Abstract
The 1,25-dihydroxyvitamin D3(1,25- [OH]2VD3) modulates the differentiation of monocytic cell lines and monocytes (MOs) in vitro. Up to now several target genes of 1,25(OH)2VD3have been described in monocytic cell lines; however, little is known about target genes in primary MOs. With the Differential Display technique, we found a transcript up-regulated by 1,25(OH)2VD3 in short-term cultured human blood MOs, which we called MADDAM (metalloprotease and disintegrin dendritic antigen marker; EMBL/GenBank/DDBJ accession no. Y13786). Northern blot analysis confirmed this result and revealed a signal of MADDAM messenger RNA (mRNA) at about 7.5 kilobases (kb). Long-term culture (more than 20 hours) of MOs during macrophage (MAC) differentiation led to a rapid and complete down-regulation of MADDAM expression. In contrast, MADDAM expression was maintained in MOs differentiated along the dendritic cell (DC) pathway and induced in CD34+-derived DCs. In addition, in situ hybridization revealed signals of MADDAM mRNA in follicles of human lymph nodes and MADDAM mRNA was detected in freshly isolated human blood-DCs by reverse transcription-polymerase chain reaction (RT-PCR). By means of a database search, we found that MADDAM is a member of the ADAM (a metalloprotease and disintegrin) family, the human homologue to murine meltrin-β (ADAM 19). From these data, we conclude that MADDAM is an important marker for the differentiation and characterization of DCs and the distinction between MACs and DCs.
Cells of the mononuclear phagocyte system, like monocytes (MOs), macrophages (MACs), and dendritic cells (DCs), are important effector cells of the immune response. Beside their nonspecific activity against microorganism and tumor cells1,2 MOs and MACs are involved in antigen presentation, thereby leading to a specific immune response.3 DCs, in contrast, are professional antigen-presenting cells,4-6 and much more potent than MOs/MACs in stimulating primary immune response.7 In vitro MACs as well as DCs can be generated from human blood MOs, depending on the culture conditions.8 The in vitro differentiation process of MOs to MACs is induced by human serum,9 but can also be initiated by the active form of vitamin D3, 1α,25-dihydroxyvitamin D3(1,25[OH]2VD3),10,11 that also influences the functions of other hematopoetic cells like T and B lymphocytes.12-16 Most of the 1,25(OH)2VD3 effects are mediated by the vitamin D receptor (VDR), which is a member of the superfamily of nuclear steroid, thyroid, and retinoic acid receptors, acting as a transcription factor in the form of a homodimer or heterodimer.17-19 The distribution of the VDR is ubiquitous,20-22 and MOs/MACs also express this receptor.23 Nevertheless, only a few genes in MOs/MACs are known to be regulated by 1,25(OH)2VD3, eg, CD14, a monocytic differentiation marker,24 and macrophage colony-stimulating factor (M-CSF), a survival factor of MOs/MACs.25 26 For a better understanding of the effects of 1,25(OH)2VD3 on the differentiation process of human MOs to MACs, we were interested in 1,25(OH)2VD3-regulated genes during short- and long-term culture.
The in vitro differentiation of human blood MOs along the DC pathway is regulated by fetal calf serum (FCS), granulocyte-macrophage colony-stimulating factor (GM-CSF), and interleukin-4 (IL-4), leading to an “immature” type of DC, 27-29 whereas the terminal differentiation of these cells can be induced by the addition of tumor necrosis factor–alpha (TNF-α), lipopolysaccharide (LPS), or CD40 ligand.29-31 Up to now the characterization of DCs at the various differentiation and activation stages is difficult, because only few specific marker are available. As several clinical trials use DCs in different protocols as cellular vaccines against malignant diseases,32-34 an exact characterization of this cell type and its functions is of great importance.
In this report we describe the isolation and cloning of metalloprotease and disintegrin dendritic antigen marker (MADDAM), a novel human metalloprotease disintegrin belonging to the ADAM family35-38 that is up-regulated during DC differentiation in vitro, but not expressed in MO-derived MACs. Therefore, MADDAM can serve as a marker for the early distinction between MACs and DCs.
Materials and methods
Chemicals
All chemical reagents used were purchased from Sigma (Deisenhofen, Germany), unless otherwise noted. The 1,25(OH)2VD3 was kindly provided by Hoffmann-LaRoche (Basel, Switzerland), and recombinant human IL-4 by Schering-Plough (Kenilworth, NJ).
Cell separation and culture
Peripheral human blood mononuclear cells (MNCs) were isolated by leukapheresis of healthy donors, followed by density gradient centrifugation over Ficoll/Hypaque (Pharmacia, Freiburg, Germany). MOs were obtained from these cells by countercurrent centrifugation in a J6M-E centrifuge (Beckmann, Munich, Germany), as described previously.39 They were more than 90% pure, as determined by morphology and expression of the CD14 antigen. MOs of single donors were cultured under serum-free conditions at a density of 106 cells/mL in petri dishes with RPMI (Biochrom, Berlin, Germany), supplemented with 50 mmol/L mercaptoethanol, antibiotics (50 U/mL penicillin and 50 mg/mL streptomycin), 1 mmol/L pyruvate, 1 × MEM nonessential amino acids (GIBCO BRL, Eggenstein, Germany), 1 × MEM vitamins (GIBCO BRL), and 0.22 mg/mL L-glutamin (GIBCO BRL). For control experiments different elutriation fractions containing lymphocytes were used.
MACs were generated in petri dishes by culturing MOs for the indicated time with 2% AB-serum (Sigma, Deisenhofen, Germany) or under serum-free conditions with 10−7 mol/L 1,25(OH)2VD3.
DCs were generated by 3 different methods: (1) For isolation of blood-DCs, we used a pool of 1 × 109 cells of the blood-DC–enriched fractions IIc (74 mL/min.), IIb (82 mL/min.), and IIc (92 mL/min.) obtained during monocyte elutriation. Starting with these fractions (2%-4% blood-DCs) the blood-DCs were isolated with the Blood Dendritic Cell Isolation Kit and the SuperMACS II system (Miltenyi, Bergisch Gladbach, Germany), following the manufacturer's instructions. The purity of the blood-DCs (90%-98%) was determined by FACS analysis (FACS Calibur, Becton Dickinson) by using the 4-color assay containing anti-lin-1-FITC, anti-CD123-PE, anti-HLA-DR-PerCP, and anti-CD11c-APC (Becton Dickinson, San Jose, CA). (2) DCs were obtained by culturing MOs for the indicated time, either under serum-free conditions (CellGrow DC medium, Cell Genix, Freiburg, Germany) or with RPMI medium plus 10% FCS (c.c.pro, Karlsruhe, Germany), respectively, and with 35 ng/mL GM-CSF (Sandoz-Essex, Munich, Germany) and 500 U/mL IL-4 (Schering-Plough) in culture flasks.27-29 In some experiments, 10 ng/mL TNF-α (BASF, Knoll AG, Ludwigshafen, Germany) was added after 5 days of culture. (3) DCs were also generated from CD34+ cells. CD34+ cells were isolated from the blood after in vivo mobilization with G-CSF and cultured with 10% FCS, 200 ng/mL GM-CSF, 10 ng/mL stem cell factor (R&D Systems, Minneapolis, MN), and 10 ng/mL TNF-α for 2 weeks.40-43
The human cell lines, HL-60, HT29, Daudi, Jurkat, K562, HaCat, MelEi, N1 (fibroblasts), and HepG2 were cultured in RPMI supplemented with the additives described above and 10% FCS. For the induction of dendritic cell differentiation, HL-60 were cultured for up to 48 hours with 180 ng/mL calcium ionophore A23187 (Sigma).
RNA preparation
Total RNA was isolated from primary cells and cell lines by the method of Chomczynski and Sacchi.44 Only total RNA of blood-DCs was obtained by using RNeasy Mini Kit (Qiagen, Hilden, Germany).
Northern blot analysis
Total RNA (10 μg per lane) was electrophoretically separated on a 1% agarose formaldehyde gel, transferred to nylon membranes (Magna NT, MSI, Westborough, MA) and ultraviolet (UV) cross-linked. The complementary DNA (cDNA) fragment of MADDAM obtained by the Differential Display technique (DD) was phospholabeled with gene-specific primers and Klenow enzyme (Boehringer, Mannheim, Germany). Hybridization was performed overnight at 65°C in Church buffer (0.5 mol/L sodium phosphate buffer, pH 7.2, 7% SDS, 1 mmol/L EDTA, and 150 μg/mL transfer RNA [tRNA]).45,46 In control experiments, membranes were rehybridized with an 18S ribosomal RNA (rRNA)-oligonucleotide (5′-ACG GTA TCT GAT CGT CTT CGA ACC-3′) or a glyceraldehyde-3-phosphate dehydrogenase (GAPDH)-oligonucleotide complementary to the nucleic acids +1101 base pairs (bp) to +1187 bp of the published sequence labeled by T4 Kinase (5′-End Labelling Kit, Amersham, Buckinghamshire, UK).47
For nonradioactive Northern blot analysis the Digoxigenin (DIG)-System (Boehringer) was used according to the manufacturer's protocol. The DD cDNA-fragment of MADDAM was labeled with gene-specific primers and DIG-dUTP using the “DIG DNA Labelling and Detection Kit“ by PCR with the conditions: 2 minutes, 95°C, 10 cycles: 10 seconds, 95°C; 30 seconds, 60°C; 2 minutes, 72°C and 20 cycles: 10 seconds, 95°C; 30 seconds, 60°C; 2 minutes (+20 seconds extension per cycle), 72°C and afterwards 7 minutes, 72°C.
Messenger RNA differential display technology
This method was performed using the RNAmap-system (GenHunter, Nashville, TN).48 Total RNA (30 μg) of MOs cultured under different conditions was digested with 10 units DNAse I (Boehringer) as described previously.49 The 0.3 μg RNA was reverse transcribed with the 2 base-anchored oligo-dT primer T12MC and amplified by PCR using AmpliTaq (Perkin Elmer, Weiterstadt, Germany) and the primer combination T12MC and AP1 (5′-AGC CAG CGA A-3′) in the presence of (33P) deoxycytidine triphosphate. The amplified cDNA was separated on a 6% polyacrylamide gel. After autoradiography bands of interest were excised, the cDNA fragments were isolated, amplified, cloned by inserting into the EcoRI site of the plasmid vector pCR 2.1 (TOPO TA cloning kit, Invitrogen, San Diego, CA), and sequenced.
Screening of the complementary DNA library
Plaque screening was performed as previously described.46 In brief, C600hfl bacteria were infected with 5 × 105 plaque-forming units of a λgt10 lymph node cDNA library (Clontech) and plated on LBbroth-Agar–coated petri dishes containing 10 mmol/L MgSO4. After formation of plaques, phage DNA was transferred to NitroPlus membranes (MSI) and denatured (0.5 mol/L NaOH, 1.5 mol/L NaCl). After neutralization (0.5 mol/L Tris/HCl, pH 8.0, 1.5 mol/L NaCl), membranes were UV cross-linked and hybridized with 2 different 32P-labeled cDNA-fragments of MADDAM (1: +6186 bp to +6489 bp; 2: +2787 bp to +3214 bp). Positive plaques were excised, resuspended in phage-buffer, and subjected to a second round of screening. The inserts of isolated λgt10 phages were PCR-amplified using vector-specific primers (λgt 1: 5′-TGG GTA GTC CCC ACC TTT TGA GCA AGT TCA G-3′, λgt 2: 5′-CAG AGG TGG CTT ATG AGT ATT TCT TCC AGG GT-3′), and the KlenTaq-system (Clontech). The PCR conditions were 5 minutes, 95°C; 2 minutes, 72°C; adding the enzyme and 35 cycles with 30 seconds, 94°C; 30 seconds, 65°C; and 6 minutes, 72°C. After separation on an agarose gel cDNA inserts were excised, extracted, and sequenced using the vector primers (λgt 1 and λgt 2) and gene-specific primers.
Sequence analysis
The cDNA were sequenced by Dye Deoxy Terminator Cycle Sequencing (Applied Biosystems, Weiterstadt, Germany) according to the manufacturer's instructions on the Applied Biosystems DNA Sequencing System (model 373 A).
Flow cytometry
In brief, the cells were harvested, washed twice with cold phosphate-buffered saline (PBS) containing 0.1% sodiumazide and 0.6 mg/mL immunoglobulin, and incubated for 30 minutes at 4°C with specific fluorescein isothiocyanate (FITC)- or phycoerythrin (PE)-conjugated monoclonal antibodies, respectively. The following antibodies were used: CD1a-PE (Coulter, Krefeld, Germany), CD14-FITC (Coulter), CD83-PE (Immunotech, Marseille, France), HLA-DR-FITC (Pharmingen, Hamburg, Germany), and CD86-FITC (Pharmingen). After 2 additional washes, the cells were fixed with 1% paraformaldehyde in PBS and analysis was performed using a FACScan (Becton Dickinson, Mountain View, CA).
In situ hybridization
One microgram of the linearized pBluescript plasmid containing the 1019-bp cDNA fragment of MADDAM (+5470 bp to +6489 bp) was transcribed using T3 and T7 RNA-polymerase (Stratagene, Heidelberg, Germany) and33P-UTP (Amersham) to obtain sense or antisense cRNA probes. Paraffin-embedded tissue sections from human lymph nodes were generated by fixation in 4% paraformaldehyde/0.5% glutaraldehyde overnight and in situ hybridization was performed as previously described.50 Briefly, proteinase K–treated slides were prehybridized for 4 hours at 50°C (50% formamide, 10% dextran sulfate, 10 mmol/L Tris (pH 8.0), 10 mmol/L sodium phosphate (pH 7.0), 2 × SSC, 5 mmol/L EDTA (pH 8.0), 150 μg/mL tRNA, 10 mmol/L DTT, 10 mmol/L β-mercaptoethanol) and hybridized at 50°C overnight with a 5 × 104 cpm/μL antisense probe. Slides were washed (50% formamide, 2 × SSC, 20 mmol/L mercaptoethanol) and single strand RNA was digested with RNase A (20 μg/mL) for 30 minutes at 37°C. After further washing and dehydrating, slides were coated with Kodak NTB2 emulsion (Rochester, NY) and exposed for 2 weeks.
Reverse transcriptase-polymerase chain reaction
Total RNA (250 ng) of blood-DCs, MACs, and DCs were reverse transcribed for 60 minutes at 42°C using 5 μmol/L oligo(dT12) primer (Boehringer) and 200 units of Superscript Reverse Transcriptase (Life Technologies, Eggenstein, Germany). For amplification of MADDAM cDNA, we used the primers: 5′-GGC CGT GTG GTG CTT TGC TAG-3′ (sense) and 5′-AAG GAT GAC CCA CGG CAA GGA C-3′ (antisense), and for β-actin cDNA the primers: 5′-TGA CGG GGT CAC CCA CAC TGT GCC CAT CTA-3′ (sense) and 5′-CTA GAA GCA TTT GCG GTG GAC GAT GGA GGG-3 (antisense). The amplification was performed with 4 μL (MADDAM) or 2 μL (β-actin) of the reverse transcription reaction, 1 μmol/L sense and antisense primer, 25 μmol/L of each deoxynucleotide triphosphate and 5 units Taq DNA polymerase (Boehringer) in a total volume of 50 μL and the conditions: 94°C, 30 seconds; 65°C, 30 seconds; and 72°C, 2 minutes for 18 and 22 cycles (β-actin) or 26 and 30 cycles (MADDAM). Six microliters of the PCR products were analyzed in a 1.5% agarose gel with 0.5 μg/mL ethidium bromide.
Results
Identification of MADDAM as a 1,25-dihydroxyvitamin D3–regulated gene in monocytes
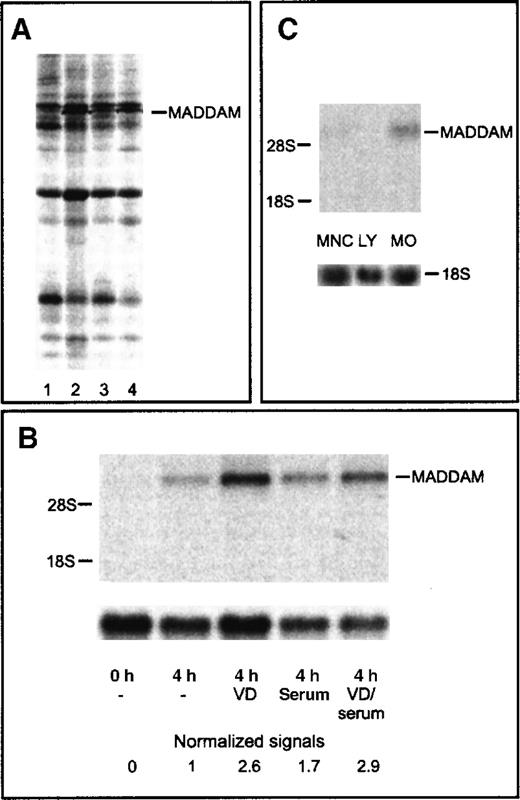
To identify 1,25(OH)2VD3-responsive genes, we cultured freshly isolated human blood MOs with or without 10−7 mol/L 1,25(OH)2VD3 for 4 hours in the absence or presence of 2% serum. With the use of the mRNA DD technology, we detected a stronger signal of a cDNA fragment in MOs cultured with 1,25(OH)2VD3 compared with unstimulated MOs (Figure 1A). The cDNA fragment (290 bp) was excised, reamplified, cloned into a plasmid vector, and sequenced.
Differential display and Northern blot analysis of the MADDAM cDNA fragment.
Differential Display (DD) of cDNA of different short-term cultured MOs (A) and Northern blot analysis of MADDAM expression in human blood MOs, lymphocytes, and mononuclear cells (B,C). MOs were stimulated for 4 hours with 1,25(OH)2VD3in the absence (2) or presence (4) of serum. Control cells were cultured for 4 hours without 1,25(OH)2VD3 under serum-free conditions (1) or with serum (3). RNA was extracted and cDNA amplified with the primer set AP1 and T12MC. The cDNA fragment of MADDAM is marked (A). Total RNA of freshly isolated MOs (0 hours) or MOs cultured for 4 hours in the presence (serum) or absence (−) of serum with or without 10−7 mol/L 1,25(OH)2VD3 (VD) was isolated and Northern blot was performed according to “Materals and methods.” As a control for RNA loading, the membrane was rehybridized with an 18S rRNA-oligonucleotide and MADDAM signals were normalized to the corresponding 18S signals (B). In (C), the mRNA expression of MADDAM in mononuclear cells (MNCs), an elutriated lymphocyte fraction (LY), and elutriated MOs, stimulated for 4 hours with 10−7 mol/L VD under serum-free conditions, was compared by Northern blot analysis.
Differential display and Northern blot analysis of the MADDAM cDNA fragment.
Differential Display (DD) of cDNA of different short-term cultured MOs (A) and Northern blot analysis of MADDAM expression in human blood MOs, lymphocytes, and mononuclear cells (B,C). MOs were stimulated for 4 hours with 1,25(OH)2VD3in the absence (2) or presence (4) of serum. Control cells were cultured for 4 hours without 1,25(OH)2VD3 under serum-free conditions (1) or with serum (3). RNA was extracted and cDNA amplified with the primer set AP1 and T12MC. The cDNA fragment of MADDAM is marked (A). Total RNA of freshly isolated MOs (0 hours) or MOs cultured for 4 hours in the presence (serum) or absence (−) of serum with or without 10−7 mol/L 1,25(OH)2VD3 (VD) was isolated and Northern blot was performed according to “Materals and methods.” As a control for RNA loading, the membrane was rehybridized with an 18S rRNA-oligonucleotide and MADDAM signals were normalized to the corresponding 18S signals (B). In (C), the mRNA expression of MADDAM in mononuclear cells (MNCs), an elutriated lymphocyte fraction (LY), and elutriated MOs, stimulated for 4 hours with 10−7 mol/L VD under serum-free conditions, was compared by Northern blot analysis.
Northern blot analysis with the cloned 290 bp DD cDNA fragment, which we called MADDAM, revealed weak signals at about 7.5 kilo bases (kb) in MOs cultured for 4 hours in the presence or absence of serum without 1,25(OH)2VD3. Both signals were further up-regulated by stimulation with 10−7 mol/L 1,25(OH)2VD3. In freshly isolated MOs, no signal was detected (Figure 1B). To exclude the possibility that nonmonocyte contaminants are responsible for the MADDAM expression, we compared the expression of MADDAM in mononuclear cells, lymphocytes, and elutriated MOs. We found no MADDAM mRNA expression in the lymphocyte fraction (less than 1% MOs), a weak signal in MNCs (10%-15% MOs), and a strong expression in MOs (Figure 1C).
Molecular cloning of MADDAM
We obtained the full-length sequence of the mRNA transcript (MADDAM) by screening of a lymph node library (Clontech) with different probes of MADDAM. The combined sequence of 6489 kb contains a putative open reading frame of 2757 bp and an unusual polyadenylation site (ATTAAA) 34 nucleotides upstream of the poly(A)+tail (data not shown). The open reading frame encoded for a putative protein with 919 amino acids (101 kd), that showed a strong homology of 84% to the murine meltrin-β51 belonging to the ADAM-family, which is also named MDC (metalloprotease disintegrin cysteine-rich) family (Figure2).35,37 Further comparison of MADDAM with sequences of the database (EMBL/GenBank/DDBJ) revealed strong homologies to other members of this family, eg, 48% to human ADAM 12 and 41% to Xenopus laevis ADAM 13. In correlation with these homologies, the deduced amino acid sequence of MADDAM contains a metalloprotease domain with a zinc-binding active site, a disintegrin domain, and cysteine-rich domain. In addition, the putative cysteine switch (at amino acid position 174) and the furin cleavage site (at amino acid position 200-204) were present in the protein sequence of MADDAM, and they are very important for the protease activity.52
Comparison of the amino acid sequences of MADDAM and murine meltrin-β.
The putative cysteine switch, furin cleavage site, and zinc-binding active site are marked. The sequence data are available from EMBL/GenBank/DDBJ under accession number Y13786.
Comparison of the amino acid sequences of MADDAM and murine meltrin-β.
The putative cysteine switch, furin cleavage site, and zinc-binding active site are marked. The sequence data are available from EMBL/GenBank/DDBJ under accession number Y13786.
Expression of MADDAM messenger RNA during the in vitro differentiation of monocytes
During the in vitro differentiation of MOs to MACs, a transient mRNA expression of MADDAM was detected after 4 hours, which was completely down-regulated after 1 day of culture. However, during the differentiation of MOs to dendritic cells (DCs), MADDAM mRNA expression was induced in short-term cultured cells and maintained at a high level (data not shown). Therefore, we investigated in particular the MADDAM expression in dendritic cells.
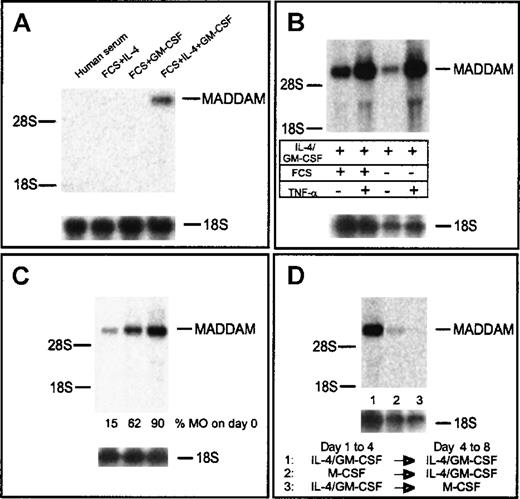
First, we analyzed the influence of different medium supplements normally used for generation of DCs from MOs. Only in DCs cultured with IL-4 in combination with GM-CSF did we observed signals of MADDAM mRNA after 7 days of culture (Figure3A). To exclude a role of FCS for the regulation of MADDAM, MOs were also cultured for 7 days in serum-free DC medium (Cell Genix, Freiburg, Germany) with IL-4 and GM-CSF. As expected, MADDAM mRNA was expressed under serum-free conditions as well. For the induction of terminal DC maturation, we added 10 ng/mL TNF-α after 5 days of culture for another 2 days. We analyzed the terminal differentiated DCs on day 7 by flow cytometry analysis, which revealed a strong expression of CD1a and HLA-DR and an up-regulation of CD83 by TNF-α (data not shown). Concerning the MADDAM expression, we found a strong increase by culture of immature DCs with TNF-α either under serum-free or serum-containing culture conditions (Figure 3B).
MADDAM mRNA expression in MAC compared with immature and mature DCs on day 7 of culture.
MOs were cultured with either human serum to induce MAC differentiation or FCS + IL-4, FCS + GM-CSF, and FCS + IL-4 + GM-CSF, respectively, to induce DC differentiation (A). In addition, MOs were cultured under serum-free conditions with IL-4 + GM-CSF with or without the addition of TNF-α on day 5 (B). In (C), DCs were generated from either MNCs or different elutriation fractions containing MOs with IL-4 + GM-CSF + FCS and the addition of TNF-α on day 5. In (D), MOs into MAC differentiation under serum-free conditions with 100 ng/mL M-CSF was switched to the DC differentiation pathway on day 4 by culture with FCS, IL-4, and GM-CSF or the other way around. Total RNA was extracted and the Northern blot analysis was performed as described in “Materials and methods.” As a control for RNA loading, the membrane was rehybridized with an 18S rRNA-oligonucleotide (A-D).
MADDAM mRNA expression in MAC compared with immature and mature DCs on day 7 of culture.
MOs were cultured with either human serum to induce MAC differentiation or FCS + IL-4, FCS + GM-CSF, and FCS + IL-4 + GM-CSF, respectively, to induce DC differentiation (A). In addition, MOs were cultured under serum-free conditions with IL-4 + GM-CSF with or without the addition of TNF-α on day 5 (B). In (C), DCs were generated from either MNCs or different elutriation fractions containing MOs with IL-4 + GM-CSF + FCS and the addition of TNF-α on day 5. In (D), MOs into MAC differentiation under serum-free conditions with 100 ng/mL M-CSF was switched to the DC differentiation pathway on day 4 by culture with FCS, IL-4, and GM-CSF or the other way around. Total RNA was extracted and the Northern blot analysis was performed as described in “Materials and methods.” As a control for RNA loading, the membrane was rehybridized with an 18S rRNA-oligonucleotide (A-D).
In addition, we analyzed the influence of non-MO contaminants on MADDAM expression in mononuclear cells (10%-15% MOs), an elutriation fraction containing about 50% lymphocytes and 50% MOs and elutriation purified MOs (more than 90% MOs). Dependent on the MO content in the starting cell preparation, MADDAM expression increased (Figure 3C). In addition, DCs derived from MOs purified by MACs sorting (more than 95%) with anti-CD14 superparamagnetic beads (Miltenyi) showed a comparable MADDAM mRNA expression (data not shown).
As recently described by Palucka and colleagues,53 the differentiation of MOs into DCs can be switched on day 4 into the MAC differentiation pathway by cultivation of the cells with M-CSF for another 4 days. In line with our results, we found that the MADDAM expression was completely shut down after M-CSF culture. However, the switch of the MACs to the DC differentiation pathway led to an induction of MADDAM expression (Figure 3D).
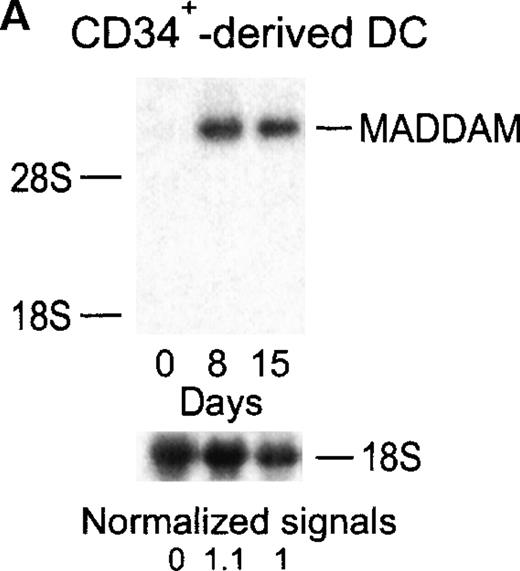
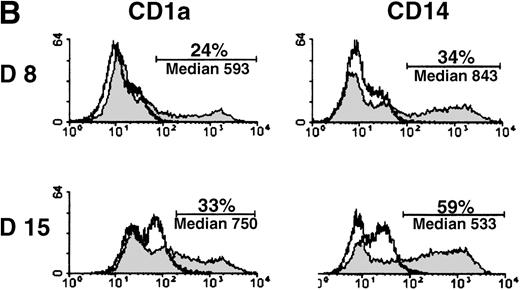
MADDAM expression was independent on the progenitor cells used for DC generation, because MO-derived DCs, as well as DCs generated from CD34+ cells, expressed MADDAM. No MADDAM mRNA expression was found in CD34+ cells on day 0 of culture, but after 8 and 15 days, strong MADDAM signals were detected (Figure4A). During this time, the number of CD1a+ and CD14+ cells slightly increased (Figure 4B).
Expression of MADDAM mRNA during the differentiation process of CD34+ cells into DCs in vitro.
CD34+ cells were cultured up to 15 days with FCS, stem cell factor, TNF-α, and GM-CSF (A). Total RNA was isolated and Northern blot analysis was performed as described in “Materials and methods.” As a control for RNA loading, the membrane was rehybridized with an 18S rRNA-oligonucleotide and MADDAM signals were normalized to the corresponding 18S signals. In parallel, flow cytometry analysis was performed with anti-CD1a-PE, anti-CD14-FITC, and isotype control antibodies at day 8 and day 15 of culture. The median fluorescence and the number of positive cells are shown (B).
Expression of MADDAM mRNA during the differentiation process of CD34+ cells into DCs in vitro.
CD34+ cells were cultured up to 15 days with FCS, stem cell factor, TNF-α, and GM-CSF (A). Total RNA was isolated and Northern blot analysis was performed as described in “Materials and methods.” As a control for RNA loading, the membrane was rehybridized with an 18S rRNA-oligonucleotide and MADDAM signals were normalized to the corresponding 18S signals. In parallel, flow cytometry analysis was performed with anti-CD1a-PE, anti-CD14-FITC, and isotype control antibodies at day 8 and day 15 of culture. The median fluorescence and the number of positive cells are shown (B).
Distribution of MADDAM in tissues and cell lines
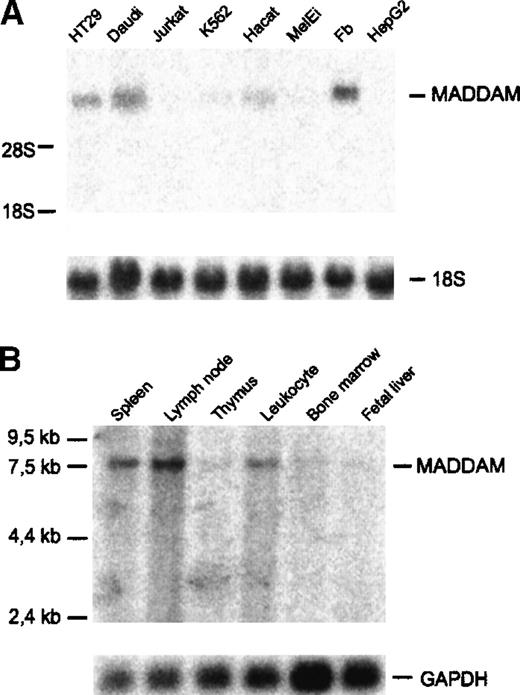
Next, we investigated the expression of MADDAM in various human cell lines and tissues by Northern blot analysis. MADDAM mRNA was detected in HT29, a colon adenocarcinoma cell line, in Daudi, a B-lymphoma cell line, and fibroblasts (N1), but not in Jurkat, a T-lymphoma cell line, and in MelEi, a melanoma cell line. Only weak signals were found in K562, a chronic myeloid cell line, and in HaCat, a keratinocyte cell line (Figure 5A). It is known that 1,25(OH)2VD3 induces differentiation of many cell types, especially myelomonocytic cell lines. Therefore, we cultured different myelomonocytic cell lines, HL-60, U937, and THP-1 cells either with 10−7 mol/L 1,25(OH)2VD3 or 10−8 mol/L PMA in the presence of 10% FCS, up to 3 days to induce MAC differentiation and examined the expression of MADDAM by Northern blot analysis of total RNA. In none of the myelomonocytic cell lines, MADDAM mRNA was expressed neither constitutively nor after induction of differentiation with 1,25(OH)2VD3 or PMA for 3 days (data not shown). Koski and colleagues54 demonstrated that HL-60 cells cannot only differentiate into MACs, but also develop DC morphology after culture for 2 days with calcium ionophore. Therefore, we cultured HL-60 for up to 2 days with 180 ng/mL calcium ionophore A23187 and found a weak induction of the expression of MADDAM mRNA after stimulation with calcium ionophore (data not shown).
Expression of MADDAM mRNA in human cell lines and tissues.
(A) Human cell lines; (B) tissues. The commercially available tissue blot contained poly(A)+RNA of the indicated tissues (Multiple tissue Northern blot II; Clontech). Northern blot analysis was performed as described “Materials and methods.” As a control for RNA loading, membranes were rehybridized with an 18S rRNA- (A) or GAPDH-oligonucleotide (B).
Expression of MADDAM mRNA in human cell lines and tissues.
(A) Human cell lines; (B) tissues. The commercially available tissue blot contained poly(A)+RNA of the indicated tissues (Multiple tissue Northern blot II; Clontech). Northern blot analysis was performed as described “Materials and methods.” As a control for RNA loading, membranes were rehybridized with an 18S rRNA- (A) or GAPDH-oligonucleotide (B).
Investigation of the MADDAM mRNA distribution in different tissues revealed a strong expression in lymph nodes, spleen, and blood. A weak expression was detected in thymus, bone marrow, and fetal liver (Figure5B).
In situ expression of MADDAM mRNA in human lymph nodes and in blood dendritic cells
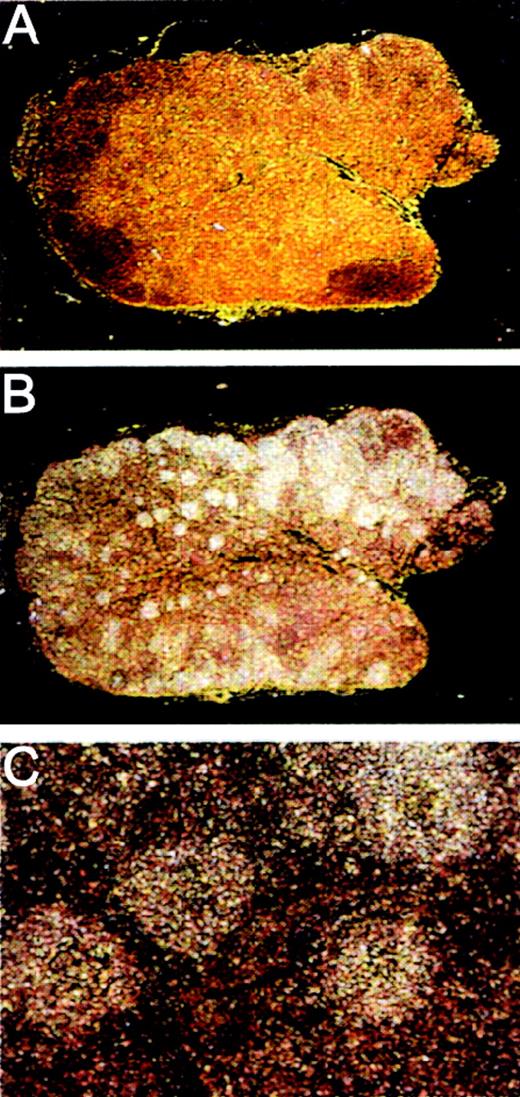
On the basis of the Northern blot analysis of human tissues of the immune system, we were interested in the localization of the MADDAM mRNA expression in human lymph nodes and performed in situ hybridization with 33P-labeled sense (Figure6A) and antisense (Figure 6B,C) cRNA probes of MADDAM. We found specific MADDAM mRNA signals distributed over the whole follicle (Figure 6B,C).
Localization of MADDAM mRNA in human lymph nodes by in situ hybridization.
Human lymph node paraffin embedded series sections were generated and hybridized with 33P-labeled sense (A) or antisense (B,C) probes of MADDAM as described in “Materials and methods.” In A and B, a 10 × magnification, in C a 100 × magnification is shown.
Localization of MADDAM mRNA in human lymph nodes by in situ hybridization.
Human lymph node paraffin embedded series sections were generated and hybridized with 33P-labeled sense (A) or antisense (B,C) probes of MADDAM as described in “Materials and methods.” In A and B, a 10 × magnification, in C a 100 × magnification is shown.
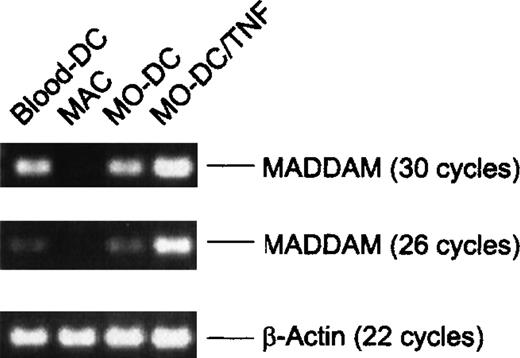
To further confirm the expression of MADDAM in DCs in vivo, we isolated blood-DCs by using the “Magnetic Cell Sorting” technique. Instead of the Northern blot analysis, we performed RT-PCR with the blood-DC RNA because only a small amount of cells (1-2 106 blood-DCs per isolation) could be obtained. As negative and positive controls, respectively, we used RNA of MO-derived MACs and DCs. According to the Northern blot analysis, we found signals of MADDAM in MO-derived DCs, but not in MO-derived MACs. Interestingly, blood-DCs showed a strong expression of MADDAM similar to the expression in MO-derived DCs. The β-actin cDNA was amplified as a control and after either 18 or 22 cycles, the amount of amplified cDNA was equal in all samples (Figure 7).
Expression of MADDAM in freshly isolated blood-DCs detected by RT-PCR.
For comparison MADDAM expression in MO-derived MACs and DCs generated from MOs with or without TNF-α is shown. As a control β-actin was amplified. No signals were detected by amplification of the corresponding amount of RNA omitting the RT step.
Expression of MADDAM in freshly isolated blood-DCs detected by RT-PCR.
For comparison MADDAM expression in MO-derived MACs and DCs generated from MOs with or without TNF-α is shown. As a control β-actin was amplified. No signals were detected by amplification of the corresponding amount of RNA omitting the RT step.
Discussion
MOs are common precursor cells of MACs and DCs in vitro, but little is known about the respective differentiation pathways in vivo.8 The aim of this study was to identify genes involved in the differentiation process of MOs to MACs. We identified a transcript, named “MADDAM,” that was up-regulated in short-term cultured human blood MOs by 1,25(OH)2VD3. Surprisingly, beside the expression of MADDAM in short-term cultured MOs, but not in MACs, MADDAM was expressed in DCs derived from MOs or CD34+ cells, indicating a role of MADDAM in DC differentiation.
The MADDAM sequences revealed strong homology to members of the ADAM (a disintegrin and metalloprotease) family, which is also known as MDC (metalloprotease/disintegrin/ cysteine-rich) family. The strongest homology was obtained to murine meltrin-β, which has been cloned recently by Inoue and colleagues.51 Proteins of the ADAM family contain several distinct protein modules, including a prodomain and metalloprotease domain, a disintegrin domain, a cysteine-rich domain, and an EGF repeat domain.35 They are involved in sperm-egg fusion,38,55 cell-cell fusion of myoblasts,56 and in processing of cytokines such as TNF-α and TRANCE.57-59 Meltrin-β is expressed in mouse fibroblasts and regulated by 1,25(OH)2D3 in mouse osteoblasts.51 Accordingly, we found MADDAM mRNA in human fibroblasts and the expression was up-regulated by 1,25(OH)2D3 in human MOs. In addition, murine meltrin-β is strongly expressed in various tissues such as bone marrow, heart, spleen, and brain, whereas the expression of MADDAM seems to be more restricted and was only weakly detected in bone marrow, fetal liver, or thymus by Northern blot analysis. Despite the fact of some differences in the expression pattern, we suggest, on the basis of the strong homology of 84% to the amino acid sequence of murine meltrin-β, that MADDAM is the human homologue of meltrin-β. As MADDAM contains a putative transmembrane region and a signal peptide, it is likely to be expressed on the surface of the respective cell. In contrast, decysin, a metalloprotease, which has recently been described in CD4+CD11c+dendritic cells isolated from tonsils, lacks a transmembrane region and therefore is possibly a secreted protein.60 In addition, there is also evidence that another metalloprotease, gelatinase B, is expressed in dendritic cells.61
MADDAM was not expressed in freshly isolated blood MOs and rapidly down-regulated during MO to MAC differentiation, but was induced during short-term culture of blood MOs by adherence and 1,25(OH)2D3. The role of 1,25(OH)2D3 for the expression of MADDAM in human MOs is unclear. The 1,25(OH)2D3 could either directly regulate MADDAM gene expression via the 1,25(OH)2D3 receptor and/or regulate the expression indirectly by the up-regulating of MO adhesion.10 In contrast to MOs, where 1,25(OH)2D3 is a known differentiation agent,10,11 1,25(OH)2D3 has been reported to suppress the differentiation of DCs.62 Our preliminary data indicate no influence of 1,25(OH)2VD3 on the regulation of MADDAM during the differentiation process of DCs.
The time course of MADDAM expression in MOs on the one hand and the possible function as metalloprotease and disintegrin on the other hand supports the hypothesis that MADDAM may be important for the early adhesion and migration process of MOs in vivo during their extravasation from the blood through the endothelium into the tissue. If MOs reach the tissue, adhere, and differentiate into MACs, they may become independent of MADDAM function and therefore the expression of this protein is silenced. However, the expression of another metalloprotease in human MOs/MACs, gelatinase B (type IV collagenase), is up-regulated during the final maturation of MOs to MACs in vitro,46 also suggesting a role of matrix proteins for the function of mature macrophages as described for fibronectin by Perri and colleagues.63
In contrast to MACs that are a more sessile type of immune cell, DCs have to be very mobile. During their early differentiation process, they migrate to different tissues, eg, skin, where they encounter and uptake antigens. Then, during the next step of differentiation, they mature and have to find their way to the lymph nodes, where they present the processed antigen to T lymphocytes, thereby inducing a specific immune response.7 These specific migration processes of DCs require the capacity to digest matrix proteins, eg, need to express metalloproteases such as MADDAM. Recently, it has been shown by Kobayashi64 that matrix metalloproteinase-9, but not matrix metalloproteinase-2, is involved in the migration and differentiation of murine Langerhans cells. MADDAM was expressed during the whole differentiation process of DCs in vitro, independent of whether we analyzed DCs generated from MOs or CD34+stem cell preparations. The final maturation of MO-derived DCs in vitro induced by TNF-α29,30,65 led to an increased MADDAM expression and therefore perhaps to an up-regulated migration. In addition, freshly isolated blood-DCs also showed a significant MADDAM expression. In the light of the recent publication of Randolph and colleagues,66 the capacity of MOs to reverse transmigrate after reaching the subendothelial compartment is a key event for the differentiation of MOs to DCs. MOs unable to reverse transmigrate become MACs. If MADDAM is involved in MO migration, the regulation of this metalloprotease may determine the fate of MOs, eg, differentiation into either DCs or MACs.
As other members of the ADAM family, such as the metalloprotease “TNF-α converting enzyme” (TACE/ADAM 17) and ADAM 10, are involved in TNF-α processing,57,58 67 MADDAM may also play a role in the autocrine regulation of cytokines, regulating DC differentiation to a more mature type of antigen-presenting cell.
In situ hybridization revealed expression of MADDAM in lymph follicles, but the assignment of MADDAM expression to a certain cell type of the follicle is difficult. Our data showed the expression of MADDAM in Daudi, a B-lymphoma cell line, and in freshly isolated blood-DCs, which are probable progenitor cells of the CD4+CD11c+DC subpopulation in germinal centers of lymph follicles.68This may point out that either the CD4+CD11c+germinal center DC population or B cells may be responsible for the respective signals of MADDAM in the lymph nodes. Most likely both cell types express MADDAM.
In conclusion, we isolated a 1,25(OH)2VD3–regulated mRNA transcript (MADDAM) in human blood MOs. On the basis of the GenBank data and the analysis of the amino acid sequence of the putative protein, we identified the human homologue to murine meltrin-β (ADAM 19), a new human member of the ADAM family. Because of the different expression pattern of MADDAM in MACs and DCs, we suggest that MADDAM may serve as a novel marker for the distinction of DCs from other cells of the myeloid pathway and may be directly involved in the divergence along the 2 distinct pathways toward DCs or MACs. Further investigations will clarify the possible role of MADDAM for the function of DCs and other cell types, such as B lymphocytes, in vitro and vivo.
Acknowledgments
We thank Prof Dr C. Bogdan for the helpful criticism in the preparation of the manuscript, Dr S. W. Krause for carrying out the FACS analysis of blood-DC, A. Pietryga-Krieger for performing the cDNA sequencing and S. Seegers for excellent technical assistance.
Supported by Deutsche Forschungsgemeinschaft (An 111/6-6).
Reprints:M. Kreutz, Department of Hematology/Oncology, University of Regensburg, Franz-Josef-Strauß-Allee 11, D-93042 Regensburg, Germany; e-mail:marina.kreutz@klinik.uni-regensburg.de.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal