mutations may deregulate BCL-6gene expression,24 we examined the possible role ofBCL-6 gene mutations in the process of FCL transformation to DLBCL. Our findings demonstrate that histologic transformation of FCL may be associated with the accumulation of new mutations in the 5′ noncoding regulatory region of BCL-6 oncogene. Clustering of the new acquired mutations in specific parts of theBCL-6 5′ noncoding regulatory region suggests the presence of currently unknown regulatory factors, which may effectBCL-6 gene expression. While our work was under review, Capello et al26 reported on accumulation of new BCL-6 gene mutations in 2 of 5 transformed cases, similar to our data.
Several previous studies have found evidence that heterogeneous secondary genetic changes may be associated with histologic transformation of FCL; nonetheless, none of them was a predominant genetic lesion responsible for the transformation.4-13 The most common secondary cytogenetic findings seen in transformed FCL comprised of del(6q) and acquisition of additional copy of chromosome 7.4-6 New chromosomal translocations involving chromosome 3q27, on which BCL-6 gene is located, were not among the commonly reported secondary changes observed during or after FCL transformation. Therefore, if the BCL-6 oncogene has a role in FCL transformation, it should be deregulated by a mechanism different from gross chromosomal aberration.
BCL-6 gene may be altered by somatic mutations of the 5′ noncoding regulatory region. These mutations are regarded as a marker of B-cell transit through the germinal center, because they are absent in pregerminal center naı̈ve lymphocytes or B-cell malignancies originating from pregerminal center B cells.22,26 Previous studies showed that some of the mutations in the BCL-6 5′ regulatory region could deregulate its expression. Pasqualucci et al24 evaluated the effect of regulatory region mutations onBCL-6 expression in 11 lymphoma cases (6 DLBCLs and 5 Burkitt lymphomas [BL], corresponding to 20 mutated alleles) and normal germinal center (GC) cells (20 mutated alleles). Significant overexpression of the reporter gene was observed in 33% of DLBCL mutant alleles derived from 3 of 6 DLBCL cases. Conversely, none of the alleles derived from the BL cases or normal GC cells displayed an altered transcriptional activity.
In this study, we demonstrate association between FCL transformation and accumulation of new mutations in the 5′ noncoding regulatory region of the BCL-6 oncogene. New mutations were observed in transformed cases but not in cases relapsing without transformation. The time intervals between the diagnosis and the subsequent relapse or transformation were overlapping, thus excluding the possibility that accumulation of new mutations was predominantly time dependent. The relatively small number of relapsed cases evaluated in this study precludes the general conclusion of the lack of association between FCL relapse and accumulation of new BCL-6 gene mutations. However, our finding in the same patient of new mutations only in the biopsy obtained at the time of transformation but not at the time of FCL relapse (Figure 2) suggests their potential role in transformation. In addition, our finding of identical mutations in transformed DLBCL specimens from 2 different patients and clustering of the mutations in a specific part of 5′ regulatory region of BCL-6 gene may suggest functional significance of these mutations and the presence ofBCL-6 regulatory factors that may bind to these regions. Significant progress has been made in elucidation of BCL-6function and its interaction with other cellular factors,15 27-31 but almost no data exist on the potential transcription factors involved in the regulation of BCL-6expression. Clustering of the new confirmed mutations in the region of GATA 2 binding site suggests a potential role for this transcription factor in BCL-6 regulation. It is likely that additional, presently unknown transcription factors may be involved in the regulation of BCL-6 expression. Further studies mapping functionally significant mutations will help to clarify the mechanisms of BCL-6 regulation.
In our previous work25 and in the current study, we demonstrated that mutations in the 5′regulatory region of theBCL-6 gene are ongoing. It is possible that as a result of ongoing BCL-6 gene somatic mutations, the lymphoma cell population becomes heterogeneous, and a mutational variant having a selective growth advantage because of BCL-6 overexpression compared with parental clones gives rise to the higher-grade NHL lymphoma cell population. Indeed, the finding of some of the mutations in FCL subclones at the time of diagnosis but not at the time of transformation to DLBCL (Table 1) and appearance of new mutations not observed at the time of diagnosis confirm the concept of clonal selection during transformation.
In conclusion, this study shows an association between somatic mutations in the 5′ regulatory region of BCL-6 gene and FCL transformation. Furthermore, our data suggest the presence of particular subregions in the 5′ noncoding regulatory region of the BCL-6 gene that may have a specific role in its regulation. Further studies are needed to determine the functional significance of these new accumulated mutations and identification of transcription factors involved in BCL-6 gene transcription regulation.
Supported by grants CA33399 and CA34233 from the USPHS-NIH.
Reprints:Ronald Levy, Stanford University School of Medicine, Division of Oncology M207, Stanford, CA 94305-5306.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal