Abstract
Acute promyelocytic leukemia (APL) is typified by the t(15;17) translocation, which leads to the formation of thePML/RARA fusion gene and predicts a beneficial response to retinoids. However, approximately 10% of all APL cases lack the classic t(15;17). This group includes (1) cases with cryptic PML/RARA gene rearrangements and t(5;17) that leads to the NPM/RARA fusion gene, which are retinoid-responsive, and (2) cases with t(11;17)(q23;q21) that are associated with the PLZF/RARA fusion gene, which are retinoid-resistant. A key issue is how to rapidly distinguish subtypes of APL that demand distinct treatment approaches. To address this issue, a European workshop was held in Monza, Italy, during June 1997, and a morphologic, immunophenotypic, cytogenetic, and molecular review was undertaken in 60 cases of APL lacking t(15;17). This process led to the development of a novel morphologic classification system that takes into account the major nuclear and cytoplasmic features of APL. There were no major differences observed in morphology or immunophenotype between cases with the classic t(15;17) and those with the cryptic PML/RARAgene rearrangements. Auer rods were absent in the t(5;17) case expressing NPM/RARA. Interestingly, this classification system distinguished 9 cases with t(11;17)(q23;q21) and, in addition, successfully identified 2 cases lacking t(11;17), which were subsequently shown to have underlying PLZF/RARA fusions. The PLZF/RARAcases were characterized by a predominance of blasts with regular nuclei, an increased number of Pelger-like cells, and by expression of CD56 in 4 of 6 cases tested. Use of this classification system, combined with an analysis for CD56 expression, should allow early recognition of APL cases requiring tailored molecular investigations.
Introduction
Acute promyelocytic leukemia (APL) was first recognized for its particularly poor clinical outcome due to a serious hemorrhagic syndrome, which is related to disseminated intravascular coagulopathy and abnormal fibrinolysis.1-5 APL is also typified by a unique differentiation response to retinoids, such as all-trans-retinoic acid (ATRA), which have transformed clinical practice for this condition.6-9 The initial morphological description of APL emphasized a specific hypergranular, promyelocyte-like, blast cell component in the bone marrow. Although the term promyelocytic leukemia has been widely accepted since 1957,1,2 it is partly misleading because APL cells are morphologically different from normal promyelocytes. In 1976, the French-American-British (FAB) cooperative group revised the morphological description of acute myeloid leukemias (AMLs); the term M3-AML was assigned to hypergranular promyelocytic leukemia characterized by blast cells with heavy azurophilic granules, bundles of Auer rods (faggots), and a reniform or bilobed nucleus.10
Major interest has focused on APL following the identification of a specific cytogenetic abnormality: t(15;17)(q22;q21) translocation.11 Although the vast majority of M3 cases fit the description of hypergranular or classical M3, a cytological hypogranular or microgranular variant form, M3v, has been identified. It is commonly associated with hyperleukocytosis,12accounts for 15%-20% of APL cases, and shares the same t(15;17). In M3v-AML, the majority of blasts have a bilobed, multilobed, or reniform nucleus and, under usual staining, are devoid of granules or contain only a few fine azurophilic granules; however, at least a few cells with all the cytoplasmic features of M3 are present.13 Rare morphological forms associated with t(15;17) have been reported. These cases are characterized by hyperbasophilic microgranular blast cells with cytoplasmic budding that mimicks micromegakaryocytes14-16 or by blast cells that exhibit toluidine blue positive basophilic granules17,18or eosinophilic granules.19 Other cases have been reported to have M1- or M2-like morphology.15 20
Immunophenotypic studies have shown a distinctive pattern in APL as compared to other AMLs: positivity for the CD33 and CD13 myeloid antigens, negativity for HLA-DR, and low frequency of CD34 expression.21-23 Positivity for this latter antigen and for CD2 and CD19 has been correlated with the M3v form.24,25 Interestingly, rare cases of HLA-DR−, CD33+ AML, whose morphology resembles M3v but lacks t(15;17) and expresses the natural killer (NK)-associated CD56 antigen, have been described.26 Infrequently, however, t(15;17)-associated APL can also exhibit positivity for this antigen.27
In 1990, t(15;17) was cloned and was found to disrupt a previously uncharacterized gene, PML, on chromosome 15 and the gene encoding the retinoic acid receptor α (RARA) on chromosome 17. The resultant PML/RARA fusion protein, which retains the retinoic acid (RA) ligand-binding domain, not only plays a key role in leukemogenesis, but it is also somewhat paradoxically implicated in mediating the response to retinoids.28 It was originally thought that all cases of APL were associated with t(15;17),29 but it is now clear that this chromosomal change is not invariably present. In some instances, fusion of thePML and RARA genes still occurs as a result of insertion events or more complex rearrangements,30,31whereas in others, RARA is fused to a gene other thanPML.28 To date, 3 alternative translocations associated with APL have been characterized: t(11;17)(q23;q21); t(5;17)(q35;q21); and t(11;17)(q13;q21). In these translocations,RARA is fused to the PLZF (promyelocytic leukemia zinc finger),32,33NPM(nucleophosmin),34-36 and NuMA (nuclear mitotic apparatus)37,38 genes, respectively. Recently, theSTAT5b gene located at 17q21 has been identified as a newRARA partner in a [der(17)] APL case.39
The nature of the fusion partner has a considerable impact on the characteristics of the disease.PML/RARA-mediated APL is sensitive to retinoids, which in combination with chemotherapy, have led to considerable improvements in outcome.9,40,41 Indeed, this condition is now one of the most prognostically favorable subtypes of AML.42 Preliminary studies suggest that APL associated with NPM/RARA and NuMA/RARA is also retinoid-responsive.38,43 In marked contrast,PLZF/RARA-associated APL, is resistant to ATRA in vitro, and patients previously treated with ATRA as a single-agent therapy had an adverse prognosis.33 Furthermore, distinct from t(15;17)-associated APL, preliminary studies suggest that patients with the PLZF/RARA fusion gene are also resistant to arsenic trioxide,44,45 but they could be sensitive to the combination of ATRA and granulocyte colony-stimulating factor (G-CSF).46 Clearly, the underlying molecular lesion associated with APL has considerable implications for an optimal therapeutic approach.
Because initiation of therapy is highly dependent upon a successful morphological diagnosis, this study sought to determine whether APL cases lacking t(15;17) could be reliably identified. This issue was addressed at a European workshop held in Monza, Italy, during June 1997, whereby a large series of cases with morphologically suspected APL, but lacking t(15;17), were referred for central morphological, immunophenotypic, cytogenetic, and molecular review. This process led to the development of a novel morphological classification system that was found to reliably distinguish APL cases with thePLZF/RARA fusion gene from those with ATRA-responsive disease, including those with underlyingPML/RARA and NPM/RARA gene rearrangements.
Materials and methods
Patient characteristics
The 90 cases referred as APL at diagnosis and lacking the classical t(15;17) were provided by 42 laboratories from 6 European countries and from Memphis, TN (see “”). For controls, we examined 20 further cases of APL with the classical t(15;17). The corresponding karyotypes and molecular data were reviewed concomitantly but separately. Morphological reviewers were ignorant of the cytogenetic and molecular data and vice versa. Full details of the molecular and cytogenetic characterization of these cases are presented by Grimwade et al.31 In a second step, the reviewed data were combined, and the immunophenotypic data were considered. Unique patient numbers were assigned to each case and are consistent with those of Grimwade et al.31
We excluded 23 cases: 4 cases (1 M1-AML and 3 M2-AML cases) were excluded as a result of morphological review, and furthermore, all were negative for the PML/RARA transcript by reverse transcriptase–polymerase chain reaction (RT-PCR); 4 cases were excluded as a result of cytogenetics because of the presence of a minor clone with the classical t(15;17); and 15 cases were excluded due to insufficient material for adequate molecular analysis. We retained 67 cases and, on the basis of molecular and cytogenetic analyses, classified them as cases with or withoutPML/RARA gene rearrangements
Cases with a PML/RARA gene rearrangement.The majority of these 49 cases were due to insertion events including 27 cases with documented formation of PML/RARAand 1 case in which RARA/PML was the sole fusion gene created. In 14 of 49 workshop cases, thePML/RARA fusion gene resulted from complex chromosomal rearrangements. In the remaining 7PML/RARA positive cases, conventional cytogenetic analysis revealed an isochromosome for the long arm of the derivative chromosome 17, ider(17)(q10)t(15;17)(q22;q21), [ider(17)]. These cases were subsequently defined as t(15;17)-positive with an additional abnormality, and they were retained to determine whether this superimposed cytogenetic abnormality influenced morphological appearances. This latter group was excluded from the accompanying molecular study,31 which considered only cases lacking t(15;17).
Cases lacking a PML/RARA gene rearrangement.
Of 18 cases lacking a PML/RARA gene rearrangement, 11 cases possessed PLZF/RARA rearrangements including 2 new cases lacking t(11;17)(q23;q21); 6 of the cases with documented t(11;17) were reported previously.26,33,44,46-49 There were 2 cases with t(5;17): a new case with t(5;17)(q34;q21), expressing NPM/RARA, and a case with an unbalanced der(5)t(5;17), which has been partially described previously,50,51 with a proven deletion of 1RARA allele.31 In the remaining 5 cases, theRARA gene was not found to be rearranged.
Morphological analysis
Bone marrow smears were reviewed by a committee of morphologists (D.S., V.L., A.C.R., D.H., and G.F.) using a Leica-ICG video system (Rueil-Malmaison, France). Peripheral blood films were reviewed, too; but according to the FAB classification,10they were not considered by themselves as sufficient for diagnosis. Therefore, each case had a minimum of 20 fields from bone marrow smears registered at a 1000-fold magnification and, according to general agreement, was classified using FAB criteria.10 13Furthermore, for each case, a total of 100 bone marrow blast cells were analyzed by this committee, thereby taking into separate account the major nuclear and cytoplasmic features considered characteristic of M3/M3v cells (Figures 1 and2). The blasts were initially classified according to the appearance of the nuclear outline, regular (round or oval) or irregular (reniform, bilobed, or folded). They were then further categorized according to the degree of granularity of the cytoplasm (hypergranular or hypogranular). Finally, the presence or absence of Auer rods was taken into account. Faggots were defined by the presence of at least 3 Auer rods per cell.
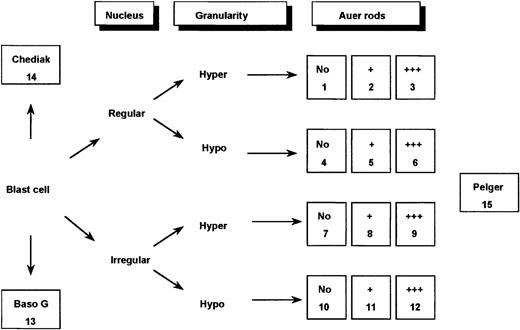
Classification system used for defining 15 categories of cells.
In the figure, Hyper indicates hypergranular cytoplasm; Hypo, hypogranular or agranular cytoplasm; +, the presence of 1 or 2 Auer rods; +++, the presence of 3 or more Auer rods or faggots; Baso G, basophilic granules; Chediak, Chediak-like inclusions; and Pelger, Pelger-like cells.
Classification system used for defining 15 categories of cells.
In the figure, Hyper indicates hypergranular cytoplasm; Hypo, hypogranular or agranular cytoplasm; +, the presence of 1 or 2 Auer rods; +++, the presence of 3 or more Auer rods or faggots; Baso G, basophilic granules; Chediak, Chediak-like inclusions; and Pelger, Pelger-like cells.
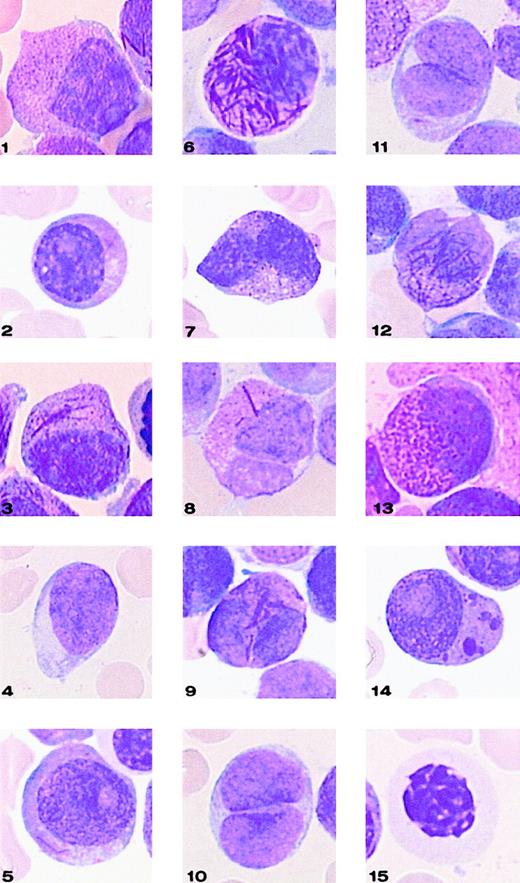
Representative cells from each cell category of the classification system.
Bone marrow smears were stained with May-Grünwald-Giemsa and registered at a 1000-fold magnification on a Leica-ICG system database.
Representative cells from each cell category of the classification system.
Bone marrow smears were stained with May-Grünwald-Giemsa and registered at a 1000-fold magnification on a Leica-ICG system database.
Application of this 3-parameter classification system (nucleus, granularity, and Auer rods) led to the distinction of 12 categories of cells (Figures 1 and 2). The classification system also allowed for the presence of a series of additional morphological changes including basophilic or Chediak-Higashi granules. The blasts with these latter features were initially assigned to a category within the 3-parameter classification system (ie, categories 1-12), then recounted and reassigned to category 13 (basophilic granules) or category 14 (Chediak granules) (Figures 1 and 2). A category initially planned for identifying hyperbasophilic cytoplasm was not used because the hyperbasophilic variant form of APL was absent from our series. Apart from blasts, a 15th category was defined by Pelger-like maturing cells (Figures 1 and 2) whose percentage was evaluated on total cells from granulocytic lineage, including blasts (n = 100), and reported on the grid. In a second step, cytogenetic and molecular data were taken into account, and a mean value was calculated for each morphological cell category within each cytogenetic/molecular group (Figure3).
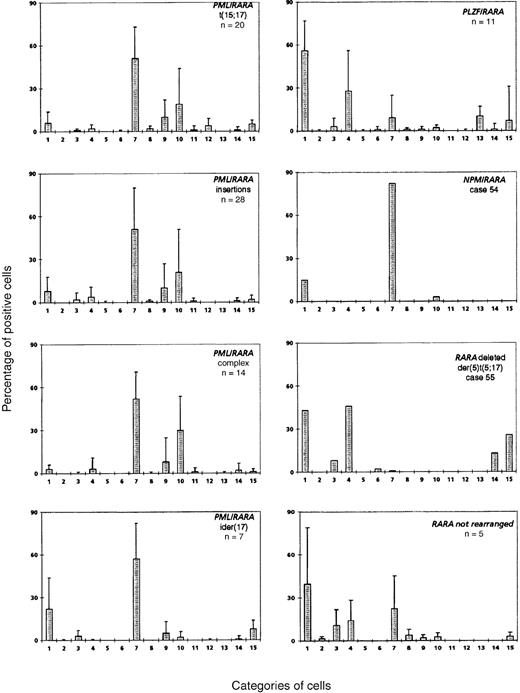
Mean percentage of positive cells in each cell category according to the cytogenetic/molecular groups.
Cell categories are noted in Figure 1, and the mean of positive cells is given as plus or minus the SD. A total of 100 blasts were analyzed for each case, assigned to categories 1-12, and then recounted and reassigned to category 13 or 14. Category 15 was defined by Pelger-like maturing cells whose percentage was evaluated on the total cells, including blasts, from the granulocytic lineage (n = 100).
Mean percentage of positive cells in each cell category according to the cytogenetic/molecular groups.
Cell categories are noted in Figure 1, and the mean of positive cells is given as plus or minus the SD. A total of 100 blasts were analyzed for each case, assigned to categories 1-12, and then recounted and reassigned to category 13 or 14. Category 15 was defined by Pelger-like maturing cells whose percentage was evaluated on the total cells, including blasts, from the granulocytic lineage (n = 100).
Immunophenotype
Immunophenotypic features of 63 of 87 APL cases, including the 20 t(15;17) controls, were available and thus reviewed. This process took into account surface antigens considered to have a characteristic expression pattern in APL: HLA-DR, CD34, CD13, and CD33. In some instances, the CD56 antigen that has been associated with APL was studied. The CD16 data were also available for some CD56+cases. Unfortunately, as limited data on the CD2, CD15, and CD19 expression were available, these antigens were not considered. Immunophenotyping was performed by cytofluorometry using direct immunofluorescence either on bone marrow samples (58 of 63 cases) after centrifugation through a Ficoll-Hypaque density gradient (Pharmacia Biotech) or on total peripheral blood (5 of 63 cases) after lysis of red blood cells, in cases where the blast cell count exceeded 50 × 109 cells per L (80%-99% blast cells).
We used fluorescein isothiocyanate (FITC)- or phycoerythrin (PE)-conjugated monoclonal antibodies (mAbs) (Immunotech International, Marseille, France; Beckman Coulter, High Wycombe, England; and Dako SA, Trappes, France, and High Wycombe, England). For each sample, a minimum of 5000 cells were gated on forward and side scatter parameters, and thus, at least 90% of the analyzed cells were blasts. In all cases, negative controls included mouse immunoglobulin (Ig) directly conjugated to FITC or PE. Expression in more than 20% blast cells was required to define antigen positivity. For the purposes of this study, the typical APL phenotype was defined as CD34−, HLA-DR−, CD13+, CD33+, and CD56−. Immunophenotypic results were compared to white blood cell (WBC) counts; cases with WBCs greater than 10 × 109 cells per L were considered as having hyperleukocytosis.
Statistical analysis
The Fisher exact test was used to test for associations between the cytogenetic/molecular subgroup, M3/M3v morphology, hyperleukocytosis, and immunophenotype.
Results
Morphological analysis of cases withPML/RARA rearrangements
Control cases with the classical t(15;17).
The mean percentage of each cell category in each cytogenetic/molecular group is reported in Figure 3. In the control group (n = 20) with classical t(15;17), the majority of blasts had an irregular nucleus and hypergranular cytoplasm, which either lacked Auer rods (category 7; range, 3%-90%; mean, 51%) or contained faggots (category 9; range, 0%-41%; mean, 10%). These 2 categories correspond to the classical M3 form of the FAB classification,10 which accounted for 16 of 20 control cases. Another peak was noted with 18% of the cells falling within category 10, ie, irregular nucleus, poorly granular cytoplasm, and no Auer rods. These cells therefore corresponded to the characteristic cells of the M3v form of the FAB classification.13 These cells were not a minor component of the classical form of M3, but all cells belonged to the M3v subtype, which accounted for the remaining 4 cases. Indeed, each of these M3v cases had a majority of blast cells classified in category 10 (range, 73%-90%) and very few blasts belonging to category 7 (range, 3%-25%) or category 9 (range, 0%-8%).
Within the whole control group, Auer rods were seen in each M3 and M3v case; faggots were absent in 4 cases (2 M3 and 2 M3v cases); only a minority of blasts with regular nuclei were observed (categories 1-6; range, 0%-27%; mean, 9%); no basophilic granules (category 13) were noted; and Chediak-like inclusions were seen in only 1 case in a few blast cells (category 14; 1%). Pelger-like cells were rarely observed (category 15; range, 0%-10%; mean, 3%) and were not hypogranular; dysplastic features were not observed in the rare cells from the erythroid and megakaryocyte lineages. Using this classification system, M3 and M3v cases can be readily distinguished; M3 cases are characterized by a majority of blasts of categories 7-9; while in M3v, the majority of blasts belong to categories 10-12.
Cases lacking the classical t(15;17) but with PML/RARA rearrangements.
No major morphological difference was seen between the control group and the PML/RARA+ groups (Figure3). In the insertion group, the proportion of M3 and M3v cases (22 M3 and 6 M3v cases) and the morphological profile were similar to the t(15;17) control group. Only 2 cases classified as M3 had more than 33% blasts with regular nuclei (cases 8 and 26), and other cases had less than 27% of these cells (overall: categories 1-6; range, 0%-75%; mean, 9%). The unique insertion case solely expressingRARA/PML31 belonged to the M3v category (category 10, 81% cells).
The complex group included a slight excess of M3v cases compared with the t(15;17) control group (8 M3 and 6 M3v cases, respectively). Only case 32 had a moderate percentage of blasts with regular nuclei, whereas other cases had less then 12% of these cells (categories 1-6; range, 0%-36%; mean, 5%). A slight difference was observed in the ider(17) group. Each of the 7 total cases was classified as classical M3, as the majority of blasts had hypergranular cytoplasm. However, the morphological profile of this group differed from that of the control group with respect to a higher percentage of category 1 cells (range, 3%-62%; mean, 22%), ie, regular nucleus and hypergranular cytoplasm without Auer rods (Figure 4). In the ider(17) group, blasts with regular nuclei (categories 1-6) accounted for a mean value of 26% (range, 3%-69%); 3 cases had more than 33%, whereas other cases had less than 20% of these cells. In addition, a slight increase in Pelger-like cells (category 15; range, 0%-17%; mean, 8%), mostly degranulated, was observed (Figure4).
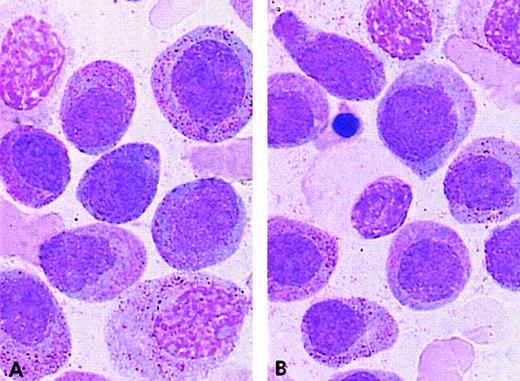
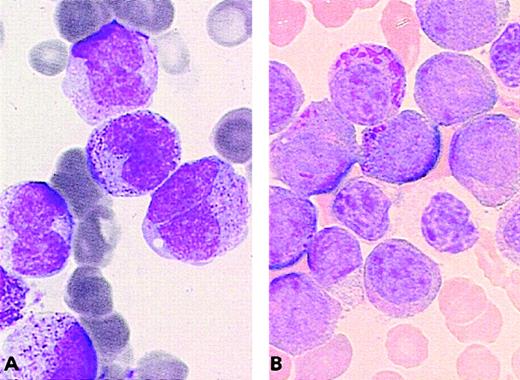
PML/RARA+ cases of the ider(17) group.
The majority of blasts with (A, B) regular nuclei and (B) a Pelger-like cell.
PML/RARA+ cases of the ider(17) group.
The majority of blasts with (A, B) regular nuclei and (B) a Pelger-like cell.
Morphological analysis of cases lackingPML/RARA rearrangements
Cases with PLZF/RARA rearrangements.
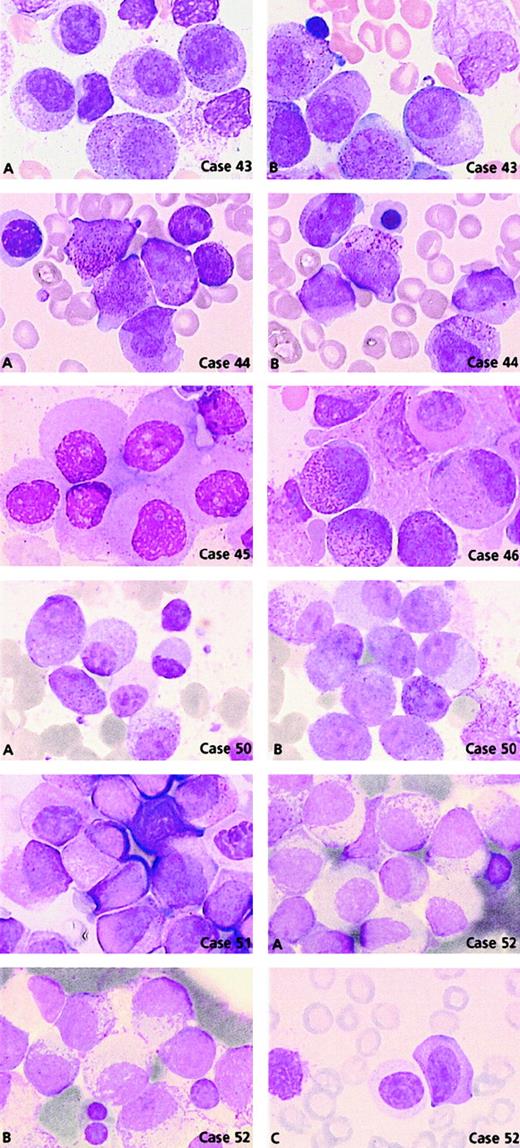
Comparison of the morphological features of t(15;17) controls with thePLZF/RARA+ group (n = 11) revealed clear differences (Figures 3 and 5). Only 2 cases, 44 and 46, could be readily classified as APL. Case 44 had 62% blasts with irregular nuclei; most of them had hypergranular cytoplasm (53% blast cells belonged to categories 7-9) and could be classified using FAB criteria as M3. In case 46, 80% basophilic granular cells (category 13) were observed, which corresponded to the basophilic M3-like form,17 but the majority (77%) of blasts corresponded to category 1 (ie, regular nucleus and hypergranular cytoplasm without Auer rods) (Figure 5). Other cases were reminiscent of M3 according to their granularity and sometimes by the presence of faggots. However, the majority of blasts had a regular round or oval nucleus that was more suggestive of FAB type M2, but these cases were retained as APL because of the presence of hypergranular cells. Indeed, in the whole PLZF/RARA group, the majority of blasts had (1) a regular nucleus (categories 1-6; range, 38%-100%; mean, 88%) and (2) an abundant cytoplasm with either coarse granules (category 1; range, 29%-83%; mean, 56%; Figure 5, case 43A, B) or, less frequently, with fine or no granules (category 4; range, 3%-67%; mean, 28%; Figure 5, cases 45, 50A, 51, and 52A). Only 2 cases, 43 and 53, exhibited faggots (category 3; 15% and 18%, respectively; Figure 5, case 43B). Case 44 was the sole case with Chediak-like granules (category 14; 12%) (Figure 5, case 44B).
PLZF/RARA cases.
Case 43: Blasts have regular nuclei with (A) hypergranular cytoplasm and (B) faggots of Auer rods. Case 44: Blasts have irregular nuclei with (A) hypergranular cytoplasm and (B) Chediak-like granules. Case 45: Blasts have regular nuclei and poorly granular cytoplasm. Case 46: Blasts with regular nuclei, basophilic granules, and a Pelger-like cell. Case 50: Blasts with regular nuclei and either (A) hypogranular or (B) hypergranular cytoplasm and (A) a Pelger-like cell. Case 51: Majority of blasts have regular nuclei and a Pelger-like cell. Case 52: Blasts with regular nuclei and either (A) hypogranular or (B) hypergranular cytoplasm with myelocyte-like features and (C) a Pelger-like cell.
PLZF/RARA cases.
Case 43: Blasts have regular nuclei with (A) hypergranular cytoplasm and (B) faggots of Auer rods. Case 44: Blasts have irregular nuclei with (A) hypergranular cytoplasm and (B) Chediak-like granules. Case 45: Blasts have regular nuclei and poorly granular cytoplasm. Case 46: Blasts with regular nuclei, basophilic granules, and a Pelger-like cell. Case 50: Blasts with regular nuclei and either (A) hypogranular or (B) hypergranular cytoplasm and (A) a Pelger-like cell. Case 51: Majority of blasts have regular nuclei and a Pelger-like cell. Case 52: Blasts with regular nuclei and either (A) hypogranular or (B) hypergranular cytoplasm with myelocyte-like features and (C) a Pelger-like cell.
Noticeably, in 5 of 11 PLZF/RARA cases, the majority of blasts had a more condensed nuclear chromatin pattern. Compared with blasts from the t(15;17) control group, the PLZF/RARA blasts had more copious and less basophilic cytoplasm, appeared more mature, and thus resembled cells that were blocked at a promyelocyte-myelocyte stage (Figure 5, cases 43A, 44, 50A, 51, and 52B). Our classification system revealed an increased number of Pelger-like cells (category 15; range, 0%-22%; mean, 10%) which were hypogranular and seen in all cases except case 49 (Figures 3 and 5, cases 46, 50A, 51, and 52C). Such appearances were reminiscent of a myelodysplastic syndrome (MDS); however, in contrast to MDS, the erythroid and megakaryocyte lineages were absent or very reduced, impairing qualitative analysis, and the majority of cells appeared blocked at the promyelocytic-myelocytic stage.
Two cases lacking t(11;17) had morphological features typical of this group; they were subsequently shown to have underlyingPLZF/RARA rearrangements.31 Case 50 was associated with del(11)(q23) terminal deletion with a breakpoint at 11q23 and case 52 with a normal karyotype. The latter case was shown to have a submicroscopic insertion of RARAinto band 11q23, thereby leading to the expression ofPLZF/RARA as the sole fusion transcript. Both cases had a similar morphology (Figure 5, cases 50A, B and 52A-C): blasts with regular nuclei and either hypergranular cytoplasm (category 1; 57% and 52%, respectively) or hypogranular cytoplasm (category 4; 43% and 48%, respectively) associated with a chromatin pattern similar to that observed in cases with documented t(11;17). In these 2 cases, Pelger-like cells were also noted (category 15; 8% and 2%, respectively) (Figure 5, cases 50A and 52C).
Cases with t(5;17).
There were no morphological similarities between the 2 APL workshop cases (54 and 55) associated with t(5;17), and they were subsequently shown to be molecularly different.31 In case 54 (a 9-year old child), t(5;17)(q34;q21) led to the formation of NPM/RARAand RARA/NPM fusion genes. In case 55, with an unbalanced der(5)t(5;17), molecular analysis revealed no evidence of anNPM/RARA rearrangement, and 1 RARA allele was found to be deleted.31 The morphology of case 54 fitted with M3, and the majority of blasts (97%) were hypergranular, with an irregular or less frequently regular nuclear outline (category 7, 82%; category 1, 15%). In contrast with the t(15;17) control group, no Auer rods were seen (Figures 3 and 6A).
Chromosome transcription t(5;17) cases.
(A) The NPM/RARA case: Majority of blasts have irregular nuclei and hypergranular cytoplasm without Auer rods. (B) The der(5)t(5;17) case: Blasts with regular nuclei, hypergranular cytoplasm with Chediak-like granules, and Pelger-like cells.
Chromosome transcription t(5;17) cases.
(A) The NPM/RARA case: Majority of blasts have irregular nuclei and hypergranular cytoplasm without Auer rods. (B) The der(5)t(5;17) case: Blasts with regular nuclei, hypergranular cytoplasm with Chediak-like granules, and Pelger-like cells.
The unbalanced der(5)t(5;17) case 55 had hyperleukocytosis (Table1) and a very peculiar morphology: the high percentage of blasts with hypergranular cytoplasm and the presence of faggots were reminiscent of M3, but the predominance of blasts with regular nuclei (categories 1-6, 99%) was more suggestive of M2. About half of the blasts possessed hypergranular cytoplasm (category 1, 43%) and half were hypogranular (category 4, 46%). A few faggots were seen (category 3, 8%; category 6, 2%). In addition, Pelger-like cells were numerous (category 15, 26%), and a significant proportion of blasts contained Chediak-like granules (category 14, 13%) (Figure 6B). This profile (Figure 3) was thus reminiscent ofPLZF/RARA-associated APL; however, an underlyingPLZF/RARA fusion gene was excluded by molecular analysis.50
WBC count and immunophenotypic characteristics of APL cases
| Groups (cases) . | WBC count, ×109 cells per L . | HLA-DR (%) . | CD34 (%) . | CD13 (%) . | CD33 (%) . | CD56 (%) . |
|---|---|---|---|---|---|---|
| PML/RARA+(50/69)* | ||||||
| t(15;17) control group (20/20) | (20/20)† | − (19/20)‡ | − (19/20) | + (19/20) | + (16/20) | − (20/20) |
| Range | 0.5-86.1 | 0-371-153 | 0-46 | 8-98 | 0-98 | 0-8 |
| Median | 1.7 | 0 | 0 | 79 | 74 | 0 |
| Insertions (16/28) | (27/28) | − (13/14) | − (12/15) | + (15/16) | + (14/14) | − (4/5) |
| Range | 0.2-257 | 0-25 | 0-74 | 0-98 | 30 -100 | 0-30 |
| Median | 4.4 | 7 | 4 | 85 | 91 | 1 |
| Complex (10/14) | (13/14) | − (9/10) | + (6/10)1-155 | + (9/9) | + (8/9) | − (2/2) |
| Range | 0.7-60 | 0-84 | 0-91 | 30-94 | 20-97 | 0-2 |
| Median | 3.1 | 6 | 55 | 60 | 53 | 1 |
| ider(17) (4/7) | (7/7) | − (3/4) | − (4/4) | + (4/4) | − (3/4) | − (1/1) |
| Range | 0.8-10.8 | 0-43 | 0-3 | 46-97 | 0-85 | 0 |
| Median | 2.8 | 0 | 1 | 69 | 5 | 0 |
| PLZF/RARA+(7/11) | (11/11) | − (7/7) | − (7/7) | + (7/7) | + (6/7) | + (4/6)1-154 |
| Range | 2-69.5 | 0-13 | 0-3 | 77-100 | 0-100 | 0-100 |
| Median | 9.9 | 1 | 0 | 96 | 93 | 70 |
| t(5;17) (2/2) | ||||||
| NPM/RARA(1/1) | 17 | − 0 | − 0 | − 6 | + 97 | − 6 |
| der (5) t(5;17) | 43.1 | − 9 | − 0 | − 13 | + 83 | − 0 |
| RARA not rearranged (4/5) | (5/5) | − (4/4) | − (3/4) | + (3/4) | + (3/4) | − (2/2) |
| Range | 3.3-84 | 0-7 | 0-22 | 1-98 | 1-99 | 1-9 |
| Median | 11.3 | 2 | 1 | 82 | 89 | 5 |
| Groups (cases) . | WBC count, ×109 cells per L . | HLA-DR (%) . | CD34 (%) . | CD13 (%) . | CD33 (%) . | CD56 (%) . |
|---|---|---|---|---|---|---|
| PML/RARA+(50/69)* | ||||||
| t(15;17) control group (20/20) | (20/20)† | − (19/20)‡ | − (19/20) | + (19/20) | + (16/20) | − (20/20) |
| Range | 0.5-86.1 | 0-371-153 | 0-46 | 8-98 | 0-98 | 0-8 |
| Median | 1.7 | 0 | 0 | 79 | 74 | 0 |
| Insertions (16/28) | (27/28) | − (13/14) | − (12/15) | + (15/16) | + (14/14) | − (4/5) |
| Range | 0.2-257 | 0-25 | 0-74 | 0-98 | 30 -100 | 0-30 |
| Median | 4.4 | 7 | 4 | 85 | 91 | 1 |
| Complex (10/14) | (13/14) | − (9/10) | + (6/10)1-155 | + (9/9) | + (8/9) | − (2/2) |
| Range | 0.7-60 | 0-84 | 0-91 | 30-94 | 20-97 | 0-2 |
| Median | 3.1 | 6 | 55 | 60 | 53 | 1 |
| ider(17) (4/7) | (7/7) | − (3/4) | − (4/4) | + (4/4) | − (3/4) | − (1/1) |
| Range | 0.8-10.8 | 0-43 | 0-3 | 46-97 | 0-85 | 0 |
| Median | 2.8 | 0 | 1 | 69 | 5 | 0 |
| PLZF/RARA+(7/11) | (11/11) | − (7/7) | − (7/7) | + (7/7) | + (6/7) | + (4/6)1-154 |
| Range | 2-69.5 | 0-13 | 0-3 | 77-100 | 0-100 | 0-100 |
| Median | 9.9 | 1 | 0 | 96 | 93 | 70 |
| t(5;17) (2/2) | ||||||
| NPM/RARA(1/1) | 17 | − 0 | − 0 | − 6 | + 97 | − 6 |
| der (5) t(5;17) | 43.1 | − 9 | − 0 | − 13 | + 83 | − 0 |
| RARA not rearranged (4/5) | (5/5) | − (4/4) | − (3/4) | + (3/4) | + (3/4) | − (2/2) |
| Range | 3.3-84 | 0-7 | 0-22 | 1-98 | 1-99 | 1-9 |
| Median | 11.3 | 2 | 1 | 82 | 89 | 5 |
The positivity of each immunophenotype is given on the left, the indicated characteristics (number of cases and percentages) on the right.
Indicates the number of cases with available phenotype/total number of cases.
Indicates the number of cases with available WBC/total number of cases.
Indicates the number of positive (+) or negative (−) cases/number of tested cases, the cut-off is more than 20% positive cells.
Indicates the percentage of positive cells.
P = .002, comparing CD34 positivity between complex cases and noncomplex cases in the PML/RARAgroup; CD34 positivity among complex cases reflected an association with M3v morphology.
P = .001, comparing CD56 positivity between PLZF/RARA and PML/RARA cases; P = .0008, comparing CD56 positivity between PLZF/RARA and all non-PLZF/RARA cases.
APL cases lacking evidence of a RARA rearrangement.
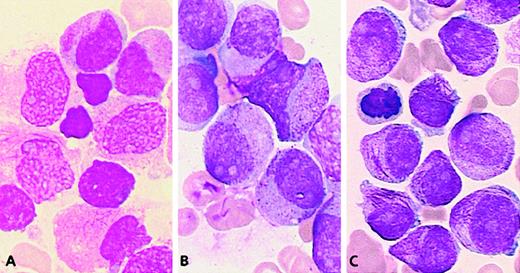
In 5 workshop cases, molecular analysis revealed no evidence of an underlying RARA rearrangement.31 Two cases (59 and 60) could be classified as M3 (Figure7A) because they exhibited a predominance of blasts with irregular nuclei. The remaining cases (56, 57, and 58) were difficult to classify as M3 or M3v according to FAB criteria as they presented with a majority of blasts with regular nuclear outline (categories 1-6, more than 67% in each case). But they were nonetheless retained as APL because of the presence of hypergranular blasts (Figure 7B,C). All cases in the whole group possessed more than 36% blasts with regular nuclei (categories 1-6; range, 37%-93%; mean, 66%). This profile (Figure 3) was close to that ofPLZF/RARA+ APL. In contrast to the typical features observed in the latter subtype of APL, case 56 exhibited a high number of blasts with faggots (category 3, 55%) (Figure 7C), and Pelger-like cells were rarely observed (category 15; range, 0%-13%; mean, 3%). The ATRA responsiveness of cases lacking RARArearrangements could not be formally established due to the lack of material available for in vitro differentiation assays. Nevertheless, in case 58, there was no observed response after 14 days of ATRA treatment. Sensitivity to retinoids could not be determined in the remaining patients because 3 patients received ATRA in combination with chemotherapy, and 1 patient died prior to therapy.
Cases lacking RARA rearrangements.
(A) Case 60: The majority of blasts have irregular nuclei, hypergranular cytoplasm, and faggots. (B) Case 57: The blasts have regular nuclei and hypergranular cytoplasm. (C) Case 56: Blasts with the same nuclear features and a high number of faggots.
Cases lacking RARA rearrangements.
(A) Case 60: The majority of blasts have irregular nuclei, hypergranular cytoplasm, and faggots. (B) Case 57: The blasts have regular nuclei and hypergranular cytoplasm. (C) Case 56: Blasts with the same nuclear features and a high number of faggots.
Immunophenotypic analysis of cases withPML/RARA rearrangements
Details of the presenting WBC count and immunophenotype of the t(15;17) control group and the cases withPML/RARA rearrangements defined by molecular methods are presented in Table 1. No significant differences were observed between the immunophenotypic profile or the WBC count of the molecularly defined group and the t(15;17) controls. Overall, there were 69 cases with PML/RARA rearrangements (53 M3 and 16 M3v cases). The median WBC count was low for the whole group (median, 3.1 × 109 cells per L; range, 0.2-257 × 109 cells per L), reflecting a preponderance of M3 cases (median, 2.4 × 109 cells per L; range, 0.5-97 × 109 cells per L), whereas the median WBC count of the M3v cases was significantly higher (median, 32.9 × 109 cells per L; range, 0.2-257 × 109 cells per L;P = .0004).
In 50 cases it was possible to correlate immunophenotype with morphological features. There were no differences observed between M3 and M3v in the expression pattern of HLA-DR (M3: median, 3.5%; range, 0%-84%; and M3v: median, 0%; range, 0%-13%); CD13 (M3: median, 82%; range, 0%-98%; and M3v: median, 70%; range, 30%-97%); or CD33 (M3: median, 78%; range, 0%-100%; and M3v: median, 83%; range, 5%-98%). However, expression of CD34 was correlated with M3v morphology (M3: median, 0%; range, 0%-74%; and M3v: median, 23%; range, 0%-91%; P = .002) and with hyperleukocytosis (CD34+: median WBC, 29 × 109 cells per L; range, 0.8-133 × 109 cells per L; and CD34−: median WBC, 1.9 × 109 cells per L; range, 0.5- 257 × 109 cells per L; P = .023). CD56 expression was found to be rare in APL with PML/RARArearrangements, with only 1 positive case among 28 cases tested. The PML breakpoint was determined in 41 cases including 13 controls; 23 cases had a 3′-bcr1-2 breakpoint (19 M3 and 4 M3v cases), and 18 cases had a 5′-bcr3 breakpoint (11 M3 and 7M3v cases; P = .16).
Immunophenotypic analysis of cases lackingPML/RARA rearrangements
Cases with PLZF/RARA rearrangements.
In this group, 5 of 11 cases had hyperleukocytosis, and the median WBC count was moderate (9.9 × 109 cells per L). Immunophenotyping performed in 7 of 11 cases showed that the blasts were typically HLA-DR−, CD34−, CD13+, and CD33+ (Table 1). Interestingly, the CD56 antigen was found to be strongly expressed in 4 of 6 tested cases (case 47, 100%; case 49, 53%; case 51, 90%; and case 52, 86%) and weakly in one case (case 50, 16%), with a median of 70%. In 2 cases strongly expressing CD56, CD16 expression was negative in case 47 and positive in case 51.
Cases with t(5;17).
Cases 54 and 55 had hyperleukocytosis. With the exception of the lack of CD13 expression, their immunophenotype was typical of APL (Table 1), with negativity for CD56 expression.
APL cases lacking evidence of a RARA rearrangement.
In this group, 3 of 5 cases had hyperleukocytosis, and the median WBC count was higher (11.3 × 109 cells per L). Immunophenotyping performed in 4 of 5 cases revealed no major differences from t(15;17) APL (Table 1). CD34 was weakly expressed in case 60, which had hyperleukocytosis, and CD56 was negative in the 2 cases tested (56 and 58).
Discussion
We performed morphological review of 90 cases referred as APL and lacking the classical t(15;17); using the FAB criteria, we were able to formally exclude 4 cases (1 M1 and 3 M2 cases) and to classify the majority of cases as either M3 or M3v. Regarding morphology, WBC count, or immunophenotype, there was no major difference seen between the t(15;17) control group and the cases lacking t(15;17) with molecular evidence of PML/RARA rearrangements. All cases with ider(17) possessed M3 morphology, which is in accordance with 9 previously published cases.52,53 If this association is confirmed in a larger series, it could be related to duplication of theRARA/PML gene and/or loss of the p53 gene on the ider(17). In the whole PML/RARA group, hyperleukocytosis was associated with M3v morphology and CD34 expression, in accordance with previous studies.24 25
In contrast to the PML/RARA cases, all cases with PLZF/RARA rearrangements were reminiscent of M3 with regard to the hypergranular cytoplasm. Thus they were retained in this study, but in 10 of 11 cases, the majority of blasts lacked the bilobed or folded nuclei characteristic of M3 or M3v. The chromatin pattern was also more condensed than typically observed in AML blasts, mimicking a block at the promyelocytic-myelocytic stage, and in addition, hypogranular Pelger-like cells were observed in the majority. Hence, these cases possessed morphological features associated with FAB types M2 and M3 as well as MDS. Previous reports of t(11;17)-associated APL indicate a wide range of morphological appearances. In a review of 6 cases,33 common morphologic features distinct from M3 or M3v were found: the blasts were more granular than typical M2-AML cases, but less granular than usually observed in M3-AML (hypergranular). Furthermore, the authors focused on the: (1) loss of the usual reddish color shift of M3 granules, (2) absence of the obscured nuclear outline typically observed in M3, (3) absence of faggot cells, (4) loss of the bilobed grooved nucleus, and (5) presence of fine granules reminiscent of M3v-AML. We agree with regard to the rarity of Auer rods and faggot cells and the loss of the bilobed grooved nucleus.33 Furthermore, our classification system demonstrates that this latter feature (regular nuclear outline) is the key criterion in distinguishing this subtype of APL; in addition, we have identified a number of other features of value including a more condensed chromatin pattern in blasts and the presence of Pelger-like cells.
Identification of these features allowed us to focus molecular analysis on 2 cases lacking t(11;17), which were subsequently found to harbor cryptic PLZF/RARA rearrangements. These 2 cases had a similar morphology that was very typical of the PLZF/RARAgroup. One case has been proven to express solelyPLZF-RARA,31 suggesting that this transcript could be responsible for the peculiar morphology of this APL subtype. Indeed, this case had all the characteristic features observed in cases with t(11;17) in which both PLZF-RARA and reciprocalRARA/PLZF transcripts were detectable, namely blast cells with regular nuclei, a maturation block at the promyelocyte/myelocyte stage, the absence of Auer rods, and the presence of Pelger-like cells (Figure 5, case 52A-C). This finding would indicate that there are some differences between t(11;17) APL arising in man and recent transgenic mouse models, which have suggested that RARA/PLZF influences morphological appearances and have implied that both fusion transcripts are required for the development of APL.55
The present study has shown that the presence of a regular nuclear outline is a key feature of t(11;17)-associated APL, and we suggest that the FAB classification could be extended to include a new subtype of M3, “M3r,” which is characterized by regular nuclei. As allPLZF-RARA cases had more than 37% blasts with regular nuclei, and the majority of PML/RARA cases had less than 30% such cells (6 of 69 cases had more than 30%), we propose a 30% threshold for defining M3r-APL, keeping all the cytoplasmic FAB criteria necessary for APL diagnosis. However, it is also clear that the 5 of 5 cases lacking RARArearrangements, 3 of 7 cases with ider(17), and the sole der(5)t(5;17) case could be classified as M3r. Moreover, in common with cases withPLZF/RARA rearrangements, the latter 2 groups demonstrated an increased number of Pelger-like cells; this feature could be related to the loss of chromosome 17p material and/or the p53 gene, as reported in some myelodysplastic syndromes.56 57
In a series of 25 APL cases, Neame et al15 identified 3PML/RARA+ M2-like cases, where the majority of blasts were normal-appearing differentiated promyelocytes without evidence of folding and lobulation of the nucleus. In our classification system, these would be classified as M3r; a further category of cells (blasts with hyperbasophilic cytoplasm) could be added in order to classify the hyperbasophilic M3 described by McKenna et al.14 Our series did not include the NUMA/RARAor STAT5b/RARA rearrangements. The previously describedNUMA/RARA case37,38 had hypergranular blasts, with irregular nuclei and Pelger-like cells, and is therefore reminiscent of M3 morphology. The case expressingSTAT5b/RARA39 was classified as M1-AML, with only a minority of blast cells evocative of M3v.58
In situations in which the morphology is similar to that associated with the PLZF/RARA rearrangement, the present study suggests that immunophenotype analysis could be helpful. Indeed, the NK-associated CD56 antigen was positive in 4 of 6 PLZF/RARAcases tested. Moreover, in a case lacking t(11;17), the CD56 positivity, as well as the evocative morphology, prompted further molecular analysis, which lead to identification of a crypticPLZF/RARA rearrangement. Positivity for CD56 and myeloid antigens, with negativity for HLA-DR and CD34, has also been associated with the NK/myeloid leukemia identified by Scott et al.26This study, which included 1 case with a PLZF/RARArearrangement, highlighted the presence of a particular morphology in two-thirds of cases, whereby blasts were characterized by a deeply invaginated nuclear membrane and fine azurophilic granulations in the cytoplasm. This appearance is clearly distinct from that observed inPLZF/RARA-associated APL; indeed, such CD56+cases identified in the present study were characterized by a regular nuclear membrane and hypergranular cytoplasm. CD56 positivity was also recently described in a new t(11;17)(q23;q21) case, which was classified as M3v but had a poorly lobulated nucleus.59The detection of CD56 in t(11;17)-associated APL led to the suggestion that this form of AML could originate from a more immature progenitor than t(15;17) APL, which is common to myeloid and NK cell lineages.33 However, as CD56 corresponds to the N-CAM molecule whose gene is located at band 11q23,60 one cannot exclude an aberrant expression related to a gene-position effect at the chromosomal breakpoint.
In 12 cases that lacked a PLZF/RARA rearrangement, the morphologic appearances were reminiscent of the PLZF/RARAgroup; these included 6 PML/RARA+ cases (2/28 insertions; 1/14 complex; 3/7) ider[17]; the case with der(5)t(5;17); and 5/5 cases lacking RARA rearrangements. The CD56 antigen was tested in 5 cases: 1 PML/RARA insertion, 1 ider(17), 1 der(5)t(5;17), and 2 cases without RARArearrangement. In each case the antigen was negative, which highlights the potential value of this antigen for distinguishing subtypes of APL, although this should be confirmed in a larger number of patients. Moreover, with the exception of the PLZF/RARA group, expression of CD56 in APL was found to be rare; it was detected in only 1 of 28 PML/RARA cases analyzed (P = .001). This is in accordance with data from the literature; indeed, only 14 cases of PML/RARAAPL expressing CD56 have been reported to date including 1 M2-like case.15 27
We report a new t(5;17) case expressing NPM/RARA, which, in common with the 2 previous reported cases,34,36 occurred in a pediatric patient. Whereas the cases described by Corey et al34 and us could be classified as M3 and presented with hyperleukocytosis, the remaining case36 had an M3v morphology. Auer rods were not observed in any of these cases, a feature very unusual in APL. The immunophenotype of our t(5;17) case was typical of APL (CD13− but CD33+) and, in contrast to the case published by Corey et al, was CD56−.
Finally, in a few APL cases, rearrangements of RARA were not found31; most of them presented with hyperleukocytosis and a morphological profile close to the PLZF/RARA group, but CD56 was negative in the 2 cases tested. The identification of such cases suggests that alternative pathways could mediate the differentiation block that characterizes APL, although it remains a possibility that RARA could still be involved by mutation or epigenetic mechanisms. Such events are currently under investigation.
In conclusion, no major morphological or immunophenotypic difference exists between APL harboring the classical t(15;17) andPML/RARA cases lacking t(15;17). Interestingly, our study highlights a specific morphological pattern for the PLZF/RARA group, which was associated with the CD56 expression. These features should alert one to the possibility of an underlying PLZF/RARA rearrangement that can be cryptic. Distinguishing APL cases with the PLZF/RARA fusion from other forms of the disease is critical because they are resistant to arsenic trioxide and ATRA as a single-agent therapy. The present study underlines the importance of combining morphologic, immunophenotypic, cytogenetic, and molecular studies to distinguish patients who could benefit from tailored therapeutic strategies.
The Monza Workshop comprised 41 cytogenetic laboratories representing 6 European countries and 1 American laboratory from St Jude Children's Hospital, Memphis, TN. The participants are listed below; participating cities or the names of participating centers and the number of cases supplied are given in parentheses. The workshop was organized by Dr Andrea Biondi. The European community–sponsored concerted action, Molecular and Cytogenetic Diagnosis in Hematological Malignancies, was headed by Prof Anne Hagemeijer. *Indicates a member of the Monza working party; †indicates a member of the working committee.
A. Hagemeijer,*† H. Van Den Berghe, F. Speleman, L. Michaux, H. Vranckx, Ph. Martiat, A. Van Orshoven, J. Rhodin, J. M. Scheiff, A. Criel, G. Verhoef, D. Selleslag, J. L. Michaux, A. Delannoy, P. Meeus, Z. Berneman (Belgium: Leuven, 6; Brugge, 3; Brussels, 3; Jolimont, 1; Antwerp, 1); N. Dastugue, E. Duchayne, F. Rigal-Huguet, A. Huynh (Toulouse, France: 7); D. Head,*†W. Miller Jr, P. B. Brett, J. Bickers, and E. Magenis (Memphis, TN; New York, NY; Sacramento, CA; New Orleans, LA; Portland, OR: 6); M. Lafage-Pochitaloff,*† M. J. Mozziconacci,*† F. Fernandez, A. Restoin, J. Gabert, F. Birg,*† D. Sainty,*† C. Arnoulet, V. J. Bardou, A. M. Stoppa, D. Coso, D. Blaise (Institut Paoli-Calmettes, Marseilles, France: 6); F. Mugneret, B. Favre,* M. A. Collonge, P. M. Carli, F. Bailly, M. Manadie, E. Racadot, D. Caillot, A. Rozenbaum, B. Salles (Dijon, France: 5); D. Belotti, S. Tosi, A. Biondi,*† G. Cazzaniga,* A. Cantù-Rajnoldi,* G. Rossi, S. Cortelazzo, G. Gaipa, P. Airo, G. Borleri, A. Rambaldi, G. Dotti (Monza, Italy: 5); J. H. Jansen,*† B. van der Reijden,* A. Hagemeijer,*† H. J. C. M. Holdrinet (The Netherlands: Erasmus University, Rotterdam, 4; Breda, 1); I. Radford-Weiss,*† E. Delabesse, G. Flandrin,*†L. Benattar, F. Valensi, L. Hoang, B. Varet, E. Macintyre (Paris Necker, France: 3); M. Mancini,* D. Diverio, F. Lo Coco, M. Nanni, S. Fenu, C. Nervi, G. Alimena (University La Sapienza, Rome, Italy: 3); C. Charrin, D. Treille-Ritouet, I. Tigaud, X. Thomas (Lyon, France: 3); F. Pasquali,* L. Seghezzi, E. Maserati, R. Invernizzi, G. Basso, R. Maccario, A. Gabbas, F. Locatelli, G. Bergamashi (Italy: Pavia, 2; Nuoro, 1); J. M. Hernandez, M. Gonzalez, A. Orfao, N. C. Gutiérrez* (Hospital Universitario, Salamanca, Spain: 2); R. Berger,*† M. Busson-Le Coniat, M. T. Daniel (Paris, Saint-Louis, France: 2); J. van den Akker, C. Perot, D. Bories, J. M. Pignon, M. F. Portnoı̈, C. Cordonnier (Paris, St Antoine and Henri Mondor, France: 2); V. Eclache* (Bobigny, France: 2); S. Raynaud, I. Sudaka (Nice, France: 2); A. Aventin, M. Espadaler,* M. Carrasco (Sant Pau Hospital, Barcelona, Spain: 2); M. Neat,*† S. M. Kelsey, A. C. Newland (St Bartholomew's and Royal London Hospitals, London, England: 2); J. Waters, M. Pomfret, D Milligan (Heartlands Hospital, Birmingham, England: 1); H. Walker, F. Oliver, S. Langabeer, A. H. Goldstone, (University College Hospital, London, England: 1); P. Chipping (North Staffordshire Hospital, North Staffordshire, England: 1); P. Revell (Stafford District General Hospital, Stafford, England: 1); J. Ropner (Gloucester Royal Infirmary, Gloucester, England: 1); D. Stevenson, D. Culligan (Aberdeen Royal Infirmary, Aberdeen, Scotland: 1); K. Rodgers, J. Emmerson, G. Abrahamson, D. Samson (Charing Cross Hospital, London, England: 1); P. Gorman, D. Sheer (ICRF, London, England); D. Grimwade,*† K. Howe,*† E. Solomon (Guy's, King's & St Thomas' School of Medicine, London, England); C. Harrison, L. Secker-Walker (Royal Free Hospital, London, England); E. Vandenberghe, G. Wilson, A. Watmore (Royal Hallamshire & Sheffield Children's Hospitals, Sheffield, England); S. Sobolewski, A.V. Kiorkian (Pilgrim Hospital, Boston, UK: 1); C. Mecucci, C. Liberatore, C. Matteucci,* R. La Starza, Fr Grignani, P. G. Pelicci* (Perugia, Italy: 1); C. Bilhou-Nabera, Ph. Bernard, M. Micheau, A. Notz-Carrère, F. X. Mahon, J. Reiffers (Bordeaux, France: 1); F. Viguié, S. Ramond, D. Bouscary (Hôtel-Dieu and Cochin, Paris, France: 1); M. J. Grégoire, Ph. Jonveaux, A. Guerci, J. Buisine (Nancy, France: 1); F. Brizard, A. Brizard, A. Sadoun (Poitiers, France: 1); F. Uetwiller, F. Maloisel, M. Mark (Strasbourg, France: 1); P. Talmant, H. Avet-Loiseau, R. Garand (Nantes, France: 1); S. Taviaux, M. Dupont, J. Taib, F. Bernard, N. Sarran, G. Margueritte (Montpellier, France: 1); D. Leroux, M. Callanan, M. F. Sotto (Grenoble, France: 1); R. Casalone, M. G. Biotti, G. Pinotti (Varese, Italy: 1); C. Chomienne (Paris Saint-Louis, France) for RT-PCR experiments on 4 cases; M. Lessard, A. Hery, (Brest, France) and I. Chudoba (Altlussheim, Germany) for M-FISH and m-band analyses on 3 cases; and V. Liso*†(Bari, Italy).
A complete list of the contributors appears in the .
Supported by grant CT94-1703 from the European Community Biomed Concerted Action; Molecular Cytogenetic Diagnosis in Haematological Malignancies by the Comité des Bouches-du-Rhône de la Ligue Nationale Française contre le Cancer; the Leukaemia Research Fund of Great Britain, UK (D.G.); the Fondazione M. Tettamanti, Associazione Italiana Ricerca sul Cancro (AIRC), Italy; and MURST (A.B.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Danielle Sainty, Department of Biology, Institut Paoli-Calmettes, 232 bd Sainte Marguerite, 13009 Marseille, France; e-mail:saintyd@marseille.fnclcc.fr.