Abstract
The distinction between benign polyclonal and malignant monoclonal lymphoid disorders by morphology or immunophenotyping is frequently difficult. Therefore, the demonstration of clonal B-cell or T-cell populations by detecting identically rearranged immunoglobulin (Ig) or T-cell receptor (TCR) genes is often used to solve this diagnostic problem. Whereas the detection of rearranged Ig genes is well established, TCR gamma (γ) and beta (β) gene rearrangements often escape detection with the currently available polymerase chain reaction (PCR) assays. To establish a sensitive, specific, and rapid method for the detection of rearranged TCR-β genes, we developed a new PCR approach with family-specific Jβ primers and analyzed the resulting PCR products by high-resolution GeneScan technique. The superior efficiency of this new method was demonstrated by investigating 132 DNA samples extracted from lymph node and skin biopsy specimens (mostly formalin fixed) and blood samples of 62 patients who had a variety of T-cell lymphomas and leukemias. In all but 1 of the tumor samples (98.4%) a clonal amplificate was detectable after TCR-β PCR and the same clonal T-cell population was also found in 15 of 18 (83%) of the regional lymph nodes and in 7 of 11 (64%) of the peripheral blood samples. Direct comparison of these results with those obtained currently by the most widely applied TCR-γ PCR revealed an approximate 20% lower detection rate in the same set of samples than with the TCR-β PCR method. These results indicate that the new TCR-β PCR is most suitable for a rapid and reliable detection of clonal T-cell populations.
The distinction between malignant lymphomas and reactive lymphoproliferative lesions is often difficult or impossible to make by conventional histology alone. This holds true especially for the identification of peripheral T-cell lymphomas consisting predominantly of small cells such as mycosis fungoides, or of a mixture of small and large cells like those in angioimmunoblastic or lymphoepitheloid T-cell lymphomas. Although the availability of several monoclonal antibodies has improved the recognition of T-cell subpopulations, the differentiation between non-neoplastic and neoplastic T cells still remains a problem. The configuration of the T-cell receptor (TCR) genes has often been proposed as the most valuable tool for identifying malignant T-cell proliferation. In normal and reactively proliferating T cells, these genes are rearranged differently (ie, polyclonal), whereas in T-cell lymphomas, the neoplastic cells contain identically rearranged monoclonal TCR genes.
Initially, the Southern blot technique was used to determine the clonality of the TCR gene rearrangements.1,2 For this purpose, the TCR-β chain gene was most often analyzed, because all, or almost all, T cells harbor functionally rearranged TCR-β genes as evidenced on the protein level.3-7 Although this assay may detect clonal T-cell populations in the vast majority of T-cell lymphomas, several drawbacks such as low sensitivity, the need for large amounts of fresh frozen tissue, and of radioactively labeled probes, will exclude this technique as a general diagnostic tool. Attempts to develop a polymerase chain reaction (PCR)-based method for the detection of clonally rearranged TCR-β genes in DNA samples proved to be difficult, because primers capable of binding all different 65 variable (Vβ) and 13 joining (Jβ) segments8-13 require too many consensus positions.14,15 This leads to a low detection rate of T-cell malignancies, especially when applied to degraded DNA extracted from formalin-fixed tissue.16,17 To overcome this problem, most of the previous studies used RNA extracts in conjunction with reverse transcriptase (RT)-PCR.18 19 However, this approach is limited to the investigation of fresh frozen samples and not applicable to formalin-fixed tissue specimens.
Therefore, various PCR assays have often been designed for the detection of TCR-γ gene rearrangements at the DNA level because of the simpler configuration of this gene and the greater homology within the various Vγ and Jγ gene segments.20,21 Although the TCR-γ PCR is applicable to DNA extracted from formalin-fixed tissue specimens, it has the drawback that 10% to 50% of the T-cell lymphomas escape detection, irrespective of the proportion of tumor cells.22-30
Because of these limitations, there is a broad need for the development of a new technique that may allow the identification of clonal TCR rearrangements with a high detection rate, even in paraffin-embedded tissues. In this paper, we describe a TCR-β PCR that overcomes these disadvantages by designing new family-specific Jβ primers applied in combination with a previously published Vβ consensus primer.31 With this assay, we investigated 132 samples obtained from 53 patients with T-cell non-Hodgin's lymphoma (T-NHL) and 9 patients with acute lymphoblastic leukemia (ALL). For control, 37 samples of reactive B-cell and T-cell lesions were included. For comparison, the 169 samples studied were also analyzed by a TCR-γ PCR, covering all known Vγ segments, and the results of both assays were compared using the high-resolution GeneScan technique. Most clonal PCR products were confirmed by direct sequencing. Clonal TCR-β rearrangements were found in all but 1 of the T-cell malignancies (98.4%) investigated, whereas clonally rearranged TCR-γ genes were detectable in only 80%. The results obtained recommend the TCR-β PCR as a new valid technique for the detection of clonal T-cell populations in patients with T-lineage lymphomas and leukemias.
Patients, materials, and methods
Patient samples and cell lines
A total of 132 samples from 62 patients with malignant T-cell disease were selected from the files of the Institute of Pathology and the Department of Dermatology at the Free University of Berlin. The lymphomas were diagnosed using morphologic and immunohistologic criteria according to the revised European-American lymphoma (REAL) classification.32 The series included 96 samples (skin, blood, lymph node) from 23 patients with primary cutaneous T-cell lymphoma (CTCL) in advanced stages, 12 anaplastic large cell lymphomas of T-phenotype (ALCL-T), 11 peripheral unspecified T-cell lymphomas (PTCL), 9 acute T-lymphoblastic leukemias (T-ALL), 3 angioimmunoblastic T-cell lymphomas (AILD-TCL), and 11 samples from 4 patients with intestinal T-cell lymphoma (ITCL) (Table1). Furthermore, 16 skin biopsy specimens from patients with non-neoplastic skin disease, including 4 biopsies from subacute or chronic dermatitis and 12 biopsies from psoriasis vulgaris, were also investigated. The majority of the samples (102 of 148) were formalin-fixed and embedded in paraffin.
Results of the TCR-β and TCR-γ rearrangement analysis
| . | n . | TCR-β . | TCR-γ . |
|---|---|---|---|
| T-cell lymphomas | 62 | 61/62 (98%) | 50/62 (80%) |
| Controls | |||
| B-cell lymphomas | 15 | 0/15 (0%) | 0/15 (0%) |
| Lymphadenopathies | 11 | 0/11 (0%) | 0/11 (0%) |
| PBMCs | 10 | 0/10 (0%) | 0/10 (0%) |
| Psoriasis vulgaris | 12 | 0/12 (0%) | 0/12 (0%) |
| Dermatitis, unspecific | 4 | ||
| T-cell lines | 8 | 8/8 (100%) | 8/8 (100%) |
| TCL* | 18 | 18/18 (100%) | 18/18 (100%) |
| . | n . | TCR-β . | TCR-γ . |
|---|---|---|---|
| T-cell lymphomas | 62 | 61/62 (98%) | 50/62 (80%) |
| Controls | |||
| B-cell lymphomas | 15 | 0/15 (0%) | 0/15 (0%) |
| Lymphadenopathies | 11 | 0/11 (0%) | 0/11 (0%) |
| PBMCs | 10 | 0/10 (0%) | 0/10 (0%) |
| Psoriasis vulgaris | 12 | 0/12 (0%) | 0/12 (0%) |
| Dermatitis, unspecific | 4 | ||
| T-cell lines | 8 | 8/8 (100%) | 8/8 (100%) |
| TCL* | 18 | 18/18 (100%) | 18/18 (100%) |
TCR: T-cell receptor; PBMCs: Peripheral blood mononuclear cells of healthy donors.
TCL: T-cell lymphomas with known and sequenced clonal TCR-γ rearrangements.
Positive controls consisted of 8 T-cell lines (Hut 102, MOLT 4, Jurkat, DHL-1, PEER, Karpas, Del, MO-T) and 18 peripheral T-cell lymphomas with known and sequenced clonal TCR-γ gene rearrangements. As negative controls, 15 B-cell lymphomas, as well as 11 lymph node specimens from unspecific lymphadenopathies, and 10 peripheral blood samples from healthy donors were included (Table 1).
Immunohistology
Four-micrometer sections of paraffin-embedded tissues were immunostained using the immunoalkaline phosphatase (APAAP) method.33 The primary antibodies were directed against T-cell receptor β-chain (clone βF1), CD3 (polyclonal CD3), CD4 (1F6), CD8 (C8-144), CD103 (Ber-ACT8), CD45RO (OPD4), CD30 (Ber-H2), and TdT (terminal desoxynucleotidyltransferase; polyclonal TdT). With the exception of βF1, which was from T-Cell Sciences (Cambridge, MA) and CD4 from Novocastra (Newcastle upon Tyne, UK), all other antibodies were purchased from DAKO (Glostrup, Denmark). To unmask antigen epitopes, deparaffinized tissue sections were subjected to high-pressure cooking before applying primary antibody.
DNA extraction
DNA used for PCR was extracted from 25 μm paraffin sections after dewaxing and proteinase K digestion using a QIAGEN DNA extraction kit (Qiagen, Hilden, Germany). DNA from the mononuclear cell fraction after Ficoll-Hypaque gradient of blood samples as well as from frozen tissue samples was extracted according to standard procedures.
Polymerase chain reaction for the detection of TCR-β rearrangements
For the detection of TCR-β gene rearrangements, 200 ng of genomic DNA was subjected to a seminested PCR. The first round of amplification was performed as 2 separate reactions involving the same Vβ consensus primer31 (Vβ pan: 5′-CTCGAATTCT(T/G)T(A/T) (C/T)TGGTA(C/T)C (G/A)(T/A)CA-3′; 200 ng) and 2 different primer sets (200 ng each set) consisting of 6 (Jβ1 family; JβFS1A) and 7 (Jβ2 family; JβFS2A) Jβ family-specific primers, respectively (Table 2). Thirty cycles were carried out with a primer annealing temperature of 60°C (40 seconds) for the initial 5 cycles and 57°C (40 sec) for the remaining 25 cycles. For reamplification, an aliquot (1%) of the first 2 reactions was used as a template in 2 additional, separate PCRs, comprising 40 cycles each with the same annealing temperature profile as described above. The same Vβ primer (200 ng) was used in combination with 2 nested family-specific Jβ primer mixes (JβFS1 and JβFS2; 200 ng each set; Table 2). The conditions for denaturation (96°C, 15 seconds) and primer extension (72°C, 40 seconds) remained constant through all cycles of the first and second PCR, whereas the concentration of MgCl2 was 2.5 mmol/L in the first and 1.5 mmol/L in the second amplification. All reactions were carried out in a final volume of 100 μL with 0.8 mmol/L of dNTPs (200 μmol/L each) and 2.5 units Taq polymerase (Perkin Elmer, Weiterstadt, Germany) in a thermal cycler (TC9600, Perkin Elmer, Weiterstadt, Germany). It is worth noting that the application of high-quality high-performance liquid chromatographic (HPLC)-purified oligonucleotides is a decisive factor for successfully performing the TCR-β PCR.
Sequences of the family-specific Jβ primers used for TCR-β PCR
| . | First amplification . | Second amplification . | ||
|---|---|---|---|---|
| . | 5′3′ . | . | 5′3′ . | |
| JβFS1A | Jβ 1.1 | ACT GTG AGT CTG GTG CC | JβFS1 | TGA GTC TGG TGC CTT GTC CAA A |
| Jβ 1.2 | ACG GTT AAC CTG GTC CC | TTA ACC TGG TCC CCG AAC CGA A | ||
| Jβ 1.3 | ACA GTG AGC CAA CTT CC | TGA GCC AAC TTC CCT CTC CAA A | ||
| Jβ 1.4 | ACA GAG AGC TGG GTT CC | AGA GCT GGG TTC CAC TGC CAA A | ||
| Jβ 1.5 | ATG GAG AGT CGA GTC CC | AGA GTC GAG TCC CAT CAC CAA A | ||
| Jβ 1.6 | ACA GTG AGC CTG GTC CC | TGA GCC TGG TCC CAT TCC CAA A | ||
| JβFS2A | Jβ 2.1 | ACG GTG AGC CGT GTC CC | JβFS2 | TGA GCC GTG TCC CTG GCC CGA A |
| Jβ 2.2 | ACG GTC AGC CTA GAG CC | TCA GCC TAG AGC CTT CTC CAA A | ||
| Jβ 2.3 | ACT GTC AGC CGG GTG CC | TCA GCC GGG TGC CTG GGC CAA A | ||
| Jβ 2.4 | ACT GAG AGC CGG GTC CC | AGA GCC GGG TCC CGG CGC CGA A | ||
| Jβ 2.5 | ACC AGG AGC CGC GTG CC | CGA GCC GCG TGC CTG GCC CGA A | ||
| Jβ 2.6 | ACG GTC AGC CTG CTG CC | TCA GCC TGC TGC CGG CCC CGA A | ||
| Jβ 2.7 | ACC GTG AGC CTG GTG CC | TGA GCC TGG TGC CCG GCC CGA A | ||
| . | First amplification . | Second amplification . | ||
|---|---|---|---|---|
| . | 5′3′ . | . | 5′3′ . | |
| JβFS1A | Jβ 1.1 | ACT GTG AGT CTG GTG CC | JβFS1 | TGA GTC TGG TGC CTT GTC CAA A |
| Jβ 1.2 | ACG GTT AAC CTG GTC CC | TTA ACC TGG TCC CCG AAC CGA A | ||
| Jβ 1.3 | ACA GTG AGC CAA CTT CC | TGA GCC AAC TTC CCT CTC CAA A | ||
| Jβ 1.4 | ACA GAG AGC TGG GTT CC | AGA GCT GGG TTC CAC TGC CAA A | ||
| Jβ 1.5 | ATG GAG AGT CGA GTC CC | AGA GTC GAG TCC CAT CAC CAA A | ||
| Jβ 1.6 | ACA GTG AGC CTG GTC CC | TGA GCC TGG TCC CAT TCC CAA A | ||
| JβFS2A | Jβ 2.1 | ACG GTG AGC CGT GTC CC | JβFS2 | TGA GCC GTG TCC CTG GCC CGA A |
| Jβ 2.2 | ACG GTC AGC CTA GAG CC | TCA GCC TAG AGC CTT CTC CAA A | ||
| Jβ 2.3 | ACT GTC AGC CGG GTG CC | TCA GCC GGG TGC CTG GGC CAA A | ||
| Jβ 2.4 | ACT GAG AGC CGG GTC CC | AGA GCC GGG TCC CGG CGC CGA A | ||
| Jβ 2.5 | ACC AGG AGC CGC GTG CC | CGA GCC GCG TGC CTG GCC CGA A | ||
| Jβ 2.6 | ACG GTC AGC CTG CTG CC | TCA GCC TGC TGC CGG CCC CGA A | ||
| Jβ 2.7 | ACC GTG AGC CTG GTG CC | TGA GCC TGG TGC CCG GCC CGA A | ||
Sequence analysis of the clonal TCR-β rearrangements obtained from various T-cell lymphomas
| Diagnosis . | Case . | 3′Vβ . | N-D-N . | 5′Jβ . | Vβ . | Dβ . | Jβ . |
|---|---|---|---|---|---|---|---|
| CTCL, skin | 12 | TGTGCCAGTAGT | TCGGC | AGCAATCAGCCCCAGCATTTTGG | V7.2 | Dx | J1.5 |
| CTCL, skin | 13 | TGTGCCAGT | CAATAAAGGGAGGGTCTGATCT | CTACGAGCAGTACTTCGGCCCC | V19 | D2 | J2.7 |
| CTCL, LN | 13 | TGTGCCAGT | CAATAAAGGGAGGGTCTGATCT | CTACGAGCAGTACTTCGGGCCC | V19 | D2 | J2.7 |
| CTCL, blood | 17 | TGTTCCAGCAGC | TGGGACAGGGGTAG | CTCCTACGAGCAGTACTTCGGCCCC | V5.4 | D1 | J2.7 |
| CTCL, blood | 17 | TGTGCCAGCAGC | TGGGACAGGGGTAG | CTCCTACGAGCAGTACTTCGGGCCC | V5.4 | D1 | J2.7 |
| PTCL | 24 | TGTGCCAGCAGCTT | CGGACAGGG | TGGAAACACCATATATTTTGGA | V7.9 | D1 | J1.3 |
| PTCL | 25 | TCTGCAGCGTTGAAGA | CAGGGGTT | TCCTACAATGAGCAGTTCTTCGGGCCA | V29.1 | D1 | J2.1 |
| PTCL | 27 | TGTGCCAGCAGCTTAG | TCGGGACAGGAG | CCTACAATGAGCAGTTCTTCGGGCCA | V7.2 | D1 | J2.1 |
| PTCL | 29 | TATGCCAGTAGTAT | TGTGAATTC | GAACACTGAAGCTTTCTTTGGA | V19 | Dx | J1.1 |
| PTCL | 31 | TGTGCCAGTAGTATA | ATAGTGGGGGGCAGTGGCG | CTGAAGCTTTCTTTGGA | V19 | D1 | J1.1 |
| PTCL | 32 | TGTGCCAGCAGTTTAT | CGTGGGGTAGCGGTG | GCACAGATACGCAGTATTTTGGCCCA | V28 | D2 | J2.3 |
| PTCL | 32 | TGTGCCAGTA | TGAATAGGACAGAG | AATCGAATCAGCCCCATTTTGG | V29.1 | D1 | J1.5 |
| T-LBL | 41 | TGTACTTCTGTGCCAGC | TCCACGTGGGGG | CAGCCCCAGCATTTTGG | V27.1 | D1 | J1.5 |
| T-LBL | 42 | TCTGCAGTGCTA | TCTGGGCCGG | CTATAATTCACCCCTCCACTTTGGG | V20.1 | Dx | J1.6 |
| T-LBL | 43 | TGCAGTGCT | CAGCCTTCGAG | GGCTACACCTTCGGTTCGG | V19 | D2 | J1.2 |
| T-LBL | 45 | TCTGCAGCGTT | CCGGGACAGGGTGCACCTACGAG | CAGTACTTCGGGCCA | V29.1 | D1 | J2.5 |
| ALC-T | 50 | TGTGCCAGCAGC | CTCAGGGGTAAC | AATTCACCCCTCCACTTTGGG | V11.1 | D1 | J1.6 |
| ALC-T | 52 | TGTGCCAGCAGCTTA | AACAGGGGC | AATTCACCCCTCCACTTTGGG | V7.2 | D1 | J1.6 |
| ALC-T | 53 | TGTGCCAGTAGC | AAAGGG | AATTCACCCCTCCACTTTGGG | V19 | Dx | J1.6 |
| ALC-T | 55 | TGTGCCAGTAGTATAG | CACGCGGACCCTA | TAGCAATCAGCCCCAGCATTTTGG | V19 | D2 | J1.5 |
| ALC-T | 58 | TGTGCCAGCAG | TTTTGGGGGGGCGA | ATGAGCAGTTCTTCGGGCCA | V7.2 | D1 | J2.1 |
| ALC-T | 58 | TGTGCCAGTAG | GACACCCTCTGAGGGGGGCGCGG | GCAATCAGCCCCAGCATTTTGG | V19 | D1 | J1.5 |
| ITCL | 59 | TGTGCCAGCAGTTAC | GGACAG | AATTCACCCCTCCACTTTGGG | V6.3 | Dx | J1.6 |
| ITCL | 60 | TGTGCCAGCAGTTACT | GTGGGCCGACG | GAGACCCAGTACTTCGGGCCA | V9 | D1 | J2.5 |
| ITCL | 61 | TGTGCCAGCAGCTTGG | TTGACAGGGGGCAGC | ATGGCTACACCTTCGGTTCGGG | V6.5 | D1 | J1.2 |
| Cell line MOLT4 | TCTGCAGTGCTAG | AGAGTCGACTAGCGATCCAAAA | AATGAGCAGTTCTTCGGGCCA | V20.1 | D2 | J2.1 | |
| Cell line HUT 102 | TCTGCAGCGT | AGGCAGGAGGCGGTTGGC | GGCTACACCTTCGGTTCGG | V30 | D2 | J1.2 | |
| Cell line DHL-1 | TGTGCCAGTAGTA | CAGTAGGGGA | ACAGATACGCAGTATTTTGGCCCA | V27 | D1 | J2.3 |
| Diagnosis . | Case . | 3′Vβ . | N-D-N . | 5′Jβ . | Vβ . | Dβ . | Jβ . |
|---|---|---|---|---|---|---|---|
| CTCL, skin | 12 | TGTGCCAGTAGT | TCGGC | AGCAATCAGCCCCAGCATTTTGG | V7.2 | Dx | J1.5 |
| CTCL, skin | 13 | TGTGCCAGT | CAATAAAGGGAGGGTCTGATCT | CTACGAGCAGTACTTCGGCCCC | V19 | D2 | J2.7 |
| CTCL, LN | 13 | TGTGCCAGT | CAATAAAGGGAGGGTCTGATCT | CTACGAGCAGTACTTCGGGCCC | V19 | D2 | J2.7 |
| CTCL, blood | 17 | TGTTCCAGCAGC | TGGGACAGGGGTAG | CTCCTACGAGCAGTACTTCGGCCCC | V5.4 | D1 | J2.7 |
| CTCL, blood | 17 | TGTGCCAGCAGC | TGGGACAGGGGTAG | CTCCTACGAGCAGTACTTCGGGCCC | V5.4 | D1 | J2.7 |
| PTCL | 24 | TGTGCCAGCAGCTT | CGGACAGGG | TGGAAACACCATATATTTTGGA | V7.9 | D1 | J1.3 |
| PTCL | 25 | TCTGCAGCGTTGAAGA | CAGGGGTT | TCCTACAATGAGCAGTTCTTCGGGCCA | V29.1 | D1 | J2.1 |
| PTCL | 27 | TGTGCCAGCAGCTTAG | TCGGGACAGGAG | CCTACAATGAGCAGTTCTTCGGGCCA | V7.2 | D1 | J2.1 |
| PTCL | 29 | TATGCCAGTAGTAT | TGTGAATTC | GAACACTGAAGCTTTCTTTGGA | V19 | Dx | J1.1 |
| PTCL | 31 | TGTGCCAGTAGTATA | ATAGTGGGGGGCAGTGGCG | CTGAAGCTTTCTTTGGA | V19 | D1 | J1.1 |
| PTCL | 32 | TGTGCCAGCAGTTTAT | CGTGGGGTAGCGGTG | GCACAGATACGCAGTATTTTGGCCCA | V28 | D2 | J2.3 |
| PTCL | 32 | TGTGCCAGTA | TGAATAGGACAGAG | AATCGAATCAGCCCCATTTTGG | V29.1 | D1 | J1.5 |
| T-LBL | 41 | TGTACTTCTGTGCCAGC | TCCACGTGGGGG | CAGCCCCAGCATTTTGG | V27.1 | D1 | J1.5 |
| T-LBL | 42 | TCTGCAGTGCTA | TCTGGGCCGG | CTATAATTCACCCCTCCACTTTGGG | V20.1 | Dx | J1.6 |
| T-LBL | 43 | TGCAGTGCT | CAGCCTTCGAG | GGCTACACCTTCGGTTCGG | V19 | D2 | J1.2 |
| T-LBL | 45 | TCTGCAGCGTT | CCGGGACAGGGTGCACCTACGAG | CAGTACTTCGGGCCA | V29.1 | D1 | J2.5 |
| ALC-T | 50 | TGTGCCAGCAGC | CTCAGGGGTAAC | AATTCACCCCTCCACTTTGGG | V11.1 | D1 | J1.6 |
| ALC-T | 52 | TGTGCCAGCAGCTTA | AACAGGGGC | AATTCACCCCTCCACTTTGGG | V7.2 | D1 | J1.6 |
| ALC-T | 53 | TGTGCCAGTAGC | AAAGGG | AATTCACCCCTCCACTTTGGG | V19 | Dx | J1.6 |
| ALC-T | 55 | TGTGCCAGTAGTATAG | CACGCGGACCCTA | TAGCAATCAGCCCCAGCATTTTGG | V19 | D2 | J1.5 |
| ALC-T | 58 | TGTGCCAGCAG | TTTTGGGGGGGCGA | ATGAGCAGTTCTTCGGGCCA | V7.2 | D1 | J2.1 |
| ALC-T | 58 | TGTGCCAGTAG | GACACCCTCTGAGGGGGGCGCGG | GCAATCAGCCCCAGCATTTTGG | V19 | D1 | J1.5 |
| ITCL | 59 | TGTGCCAGCAGTTAC | GGACAG | AATTCACCCCTCCACTTTGGG | V6.3 | Dx | J1.6 |
| ITCL | 60 | TGTGCCAGCAGTTACT | GTGGGCCGACG | GAGACCCAGTACTTCGGGCCA | V9 | D1 | J2.5 |
| ITCL | 61 | TGTGCCAGCAGCTTGG | TTGACAGGGGGCAGC | ATGGCTACACCTTCGGTTCGGG | V6.5 | D1 | J1.2 |
| Cell line MOLT4 | TCTGCAGTGCTAG | AGAGTCGACTAGCGATCCAAAA | AATGAGCAGTTCTTCGGGCCA | V20.1 | D2 | J2.1 | |
| Cell line HUT 102 | TCTGCAGCGT | AGGCAGGAGGCGGTTGGC | GGCTACACCTTCGGTTCGG | V30 | D2 | J1.2 | |
| Cell line DHL-1 | TGTGCCAGTAGTA | CAGTAGGGGA | ACAGATACGCAGTATTTTGGCCCA | V27 | D1 | J2.3 |
Clonal TCR-β PCR products were directly sequenced. The representative clone-specific CDR3 proportion is shown. The corresponding Vβ germline segments are designated to the nomenclature of Rowen et al.11 The D- and J-segments are aligned according to the nomenclature of Toyonage et al12 and Tunnacliffe et al.13 Dx: D-segment not determinable.
Polymerase chain reaction for the detection of TCR-γ rearrangements
TCR-γ gene rearrangements of the VγI subgroup were detected by a seminested PCR using 200 ng of genomic DNA as a template.30The same Jγ-specific primers were used for both rounds of amplification, whereas 2 nested Vγ primers were subjected to the first and second PCR. The first amplification consisted of 2 separate reactions (25 cycles each), one using the Jγ primer JGT1/223 and the other JGT323 (200 ng each), both in conjunction with Vγ11-8 (5′-TGCAGCCAGTCAGAAATCTTCC-3′). The reamplification was carried out in 2 separate reactions using a nested Vγ primer (Vγ21-85′-ACAGCGTCTTC(AT)GTACTATGAC-3′) and the same Jγ-specific primers (JGT1/2 and JGT3). The cycle conditions remained constant through all PCRs being 96°C, 15 seconds for denaturation; 60°C, 30 seconds for primer annealing and 72°C, 40 seconds for primer extension. The buffer conditions were the same as those described for the reamplification of the TCR-β PCR.
In those T-cell lymphoma cases in which no clonal TCR-γ rearrangements were detectable with the primers for the VγI subgroup, additional PCRs for the detection of rearrangements involving V segments of the VγII (Vγ9) and VγIII (Vγ10,11) subgroups were performed as previously described23 with one exception: instead of using a conventional Taq polymerase (ie, AmpliTaq; Perkin-Elmer), we used TaqGold (Perkin-Elmer) for amplification. We found the use of TaqGold crucial for the elimination of unwanted additional PCR products (generated by the Vγ9-11 primers), which may cause erroneous detection of clonality.
GeneScan
For GeneScan analysis of the PCR products, the Vβ and Vγ primers of the reamplification were replaced by fluorescence primers of the same sequence labeled at their 5′-end with 5-carboxyflourescein (FAM). Aliquots of PCR products (1-2 μL) were mixed with loading buffer (2 μL formamide, 0.5 μL EDTA), and 0.5 μL of the internal size standard (Genescan-500) were included for precise determination of the length of the amplificates. After denaturation for 2 minutes at 90°C, the products were separated on sequencing gel and analyzed by automatic fluorescence quantification and size determination, using the computer program GENESCAN 672 (ABI 373A, Applied Biosystems, Weiterstadt, Germany).
Direct sequencing
The clonal PCR products of most T-cell lymphomas and of all T-cell lines were directly sequenced. For this purpose, reaction mixtures were separated on 6% polyacrylamid gels and stained with ethidium bromide (Figure 1). The most dominant band was cut out, and the gel slice was incubated in 25 μL distilled water for at least 24 hours. Five microliters of the supernatant were subjected to fluorescence dye terminator cycle sequencing, and the sequencing reactions were analyzed on a 377A DNA sequencer (Applied Biosystems) after removal of the unincorporated fluorescence dye. Each sequencing reaction was carried out in both directions using the primers Vβpan and JβFS1 or JβFS2.
Results
Primer testing, specificity, and sensitivity of the TCR-β and TCR-γ polymerase chain reaction
The newly designed TCR-β PCR was established and optimized by applying DNA extracted from 8 different T-cell lines. As expected, all cell lines gave rise to single distinct PCR products that harbored the expected DNA sequences, as confirmed by DNA sequencing (Table 3). Similarly, all 18 peripheral T-cell lymphomas with known TCR-γ rearrangements gave rise to 1 or 2 (biallelic) amplificates after application of TCR-β PCR (Figure2). The primers of the TCR-γ PCR were tested as described elsewhere,23 30demonstrating the presence of clonal PCR products in all T-cell lines analyzed so far.
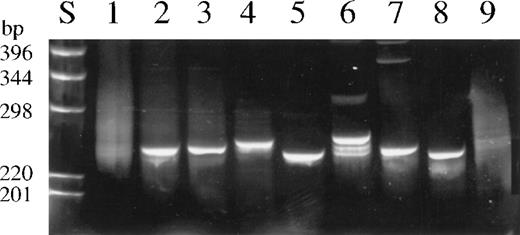
Ethidium bromide stained 6% polyacrylamid gel of representative TCR-β PCR products.
(S) DNA size marker. (1) Normal tonsillar tissue. (2) Lesional skin, CTCL (case 18). (3) Infiltrated lymph node, CTCL (case 18). (4) Peripheral T-cell lymphoma (case 29). (5) T-ALL (case 41). (6) Anaplastic large-cell lymphoma (case 58). (7) Intestinal T-cell lymphoma (case 53). (8) Cell line Hut102. (9) B-cell lymphoma. PCR products in lanes 1, 2, 8, and 9 were generated from DNA of frozen material, and those in lanes 3, 4, 5, 6, and 7, from DNA extracted from paraffin-embedded specimens.
Ethidium bromide stained 6% polyacrylamid gel of representative TCR-β PCR products.
(S) DNA size marker. (1) Normal tonsillar tissue. (2) Lesional skin, CTCL (case 18). (3) Infiltrated lymph node, CTCL (case 18). (4) Peripheral T-cell lymphoma (case 29). (5) T-ALL (case 41). (6) Anaplastic large-cell lymphoma (case 58). (7) Intestinal T-cell lymphoma (case 53). (8) Cell line Hut102. (9) B-cell lymphoma. PCR products in lanes 1, 2, 8, and 9 were generated from DNA of frozen material, and those in lanes 3, 4, 5, 6, and 7, from DNA extracted from paraffin-embedded specimens.
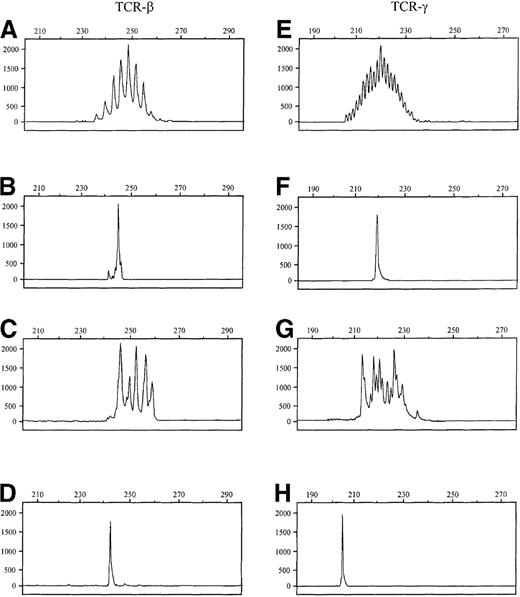
Comparison of the fluorescence-labeled TCR-β and TCR-γ PCR products by GeneScan analysis.
The x-axes represent molecular size (base pairs) and the y-axes fluorescence intensity. Samples A-D were produced by TCR-β PCR and samples E-H by TCR-γ PCR. (A, E) Normal tonsillar tissue. (B, F) Cutaneous T-cell lymphoma (case 23). (C, G) Psoriasis. (D) T-cell line HUT102 (243 bp). (H) T-cell line HUT102 (205 bp). Note: TCR-β PCR products regularly differ by 3 base pairs, indicating the presence of functional TCR-β rearrangements. TCR-γ PCR products differ by only 1 base pair, indicating frequent out-of-frame rearrangements.
Comparison of the fluorescence-labeled TCR-β and TCR-γ PCR products by GeneScan analysis.
The x-axes represent molecular size (base pairs) and the y-axes fluorescence intensity. Samples A-D were produced by TCR-β PCR and samples E-H by TCR-γ PCR. (A, E) Normal tonsillar tissue. (B, F) Cutaneous T-cell lymphoma (case 23). (C, G) Psoriasis. (D) T-cell line HUT102 (243 bp). (H) T-cell line HUT102 (205 bp). Note: TCR-β PCR products regularly differ by 3 base pairs, indicating the presence of functional TCR-β rearrangements. TCR-γ PCR products differ by only 1 base pair, indicating frequent out-of-frame rearrangements.
The specificity of the TCR-β and TCR-γ PCR technique was determined by investigating 11 normal lymph nodes, 10 peripheral blood samples from healthy donors, and 15 B-cell lymphomas. In addition, 16 inflammatory dermatoses were also investigated (Table 1, Figure 2). All normal lymph node and blood samples gave rise to a polyclonal rearrangement pattern, whereas most of the remaining samples displayed an oligoclonal pattern sometimes comprising only a few dominant clones. However, repeated independent PCR assays clearly demonstrated that the dominant clones differed from each other among the various PCRs.
The sensitivity of both PCR techniques was determined by serial dilution of T-cell lines in normal tonsillar DNA. All PCR assays were able to detect between 0.5% and 1% of clonally rearranged T-cells in a polyclonal background (0.5 ng to 1 ng T-cell line DNA in 100 ng of tonsillar DNA). The use of more PCR cycles did not increase the sensitivity.
Detection of clonal T-cell populations in T-cell lymphomas and leukemias
A total of 62 histologically and clinically well-diagnosed T-cell lymphomas and leukemias, including 23 CTCLs, 12 ALCLs, 11 unspecified PTCLs, 9 T-ALLs, 4 ITCLs, and 3 AILDs, were investigated with both the TCR-β and TCR-γ PCR techniques (Table 1). Monoclonal T-cell populations were detectable in all but 1 of the tumor samples (61 of 62; 98%) by TCR-β PCR, irrespective of the use of frozen or formalin-fixed material (Figure 1). Moreover, the same clonal TCR-β rearrangement detected in the primary skin tumor was also found in 15 of 18 (83%) and 7 of 11 (64%) of the excised regional lymph nodes and the peripheral blood samples of 23 patients with CTCL (Table4). Furthermore, multiple samples of 13 patients who had biopsies over a period of up to 3 years, all contained the same TCR-β gene rearrangement. The presence of the same clonal TCR-β rearrangement was confirmed in each case by repeated independent analysis, including PCR, GeneScan analysis, and DNA sequencing of most amplificates.
TCR-β and TCR-γ analysis in skin, lymph node, and peripheral blood samples of 23 CTCL patients
| Case . | Disease . | Stage (TNM) . | TCR-β-R . | TCR-γ-R . | ||||
|---|---|---|---|---|---|---|---|---|
| Skin . | LN . | Blood . | Skin . | LN . | Blood . | |||
| 1 | MF | T3N3M0 | + | na | na | + | na | na |
| 2 | MF | T3N0M0 | + | + | na | + | + | na |
| 3 | MF | T3N0M0 | + | na | na | − | na | na |
| 4 | MF | T3N0M0 | + | na | na | + | na | na |
| 5 | MF | T4N0M0 | + | − | − | − | − | − |
| 6 | MF | T4N0M0 | + | + | na | + | + | na |
| 7 | MF | T3N3M0 | + | + | na | + | + | na |
| 8 | MF | T3N3M0 | + | + | − | − | na | na |
| 9 | MF | T3N3M0 | + | + | − | − | − | − |
| 10 | MF | T4N3M0 | + | + | na | + | + | na |
| 11 | MF | T4N3M0 | + | + | na | + | + | na |
| 12 | MF | T4N3M0 | + | + | − | + | + | − |
| 13 | MF | T4N3M0 | + | + | + | + | na | na |
| 14 | MF | T3N3M0 | + | + | + | + | + | − |
| 15 | MF | T4N3M0 | + | na | na | + | na | na |
| 16 | MF | T2N3M0 | + | na | na | + | na | na |
| 17 | MF | T3N3M0 | + | + | + | − | − | − |
| 18 | MF | T2N3M0 | + | + | na | + | + | na |
| 19 | MF | T2N3B1M0 | + | + | na | − | − | na |
| 20 | SS | T4N3B1M0 | + | + | + | + | + | + |
| 21 | SS | T4N3B1M0 | + | + | + | + | + | + |
| 22 | SS | T4N3B1M1 | + | + | + | + | + | + |
| 23 | SS | T4N3B1M1 | + | + | + | + | + | + |
| Case . | Disease . | Stage (TNM) . | TCR-β-R . | TCR-γ-R . | ||||
|---|---|---|---|---|---|---|---|---|
| Skin . | LN . | Blood . | Skin . | LN . | Blood . | |||
| 1 | MF | T3N3M0 | + | na | na | + | na | na |
| 2 | MF | T3N0M0 | + | + | na | + | + | na |
| 3 | MF | T3N0M0 | + | na | na | − | na | na |
| 4 | MF | T3N0M0 | + | na | na | + | na | na |
| 5 | MF | T4N0M0 | + | − | − | − | − | − |
| 6 | MF | T4N0M0 | + | + | na | + | + | na |
| 7 | MF | T3N3M0 | + | + | na | + | + | na |
| 8 | MF | T3N3M0 | + | + | − | − | na | na |
| 9 | MF | T3N3M0 | + | + | − | − | − | − |
| 10 | MF | T4N3M0 | + | + | na | + | + | na |
| 11 | MF | T4N3M0 | + | + | na | + | + | na |
| 12 | MF | T4N3M0 | + | + | − | + | + | − |
| 13 | MF | T4N3M0 | + | + | + | + | na | na |
| 14 | MF | T3N3M0 | + | + | + | + | + | − |
| 15 | MF | T4N3M0 | + | na | na | + | na | na |
| 16 | MF | T2N3M0 | + | na | na | + | na | na |
| 17 | MF | T3N3M0 | + | + | + | − | − | − |
| 18 | MF | T2N3M0 | + | + | na | + | + | na |
| 19 | MF | T2N3B1M0 | + | + | na | − | − | na |
| 20 | SS | T4N3B1M0 | + | + | + | + | + | + |
| 21 | SS | T4N3B1M0 | + | + | + | + | + | + |
| 22 | SS | T4N3B1M1 | + | + | + | + | + | + |
| 23 | SS | T4N3B1M1 | + | + | + | + | + | + |
LN, lymph node; na, not analyzed; +, monoclonal; −, not monoclonal; MF, mycosis fungoides; SS, Sézary syndrome.
The application of the TCR-γ PCR using primers for the Vγ subgroups I, II, and III to the same set of samples led to the detection of monoclonally rearranged T-cell populations in 80% (50 of 62) of the tumor samples (Table 1, Figure3). The investigation of corresponding lymph nodes and blood samples of the patients with CTCL revealed monoclonality in only 63% and 44 %, respectively (Table 4). All but 2 of the clonal TCR-γ rearrangements displayed involvement of segments of the VγI subgroup (Vγ1-8), whereas 1 of the remaining 2 patients (case 12) had a clonal Vγ10, and the other (case 32), a biallelic Vγ9/Vγ10 rearrangement.
Comparison of results.
Comparison of TCR-β (A-C) and TCR-γ (D-F) PCR results obtained in 3 T-cell lymphomas with clonal TCR-β rearrangements and poly/oligoclonal TCR-γ rearrangements by GeneScan analysis. (A, D) Anaplastic large cell lymphoma (case 53). (B, E) Peripheral T-cell lymphoma, unspecified (case 34). (C, F) Intestinal T-cell lymphoma (case 50).
Comparison of results.
Comparison of TCR-β (A-C) and TCR-γ (D-F) PCR results obtained in 3 T-cell lymphomas with clonal TCR-β rearrangements and poly/oligoclonal TCR-γ rearrangements by GeneScan analysis. (A, D) Anaplastic large cell lymphoma (case 53). (B, E) Peripheral T-cell lymphoma, unspecified (case 34). (C, F) Intestinal T-cell lymphoma (case 50).
Follow-up by GeneScan analysis in a patient with cutaneous T-cell lymphoma (case 12) by TCR-β PCR.
(A) Plaque-stage at the time of first diagnosis. (B) Lymph node involvement half year later. (C) Peripheral blood at the time of lymph node involvement. (D) Skin lesion 2 years after chemotherapy; relapse. Note: The same clonal TCR-β rearrangement (250 bp) is detectable in all tissue specimens, but not in the peripheral blood sample.
Follow-up by GeneScan analysis in a patient with cutaneous T-cell lymphoma (case 12) by TCR-β PCR.
(A) Plaque-stage at the time of first diagnosis. (B) Lymph node involvement half year later. (C) Peripheral blood at the time of lymph node involvement. (D) Skin lesion 2 years after chemotherapy; relapse. Note: The same clonal TCR-β rearrangement (250 bp) is detectable in all tissue specimens, but not in the peripheral blood sample.
GeneScan analysis
Application of high-resolution separation techniques such as GeneScan analysis appeared best suited for determining the presence of clonal T-cell populations. The high resolution allowed the identification of PCR products that differed by only 1 base pair (bp) in length. Furthermore, the use of internal standards enables a precise determination of the size of the PCR products, irrespective of gel-to-gel variations or gel shift artifacts often associated with other techniques. This allows monitoring of lymphoma patients by direct comparison of the results obtained at various points in time without reanalyzing the previous samples (Figure 4).31,34 35
GeneScan analysis of normal lymphoid samples displayed a Gaussian-like distribution of the sizes of the TCR-β amplificates ranging from 234 to 261 bp (JβFS1) and 237 to 264 bp (JβFS2) and of TCR-γ PCR products ranging from 192 to 220 bp (JGT1/2 primer set) and from 210 to 238 bp (JGT3 primer set). Noteworthy, the products of the TCR-γ PCR differed by only 1 base pair in size (Figure 2E,G), indicating frequent presence of out-of-frame rearrangements, whereas the reading frame is conserved in most TCR-β rearrangements (Figure 2A,C). T-cell lymphomas or T-cell lines were characterized by 1 or 2 dominant peaks in the GeneScan profile, indicating the presence of a clonal T-cell population. Application of both TCR PCRs to reactive T-cell proliferations, especially those in extranodal origin, resulted in some cases the appearance of few dominant PCR products. However, in contrast to the monoclonal amplificates encountered in T-cell neoplasias, these products were not reproducible in independently performed PCR assays, thus representing the presence of oligoclonal T-cell populations in these lesions.
DNA sequencing of TCR-β polymerase chain reaction products
For investigation of the specificity of the TCR-β PCR, we sequenced 28 amplificates derived from various T-cell lymphomas and T-cell lines (Table 3). Sequence information was obtained by direct sequencing without additional subcloning of the PCR products. All sequences of the rearranged Vβ and Jβ segments could be identified by comparing them with databank sequences. In contrast to the TCR-γ locus the junctional TCR-β-CDR3 region showed extensive diversity due to the addition of N-region nucleotides between the Vβ-Dβ and Dβ-Jβ junctions and seemed ideally suited as clone-specific identification sequence. As an independent test of the PCR results, a comparison of the junctional regions obtained for the cell lines Jurkat, MOLT4, Hut 102, and PEER revealed complete agreement with published sequences. The sizes of the sequenced CDR3 regions were in direct correlation to the length of the PCR fragments, as demonstrated by the GeneScan analysis.
Immunohistology
In all T-cell lymphoma cases, immunohistology disclosed a T-cell phenotype with the expression of CD3 by the atypical cells. Predominance of CD4+ or CD8+ subpopulations was verified in several cases, underscoring the presence of a monoclonal T-cell population. An expression of the β-chain was detected in most cases. ALC-T cases characteristically expressed CD30. ITCL cases were characterized by the presence of atypical CD103-positive T-lymphocytes within the epithelium. T-ALL cases, in addition to T-cell antigen expression, showed an intranuclear positivity for TdT.
Discussion
The adjunct of PCR technology for the detection of clonally rearranged TCR genes has greatly contributed to the distinction between benign polyclonal and neoplastic monoclonal T-cell populations, and thus, to the diagnosis of T-cell lymphomas. However, in a significant proportion of malignant T-cell proliferations the tumor T-cell clone escapes detection with currently available TCR-γ PCR methods.24-30,36 Attempts to establish a technique for detection of clonally rearranged TCR-β genes on DNA level suitable for investigation of formalin-fixed biopsies failed so far.16-19,37,38 An interesting and promising attempt to simplify the TCR-β PCR was the use of highly degenerated Jβ consensus primers by Kneba and colleagues.15 This method detected only large T-cell clones,15 39 and, in our hands, produced high background and was difficult to reproduce when applied to routine formalin-fixed biopsy specimens. However, the results obtained encouraged us to develop a new TCR-β PCR suitable for the application in the daily diagnostic work.
To overcome the shortcomings of previous TCR-β methods, we designed family-specific Jβ primers. These primers consist exclusively of oligonucleotides with complete homology to their target genes therefore obtain several advantages over degenerated consensus primers, as already shown for the detection of clonal immunoglobulin (Ig) rearrangements.40 As a result, application of family-specific Jβ primers led to amplification of largely background-free TCR-β specific PCR products. This seems especially important for detection of small clonal T-cell populations present in early CTCL or in minimal lymph node involvement, where polyclonal background signals can superimpose clonality. Moreover, combination of TCR-β PCR with the high-resolution GeneScan technique enables a precise and reliable interpretation of the PCR data and exact monitoring between different lesions of the same patient.30,34 35
The sensitivity, specificity, and reliability of the new TCR-β PCR assay were tested by investigating a large number of histologically and clinically proven T- and B-cell lymphomas, as well as reactive lymphoid lesions and normal lymphoid tissues. Monoclonal T-cell populations were demonstrable in all but 1 of the 62 T-cell lymphomas investigated, irrespective of the use of fresh-frozen or formalin-fixed paraffin-embedded specimens. Moreover, identical TCR-β gene rearrangements were detectable in the corresponding blood and lymph node samples from 64% (7 of 11) and 83% (15 of 18) of the patients and multiple lymphoma samples from 13 patients, who had biopsies over a period of up to 3 years, contained all the same TCR-β gene rearrangement.
The investigation of the same set of T-cell lymphomas with the TCR-γ PCR technique using primers for the Vγ subgroups I, II, and III revealed a clonal TCR-γ gene rearrangement in only 80% of the cases (50 of 62). This is in line with most previous publications.24-30,41-45 No significant differences were observed between the type of the lymphoma, its location, or the proportion of tumor cells, with the exception of precursor T-lymphoblastic lymphomas, where all tumors displayed clonal a TCR-γ rearrangement. These results clearly indicate that the failure to detect clonal T-cell populations in a significant proportion of cases (12 of 62; 20%) was not due to a limited sensitivity of the TCR-γ PCR, but to a true absence of appropriate TCR-γ rearrangements. Therefore, other possibilities are required to explain the nondetectability of clonal TCR-γ rearrangements in these cases, such as a germ line configuration of the TCR-γ gene, incomplete or deleterious rearrangement within the TCR-γ locus,46 or transrearrangement between V-segments of the γ-chain and J-segments of the δ-chain.47 However, because the vast majority of the rearranged TCR-γ genes are not expressed, these nonfunctional rearrangements are without any consequence for the survival of the T-cells.
The specificity of the TCR-β and TCR-γ PCR assays was determined by investigating DNA extracted from 15 B-cell lymphomas, 11 normal lymph nodes, and 10 normal peripheral blood samples. Neither the TCR-β nor the TCR-γ PCR showed clonal rearrangements in any of these samples, confirming the specificity of both assays. Interestingly, application of both PCR systems to inflammatory skin lesions disclosed poly/oligoclonal gene rearrangements with clear dominance of 1 or only a few clones in some instances (pseudoclonal). However, these dominant PCR products were shown to possess different sizes in independently performed PCR assays. This pseudoclonality is explainable by the cellular composition of these lesions. T cells proliferating in the skin are not embedded in a background of many nonclonal lymphoid cells, leading to only a few or even single amplificates of various sizes after TCR-β or TCR-γ PCR. Especially for investigations of skin or other extranodal tissue, we recommend to perform 2 independent PCRs and to consider only those cases as clonal if complete size concordance of the dominant PCR product occurs in both assays.
In conclusion, the TCR-β PCR technique described in this article is reliable and highly suitable for the detection of small populations of clonal T cells, not only in frozen tissues but also in formalin-fixed paraffin-embedded specimens. This underscores its value as a new and highly sensitive diagnostic tool for routine biopsy material. Further, the low background of the TCR-β PCR products enables direct sequencing. This may be useful for generating clone-specific primers for investigation of minimal residual disease, thus allowing detection of tumor cell specific TCR-β rearrangements in the follow-up of T-cell lymphoma/leukemia patients.
Acknowledgments
We are particularly indebted to H. Lammert and H. Hempel for excellent technical assistance.
Supported by the Deutsche Krebshilfe, Grant 70-2202-Mü3.
This work contains parts of the doctoral thesis of C.A.
Reprints:Michael Hummel, Institute of Pathology, University Medical Centre Benjamin Franklin, The Free University of Berlin, Hindenburgdamm 30, 12200 Berlin, Germany; e-mail:hummel@ukbf.fu-berlin.de.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal