Abstract
Early B-cell factor (EBF) is a helix–loop–helix transcription factor suggested to be essential for B-cell development in the mouse. Several genetic targets for EBF have been identified in mice, among these the surrogate light chain λ5 and the signal-transducing molecules Igα (mb-1) and Igβ (B29). This article reports cloning of the human homologue of EBF, hEBF. This protein has 93% sequence and 98.8% amino acid homology with mouse EBF. The encoded protein binds DNA and is expressed in cells of the B lineage, but not in cell populations representing T lymphocytes or myeloid cells. It is also shown that EBF-binding sites are functionally conserved in the humanmb-1 and B29 promoters because hEBF interacts with these in the electrophoretic mobility shift assay (EMSA) and have the ability to increase the activity of reporter constructs under the control of these promoters in nonlymphoid HeLa cells. A third genetic target for hEBF is the promoter of the human surrogate light chain14.1. This promoter contains 5 independent binding sites capable of interacting with hEBF in the EMSA, and the activity of the promoter was induced 24-fold in co-transfection experiments. These findings suggest that the human homologue of mouse EBF displays conserved biochemical features as well as genetic targets, indicating that this protein also has an important role in human B-cell development.
Introduction
The development of a multipotent hematopoietic stem cell into a highly differentiated immunoglobulin (Ig)-producing plasma cell can be divided into several stages based on expression of certain surface markers and the recombination status of the Ig genes.1,2 The earliest human B-cell precursors express the surface molecule CD34 and carry their Ig genes in germline configuration. Subsequent differentiation results in expression of components in the pre–B-cell receptor3,4 and signal-modulating molecules such as CD19 as well as the recombination-activating genes RAG-1 and -2, allowing for recombination of the Ig heavy chain genes (IgH). The pre–B-cell receptor is composed of the signal transduction molecules Igα (mb-1) and Igβ (B29) as well as the newly recombined IgH protein in combination with the surrogate light chain.3,4 The surrogate light chain is suggested to substitute for the Ig light chain (IgL) to allow for surface expression of the IgH protein at a developmental stage when the conventional IgL has not yet been rearranged.1,2,5 In the mouse, the surrogate light chain is composed of the λ5 and VpreB proteins.6 The λ5 protein displays homology to the constant part of an IgL7 gene and has been shown to be essential for B-cell development in the mouse.8 This molecule is encoded by a single gene in the mouse,7whereas several genes—14.1, 16.1, and16.2—all have been suggested to encode the human counterpart of λ5.9-11 Interestingly, a patient with a common variable immunodeficiency displaying a B-cell developmental block similar to what is observed in the λ5-deficient mouse was shown to carry 2 defective alleles of 14.1, whereas 16.1 and 16.2 were unaffected.12 This suggests that the 14.1 gene may be the functional homologue of mouse λ5. VpreBresembles an IgL variable domain and is encoded by 2 highly homologous genes in the mouse,13,14 whereas only one gene has been found in humans.15 Surface expression of the pre–B-cell receptor has been suggested to signal that a functional IgH gene rearrangement has occurred. This results in down-regulation of surrogate light chain and Rag gene expression and allows for proliferation and further differentiation of the B-cell precursor.1,2,16 The next developmental stage involves reactivation of Rag gene expression, allowing IgL recombination and subsequent expression of the B-cell receptor on the cell surface.17 Immature B cells then leave the bone marrow to enter the circulation and peripheral lymphoid organs, where they may become IgM- and IgD-expressing mature B cells.1
Completion of this developmental pathway is dependent on stage-specific expression of certain genes, demanding a system of transcriptional activators that interact with regulatory elements controlling the expression of these genes.18,19 The importance of distinct transcription factors in the promotion of B-cell differentiation in the mouse has been shown in a number of transgenic animals.18,20,21 Early B-cell factor (EBF),22B-cell–specific activator protein (BSAP),23 and E4724,25 have all been shown to be essential for early B-cell development by targeted disruptions of the coding genes.26-31 B-cell development in humans largely resembles that in mice,1 and homologues of most of the proteins shown to be important for B-cell development in mice have also been cloned from humans. In line with this, human homologues of BSAP23 and E4725 have been characterized and suggested to play roles similar to their mouse counterparts.25,32 However, no human homologue to mouse22 and rat33EBF/Olf-1 has yet been described. The existence of such a protein located on human chromosome 5 has been suggested from fluorescence in situ hybridization (FISH) analysis using the mouse cDNA as probe34 and from the finding that an EBF-like factor appears to interact with the human CD19 promoter.35 EBF interacts with the DNA core CCCNNGGG as a homodimer via a zinc-containing DNA-binding domain.36,37 The dimerization is mediated by 2 helices with homology to helix 2 of the bh-l-h protein.22,37 EBF has been suggested to interact with and regulate the promoters of several genes in the mouse pre–B cell, ie, themb-1,38,39B29,40λ5,41,42VpreB,42,43and Pax544 45 promoters, confirming the central role of this factor in B-cell development.
We here report cloning of the human homologue of mouse EBF (hEBF) and the identification of potential target genes for this factor in human B-cell development. We show binding to and activation of the humanmb-1, B29, and 14.1 promoters by hEBF and thus suggest that hEBF plays an important role as a transcriptional activator of a number of genes defining the early stages of the B lineage in humans.
Materials and methods
Tissue culture conditions
All lymphoid cells were grown at 37°C and 5% CO2in RPMI supplemented with 7.5% fetal calf serum, 10 mmol/L HEPES, 2 mmol/L pyruvate, 50 μmol/L 2-mercaptoethanol, and 50 μg/mL gentamicin (all purchased from Life Technologies AB, Täby, Sweden). HeLa cells were grown under the same conditions but in the absence of 2-mercaptoethanol. The human pre–B-cell line Nalm6, as well as the B-cell lines Raji and Namalwa, were kindly provided by T. Leanderson (Immunology Group, CMB, Lund University). The Jurkat human T-cell line and the human epitheloid cell line HeLa were kindly provided by Pharmacia UpJohn. KM3, U937, and THP-1 cells were a gift from U. Gullberg (Hematology Research Loboratory, Lund University).
Library construction and screening
The Nalm6 cDNA library was constructed using the ZAP Express cDNA synthesis kit and the ZAP Express cDNA Gigapack III Cloning Kit (Stratagene). Briefly, 5 μg of poly A–positive Nalm6 human pre–B-cell RNA was used for cDNA synthesis with anXhoI-containing polyT primer, digested with XhoI, and ligated to an EcoRI linker. The resulting cDNAs were size fractionated and ligated into ZAP Express dephosphorylated phage arms, allowing for the propagation of the library in JM109Escherichia coli cells. A total of 500 000 plaques from the obtained library were immobilized on nitrocellulose filters and screened by 16 hours of hybridization to a random-primed full-length mouse cDNA after 2 hours of preincubation in 5× Denhardts, 6× SSC, 0.1% sodium dodecyl sulfate (SDS), and 50 μg/mL salmon sperm DNA. The membranes were washed 2 times at room temperature in 2× SSC supplemented with 0.1% SDS for 15 minutes. The filters were then subjected to autoradiography for 36 hours. Two of the obtained clones were then analyzed by dideoxy nucleotide sequencing.
Isolation and purification of bone marrow progenitors and mature peripheral blood cells
Bone marrow (BM) cells were obtained from the posterior iliac crest of healthy volunteers after informed consent, as were peripheral blood (PB) samples. The BM and PB mononuclear cells (MNCs) were isolated by Ficoll-Hypaque (Nycomed, Oslo, Norway) gradient centrifugation. Positive selection of BM CD34+ cells was performed using a magnetically activated cell sorting CD34 isolation kit (Miltenyi Biotec, Bergish Gladbach, Germany), as described previously.46 The purity of CD34+enriched cells was reproducibly greater than 80%. CD34-enriched or CD34-depleted BM cells were incubated with fluorescein isothiocyanate (FITC)-, phycoerythrin (PE)-, or allophycocyanin-conjugated monoclonal antibodies (MoAbs) against CD34, CD19, CD38, CD33, or isotype-matched irrelevant control antibodies (all from Becton Dickinson, San Jose, CA) for 15 to 20 minutes at 6 to 12°C. CD34+CD19+ (proB), CD34+CD33+ (myeloid progenitor), CD34−CD19+CD38+ (preB), and CD34−CD19+CD38− (immature B) BM cells were sorted on a FACSVantage Cell Sorter (Becton Dickinson) equipped with a 488-nm argon-ion laser (Coherent Enterprise II, Santa Clara, CA) and a 633-nm He-Ne laser (Model 127; Spectra-Physics, Mountain View, CA). PB CD15+ (mature myeloid), CD3+ (T), and CD19+IgM+ (mature B) cell populations were sorted after incubation of MNCs with MoAbs against CD3 (FITC; Becton Dickinson), CD15 (FITC; Pharmingen, San Diego, CA), CD19 (PE; Becton Dickinson), and IgM (FITC; Pharmingen) or control antibody for 15 minutes on ice. The purity of all sorted cell populations was reproducibly greater than 90% to 95%. To obtain activated PB B cells, we incubated the sorted CD19+IgM+ cells in 50 ng/mL of phorbol myristate acetate (PMA; Sigma, St. Louis, MO) and 500 ng/mL Ionomycin (Sigma) at 37°C for 4 hours.
Transient transfections and luciferase assays
A total of 500 000 cells were washed once with serum-free medium (OPTIMEM; Life Technologies), and 800 μL of the medium was added for transfection. Five microliters of Lipofectin (Life Technologies) was diluted in 100 μL of serum-free medium, incubated for 45 minutes at room temperature, and mixed with the DNA diluted in 100 μL of medium. The mixture was incubated for 25 minutes, and the combined volume of 200 μL was added to the cells. The cells were then incubated in a CO2 incubator at 37°C for 12 hours, after which the transfection medium was removed and replaced by RPMI supplemented with 10% fetal calf serum. The cells were harvested after 40 hours, and protein extracts were prepared directly in the 24-well plates by adding 80 μL of cell lysis buffer (Promega, SDS AB, Falkenberg, Sweden). This procedure results in protein extracts of even quality as judged by Western blots and repeated transfections of cytomegalovirus (CMV)-controlled reporter constructs, reducing the need to normalize for the protein content in the extracts from each transfection by co-transfection of a β-gal reporter (M.S., unpublished data). The luciferase assay was conducted with 20 μL of the obtained extracts and 200 μL of luciferase assay reagent (Promega).
Protein extracts and electrophoretic mobility shift assay (EMSA)
Nuclear extracts were prepared according to Schreiber et al.47 DNA probes were labeled with γ[32P]-adenosine triphosphate by incubation with T4 polynucleotide kinase (Boehringer), annealed with the complementary strand, and purified on a 5% polyacrylamide TBE gel. A total of 5 to 10 μg of nuclear extract or 0.5 to 2 μL of in vitro–transcribed/translated protein was incubated with labeled probe (20 000 cpm, 3 fmol) for 30 minutes at room temperature in binding buffer (10 mmol/L HEPES, pH 7.9; 70 mmol/L KCl; 1 mmol/L dithiothreitol [DTT]; 1 mmol/L EDTA; 2.5 mmol/L MgCl2; 0.05% NP40) with 0.75 μg Poly dI/dC (Pharmacia). DNA competitors were added 10 minutes before addition of the DNA probe. The samples were separated on a 6% acrylamide TBE gel, which was dried and subjected to autoradiography. Competitors based on synthetic oligonucleotides were added at molar excesses indicated in the respective figures. Full-length mb-1, B29, and 14.1 promoters were generated by polymerase chain reaction (PCR; see below) and were added at molar excesses indicated in the respective figures.
Oligonucleotides used for the EMSA were as follows:mb-1 EBF sense, 5′-AGCCACCTCTCAGGGGAATTGTGG; mb-1EBF antisense, 5′-CCACAATTCCCCTGAGAGGTGGCT; mutated mb-1 EBF sense, 5′-AGCCACCTCTCAGCCGTTTTGTGG; mutated mb-1 EBF antisense, 5′-CCACAAAACGGCTGAGAGGTGGCT; CD19 PyG sense, 5′-CGCCTTCCTCTCTGGGGGGACTGCCTG; CD19 PyG antisense, 5′-CAGGCAGTCCCCCCAGAGAGGAAGGCG; Oct binding-site sense, 5′-CATCTCAAGTGATTTGCATCGCATGAGACG; Oct binding-site antisense, 5′-CGTCTCATGCGATGCAAATCACTTGAGATC; 14.1-1sense, 5′-GCTCAAGCCCTGGGGGACTCCTGC; 14.1-1 antisense, 5′-GCAGGAGTCCCCCAGGGCTTGAGC; 14.1-2 sense, 5′-TCTCTGGACCCCAGTGAGATGCTC; 14.1-2 antisense, 5′-GAGCATCTCACTGGGGTCCAGAGA; 14.1-3 sense, 5′-CCAGGGCGCCCTCGGGGAAGTGGG; 14.1-3 antisense, 5′-CCCACTTCCCCGAGGGCGCCCTGG; 14.1-4 sense, 5′-GGACCAGCCCCGCGGGGACTCAAG; 14.1-4 antisense, 5′-CTTGAGTCCCCGCGGGGCTGGTCC; 14.1-5 sense, 5′-AGGAAAGCCCCAAGGGAGGGTCTT; 14.1-5 antisense, 5′-AAGACCCTCCCTTGGGGCTTTCCT; 14.1-6 sense, 5′-CCCAGGGCCCCAGGGGCAAGGCCA; 14.1-6 antisense, 5′-TGGCCTTGCCCCTGGGGCCCTGGG; 14.1-7 sense, 5′-GGACAGAACCCCAGGGGTAACGGT; and 14.1-7 antisense, 5′-ACCGTTACCCCTGGGGTTCTGTCC.
Plasmids and constructs
Full-length hEBF cDNA was obtained by 22 cycles of high-fidelity PCR (LaRoche) with an oligonucleotide containing the predicted translation initiation codon and a T7 promoter primer using an hEBF cDNA in the viral vector as template (hEBF 5′: 5′-AGACATATGTTTGGGATCCAGGAAAGCATCCAACGGAGTGG). The EBF expression plasmid was based on the eukaryotic expression vector pcDNA3 (Invitrogen, BV, NV Leek, The Netherlands), which places the inserted hEBF cDNA under the control of a CMV promoter. The humanmb-1 (−284 to translation start −2), B29 (−146 to +54), and 14.1 (−335 to +239) promoters were amplified by PCR with promoter-specific sense and antisense primers with genomic HeLa-cell DNA as template. The resulting PCR products were cloned in the SmaI site of the luciferase reporter plasmid pGL3 basic (Promega). All constructs were verified by sequencing.
Oligonucleotides used for promoter constructs were as follows:mb-1 PCR sense, 5′-GTGACGAGCCAGCCCTTGAACCA; mb-1PCR antisense, 5′-TCTCCCAGTGAGTCGGTTAGTTTG; B29 PCR sense, 5′-CCCAGCTGACAAAAGCCTGC; B29 PCR antisense, 5′-GGTCACTGCTCTGTCCCCGACC; 14.1 PCR sense, 5′-GAGCTCAAGCCCTGGGGGACTCCT; and 14.1 PCR antisense, 5′-CGGCCCTGACCCTCAGAGGTCCTT. The basal fos promoter construct has been described earlier.42
In vitro transcription and translation
Recombinant protein was generated by coupled in vitro transcription/translation with a reticulocyte lysate kit (Promega) in the presence or absence of [35S]methionine. Two microliters of a 15-μL reaction mix was loaded on SDS-polyacrylamide gel electrophoresis (PAGE), and 0.5 to 2 μL was used for EMSA.
Reverse transcriptase (RT) and PCR
RNA was prepared from cells with Trizol (Life Technologies), and cDNA was generated by annealing 1 μg of total RNA to 0.5 μg of random hexamers in 10 μL DEPC-treated water. RT reactions were performed with 200 U of SuperScript Reverse Transcriptase (Life Technologies) in the manufacturer's buffer supplemented with 0.5 mmol/L dNTP, 10 mmol/L DTT, and 20 U RNase inhibitor (Boehringer Mannheim, Bromma, Sweden) in a total volume of 20 μL at 37°C for 1 hour. One twentieth of the RT reaction was used in the PCR assays. PCR reactions were performed with 1 U of Taq-polymerase (Life Technologies) in the manufacturer's buffer supplemented with 0.2 mmol/L dNTP in a total volume of 25 μL. Reduced glyceraldehyde phosphate dehydrogenase (GAPDH) was amplified by 25 cycles (94°C, 30 seconds, 55°C, 30 seconds, and 72°C, 30 seconds), whereas 30 cycles were used to amplify EBF cDNA (94°C, 30 seconds, 61°C, 30 seconds, 72°C, 30 seconds). Primers were added to a final concentration of 1 mmol/L. The PCR products were blotted onto Hybond N+ nylon membranes (Amersham) using capillary blotting with 0.4 mol/L NaOH. Membranes were prehybridized in 5× Denharts, 6× SSC, 0.1% SDS, and 50 μg/mL salmon sperm DNA at 57°C for 90 minutes; they were hybridized with γ[32P]-labeled oligonucleotide for 12 hours at 57°C in the same solution. The membranes were washed at room temperature 2 times in 2× SSC supplemented with 0.1% SDS for 15 minutes.
Oligonucleotides used for RT-PCR were as follows: GAPDH sense, 5′-CCACCCATGGCAAATTCCATGGCA; GAPDH antisense, 5′-TCTAGACGGCAGGTCAGGTCCACC; GADPH hybridization, 5′-AAGATCATCAGCAATGCCTCCTGC; hEBF sense, 5′-AGACATATGTTTGGGATCCAGGAAAGCATCCAACGGAGTGG; hEBFantisense, 5′-TGAGCAAGACTCGGCACATTTCTG; and hEBFhybridization, 5′-GCCAACAGCGAAAAGACCAATAAC.
Results
Human pre–B cells and B cells express a homologue of mouse EBF
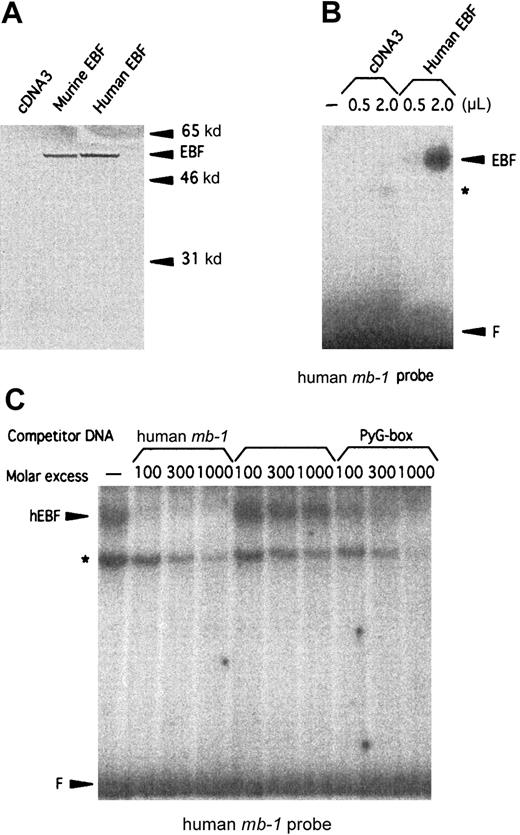
Studies of the human CD19 promoter35 as well as FISH analysis of human chromosomes with a mouse EBFprobe34 have suggested the existence of a human homologue of mouse EBF. To clone this factor, we made a λGT11 cDNA library from the human pre–B-cell line Nalm6. Low-stringency screening of 500 000 plaques with a 32P-labeled random-primed mouse EBF cDNA resulted in 12 clones that also hybridized to the cDNA in a Southern blot of phage DNA. Sequence analysis revealed a 93% overall as well as coding-region cDNA homology (GenBank accession numberAF208502) and a 98.8% amino acid homology (Figure1) to mouse EBF.22 The longest clone we obtained started at base pair 60 of the defined mouse cDNA, so to obtain the translation start site, we made a GenBank query using the hEBF sequence. This resulted in the identification of an EST fragment from a human germinal center library (GenBank accession numberAA504812) spanning the 5′ end of hEBF, including the translation initiation ATG. Using the partial hEBF cDNA as a template, we extended our cDNA by PCR with a primer containing the missing amino acids to obtain the full reading frame of hEBF. To investigate whether the cloned cDNA could be translated into a protein with biochemical properties similar to those of the mouse EBF, we made in vitro translations of hEBF. The [35S]-methionine–labeled proteins were then detected by SDS-PAGE (Figure 2A). The empty vector did not result in any translation product, whereas both the mouse EBF and the hEBF-encoding plasmids generated proteins with an apparent molecular weight of approximately 55 to 60 kd, which correlates fairly well with the predicted molecular weight of 64 kd. To investigate the ability of hEBF to interact with the suggested EBF-binding site in the human mb-1 promoter,48we performed the EMSA with either 0.5 or 2 μL of reticulocyte lysate programmed with empty expression vector or hEBF-encoding vector (Figure2B). This resulted in one prominent band with the EBF-programmed lysate that could not be detected with the lysate programmed with empty cDNA3 vector. To confirm the binding specificity between hEBF and themb-1 promoter, we performed the EMSA with in vitro–translated recombinant hEBF and with competition for complex formation by the addition of duplex oligonucleotides spanning themb-1 EBF site, a point-mutated mb-1 EBF site, or the EBF-binding PyG box from the human CD19promoter.35 49 Whereas the wild-type mb-1 EBF site as well as the CD19 PyG box competed for complex formation, the mutated EBF site did not. Hence, we conclude that the cloned cDNA encodes a protein with the ability to interact specifically with EBF-binding sites from the human mb-1 andCD19 promoters.
Human EBF (hEBF) is highly homologous to its mouse counterpart.
Amino acid alignment of human and mouse EBF (mEBF). Homologous regions are boxed.
Human EBF (hEBF) is highly homologous to its mouse counterpart.
Amino acid alignment of human and mouse EBF (mEBF). Homologous regions are boxed.
The cloned cDNA of the human homologue of EBF encodes a 57-kd protein that interacts specifically with a binding site in the human mb-1 promoter.
(A) Autoradiogram of 35S-labeled in vitro–translated mouse and human EBF. Positions of molecular-weight markers and EBF are indicated by arrows. (B) EMSA using the human mb-1 promoter EBF site in combination with either 0.5 or 2 μL of unprogrammed reticulocyte lysate (ret.) or 0.5 and 2 μL of recombinant in vitro–translated human EBF (hEBF). An unidentified DNA-binding activity present in the reticulocyte lysate is indicated by the star (*), and F indicates free probe. (C) EMSA in which competition was induced for the binding of in vitro–translated recombinant human EBF to the human mb-1 promoter EBF-binding site by the addition of duplex oligonucleotides spanning the wild-type (mb-1) and a point-mutated human mb-1 promoter EBF site (mut.mb-1), or the PyG box (PyG) from the human CD19 promoter, as indicated. The star indicates the background DNA-binding activity in the reticulocyte lysate, and F indicates free probe.
The cloned cDNA of the human homologue of EBF encodes a 57-kd protein that interacts specifically with a binding site in the human mb-1 promoter.
(A) Autoradiogram of 35S-labeled in vitro–translated mouse and human EBF. Positions of molecular-weight markers and EBF are indicated by arrows. (B) EMSA using the human mb-1 promoter EBF site in combination with either 0.5 or 2 μL of unprogrammed reticulocyte lysate (ret.) or 0.5 and 2 μL of recombinant in vitro–translated human EBF (hEBF). An unidentified DNA-binding activity present in the reticulocyte lysate is indicated by the star (*), and F indicates free probe. (C) EMSA in which competition was induced for the binding of in vitro–translated recombinant human EBF to the human mb-1 promoter EBF-binding site by the addition of duplex oligonucleotides spanning the wild-type (mb-1) and a point-mutated human mb-1 promoter EBF site (mut.mb-1), or the PyG box (PyG) from the human CD19 promoter, as indicated. The star indicates the background DNA-binding activity in the reticulocyte lysate, and F indicates free probe.
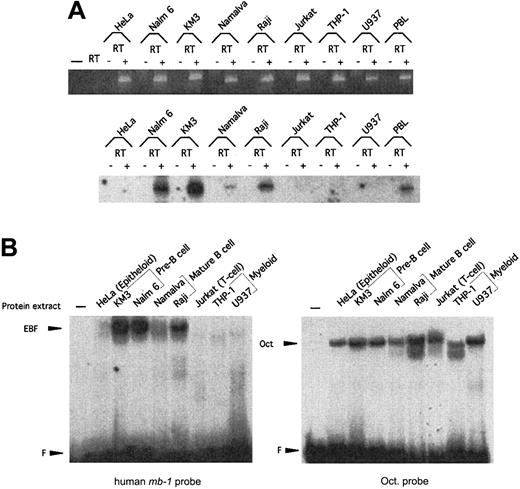
Expression of EBF in the hematopoietic system in mice is restricted to pre–B cells and B cells.22 39 To examine whether hEBF is expressed in a similar pattern, we performed RT-PCR analysis of total RNA from a set of cell lines representing different cell types (Figure3A). The presence of GADPH-encoding RNA was used as a control for the integrity of the cDNA in all of the samples. Although GADPH message could be detected in all tested cell lines, EBF message could be found only in KM3 and Nalm6 pre–B-cell lines, in Namalwa and Raji B-cell lines, and in human PB cells. No EBF message could be detected in epitheloid HeLa cells, Jurkat T cells, or U937 plasmacytoid/myeloid cells or in THP-1 myeloid cells. To investigate the presence of proteins with the ability to interact with the mb-1 promoter EBF site in the same cell lines, we performed the EMSA with nuclear extracts (Figure 3B). The integrity of the extracts was confirmed by an EMSA with a 32P-labeled Oct protein-binding site. Oct proteins could be detected in all the tested cell lines, whereas only the pre–B cell and B-cell lines contained high levels of a protein with the ability to interact with the EBF-binding site. This suggests that human pre–B cell and B-cell lines express a homologue of the mouse EBF.
Expression of hEBF is restricted to B-cell–derived cell lines.
(A) RT-PCR analysis to detect expression of hEBF in a panel of cell lines representing different lineages and B-cell developmental stages, as indicated. PBL indicates normal human peripheral blood cells, and − indicates a negative control with an RT reaction in which no RNA has been added. The upper part shows an ethidium bromide–stained agarose gel with the resulting GAPDH PCR products after 25 cycles of PCR with cDNA from the indicated cell lines. −RT indicates that RNA preparations have been included in the reaction without prior cDNA synthesis. The lower panel shows an autoradiogram of EBF RT-PCR products blotted to a nylon membrane after 30 cycles from the same cDNA preparations and hybridized with an internal oligonucleotide. (B) Autoradiograms of EMSA using approximately 5 μg of nuclear extracts from a panel of cell lines and either a32P-labeled consensus Oct protein-binding site or the EBF-binding site from the human mb-1 promoter as probes. F indicates free probe.
Expression of hEBF is restricted to B-cell–derived cell lines.
(A) RT-PCR analysis to detect expression of hEBF in a panel of cell lines representing different lineages and B-cell developmental stages, as indicated. PBL indicates normal human peripheral blood cells, and − indicates a negative control with an RT reaction in which no RNA has been added. The upper part shows an ethidium bromide–stained agarose gel with the resulting GAPDH PCR products after 25 cycles of PCR with cDNA from the indicated cell lines. −RT indicates that RNA preparations have been included in the reaction without prior cDNA synthesis. The lower panel shows an autoradiogram of EBF RT-PCR products blotted to a nylon membrane after 30 cycles from the same cDNA preparations and hybridized with an internal oligonucleotide. (B) Autoradiograms of EMSA using approximately 5 μg of nuclear extracts from a panel of cell lines and either a32P-labeled consensus Oct protein-binding site or the EBF-binding site from the human mb-1 promoter as probes. F indicates free probe.
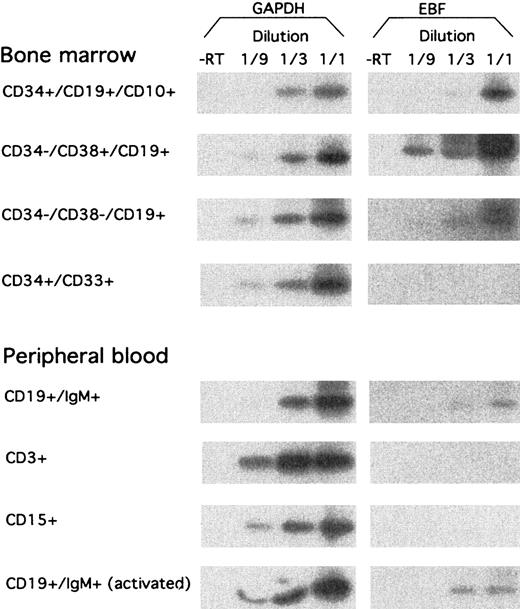
To investigate the expression pattern of EBF in primary hematopoietic cells, we sorted a set of cell populations from human BM and PB by flow cytometry. RNA was extracted from the cells, and we analyzed the expression of EBF-encoding message by RT-PCR analysis, as described earlier (Figure 4). CD34+CD19+CD10+ pro–B cells from BM expressed EBF, although at a lower level than CD34−CD19+CD38+ pre–B cells. EBF message could also be detected in CD34−CD19+CD38− BM cells, representing immature B cells, although at a lower level than in the pre–B cells. No EBF message was detected in CD34+CD33+ myeloid precursor cells from BM. EBF expression could be detected in CD19+IgM+ PB B cells, whereas neither CD3+ T cells nor CD15+myeloid cells expressed detectable levels of EBF mRNA. The presence of EBF message in CD19+IgM+ PB B cells was not significantly altered after 4 hours of stimulation with PMA and Ionomycin. This indicates to us that EBF expression is restricted to B-lineage cells and to some degree is temporarily regulated during B-cell development.
hEBF is expressed during normal B-cell development.
RT-PCR analysis of sorted primary human cells from either bone marrow or peripheral blood, as indicated. GAPDH message was detected after 27 cycles of PCR, blotting of the resulting products to a nylon membrane, and hybridization to an internal oligonucleotide; whereas 32 cycles of PCR were performed before blotting and hybridization to detect EBF-encoding mRNA. −RT indicates that RNA preparations have been included in the reaction without prior cDNA synthesis.
hEBF is expressed during normal B-cell development.
RT-PCR analysis of sorted primary human cells from either bone marrow or peripheral blood, as indicated. GAPDH message was detected after 27 cycles of PCR, blotting of the resulting products to a nylon membrane, and hybridization to an internal oligonucleotide; whereas 32 cycles of PCR were performed before blotting and hybridization to detect EBF-encoding mRNA. −RT indicates that RNA preparations have been included in the reaction without prior cDNA synthesis.
EBF-binding sites in the mb-1 and B29promoters are functionally conserved between man and mouse
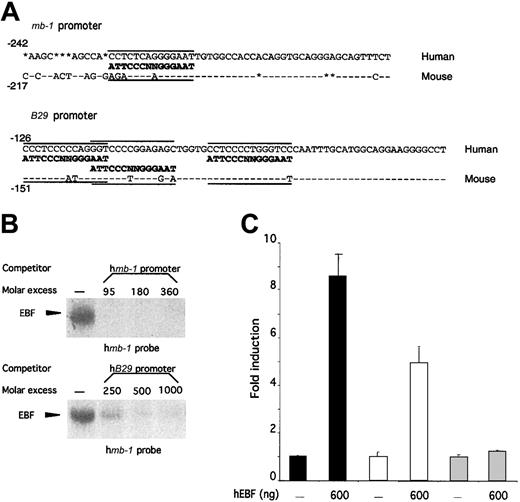
The high homology and overlapping expression patterns of human and mouse EBF suggest that the biochemical properties and the lineage specificity of EBF within the hematopoietic system are conserved between man and mouse. However, to evaluate whether EBF plays similar roles in B-cell development in these 2 species, we wanted to investigate whether the factor was able to interact functionally with the promoters of human homologues of defined genetic targets for EBF in mice. To this end, we compared the promoter structures of mouse and human mb-138,48 andB2950,51 promoters with regard to the presence of potential EBF-binding sites (Figure5A). Site-selection experiments have suggested that EBF binds to variants of the palindrome sequence ATTCCCNNGGGAAT.36 The mouse mb-1promoter contains an EBF-binding site with 12 matching base pairs out of 14.38,39 The human promoter contains a slightly different variant of this site because even though the same number of matching bases are found, 12 of 14 of the 5′ part as well as the central NN core differ (Figure 5A). The mouse B29 promoter contains 3 independent EBF-binding sites40 with 7, 8, and 10 base pairs homologous to the consensus site, whereas the corresponding EBF sites in the human promoter have 8, 9, and 9 base pairs matching the defined consensus site. This finding suggests that even though differences could be detected within the EBF-binding sites, these would probably not interfere with EBF binding. However, naturally occurring binding sites appear to be hard to predict because some sites with high homology to the consensus sequence do not interact well with EBF, whereas others with apparently lower homology bind EBF with high affinity40,42,52 (M.S., unpublished data). Thus, the ability of EBF to interact functionally with a target promoter cannot be determined solely on the basis of sequence homology, but must be addressed experimentally. To do so, we amplified the humanmb-1 and B29 promoters by PCR from genomic HeLa-cell DNA and used these fragments as competitors for hEBF–mb-1 EBF-site complex formation (Figure 5B). The humanmb-1 and B29 promoters competed efficiently (Figure 5B), whereas no efficient competition for complex formation could be seen after the addition of a CD19 promoter with a mutated EBF-binding site (data not shown). To test the function of these interactions, we cloned the promoters in front of a luciferase reporter gene and transfected these plasmids into HeLa cells together with either empty or hEBF-encoding expression plasmid (Figure 5C). This resulted in an 8-fold induction of mb-1 promoter activity and a 5-fold increase of B29 promoter activity in the presence of EBF expression plasmid, but no significant effect on the activity of a fos promoter reporter construct.42 This indicates that the human homologue of EBF shares target genes with its mouse counterpart, suggesting conserved functions for EBF in human and mouse B-cell development.
The ability of EBF to activate the mb-1 andB29 promoters is conserved between human and mouse.
(A) DNA sequences from the mouse and human mb-1 andB29 promoters. Sequence homology is indicated by − and insertions/deletions by *. The defined EBF-binding sites in the mouse promoters38-40 as well as the potential EBF-binding sites in the human promoters are aligned to a consensus EBF-binding site.36 (B) EMSA obtained by competition for binding by hEBF to an excess of mb-1 promoter EBF site by the addition of increasing amounts of PCR-amplified mb-1 orB29 promoters. The gels have been cut. (C) Diagrams indicating the relative luciferase activity obtained after transient transfections of 250 ng of mb-1 (black bars), B29(white bars), or fos (gray bars) promoter-controlled reporter constructs in the absence or presence of 600 ng of hEBF-encoding expression plasmid. The data were collected from 4 transfections in which the amount of DNA was normalized by the addition of empty expression plasmid (cDNA3). Error bars indicate standard deviation.
The ability of EBF to activate the mb-1 andB29 promoters is conserved between human and mouse.
(A) DNA sequences from the mouse and human mb-1 andB29 promoters. Sequence homology is indicated by − and insertions/deletions by *. The defined EBF-binding sites in the mouse promoters38-40 as well as the potential EBF-binding sites in the human promoters are aligned to a consensus EBF-binding site.36 (B) EMSA obtained by competition for binding by hEBF to an excess of mb-1 promoter EBF site by the addition of increasing amounts of PCR-amplified mb-1 orB29 promoters. The gels have been cut. (C) Diagrams indicating the relative luciferase activity obtained after transient transfections of 250 ng of mb-1 (black bars), B29(white bars), or fos (gray bars) promoter-controlled reporter constructs in the absence or presence of 600 ng of hEBF-encoding expression plasmid. The data were collected from 4 transfections in which the amount of DNA was normalized by the addition of empty expression plasmid (cDNA3). Error bars indicate standard deviation.
The surrogate light chain gene 14.1 is a genetic target for hEBF
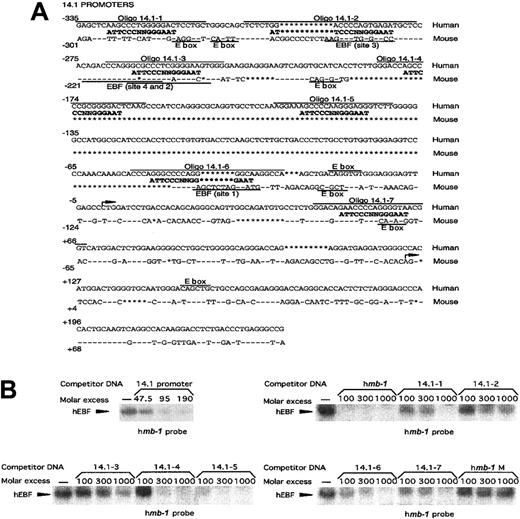
Another target gene for EBF activation in the mouse is the surrogate light chain λ5. The promoter of this gene has been suggested to be a direct target for synergistic activation by EBF and the E2A-encoded bh-l-h protein E47.42 To investigate a potential role for EBF in the regulation of the human functional homologue 14.1, we compared the sequences of the 5′ regions from the λ57 and14.153 genes (Figure6A). This showed that even though conserved regions could be observed, the overall homology was only about 40%. Furthermore, only 2 of the defined EBF-binding sites in theλ5 promoter42 appeared to be conserved, whereas several other potential binding sites were present only in the14.1 promoter. To investigate whether the 14.1promoter was capable of interacting with EBF, we performed an EMSA in which competition existed for complex formation between recombinant in vitro–translated EBF and the mb-1 promoter EBF site by the addition of PCR-amplified 14.1 promoter (−335 to +239) (Figure 6B). Inclusion of unlabeled 14.1promoter in the EMSA competed efficiently for complex formation, suggesting that EBF indeed has the ability to interact with this control element. To approximate the number of functional EBF-binding sites in the 14.1 promoter, we synthesized oligonucleotides spanning 7 potential sites and used these as competitors in EMSA experiments (Figure 6B). Oligonucleotide 14.1-1 contains a potential EBF-binding site with 10 of 14 matching base pairs and competed for binding of recombinant EBF, albeit with a low efficiency. Oligonucleotides 14.1-2 and 14.1-3, which could be considered conserved between the λ5 and 14.1promoter, both have 9 base pairs matching the EBF consensus site but appeared unable to interact efficiently with EBF. Both14.1-4 and 14.1-5 displayed a 10–base pair match to the consensus site and competed efficiently for EBF binding;14.1-6 and 14.1-7 contained 9– and 10–base pair homology with the consensus EBF site, respectively, and both competed with a low efficiency. The wild-type mb-1 promoter EBF site competed efficiently, whereas no efficient competition could be detected using the mutated mb-1 promoter EBF site (Figure2C). This suggests that the 14.1 promoter has the ability to interact with EBF through at least 5 independent sites with varying affinities.
EBF interacts with 5 independent binding sites in the promoter of the human surrogate light chain 14.1.
(A) Sequence alignment between the human 14.1 and mouseλ5 promoters. Conserved sequence is indicated by − and insertions/deletions by *. Potential EBF sites in the14.1 promoter as well as the experimentally confirmed EBF-binding sites in the λ5 promoter42 are aligned to a consensus EBF-binding site. Overlining indicates the oligonucleotides used in the EMSA analysis in panel B. E-boxes (CANNTG) in either the 14.1 or λ5 promoter are indicated by text and underlining. (B) EMSA obtained by competition for binding by hEBF to an excess of mb-1 promoter EBF site by the addition of increasing amounts of PCR-amplified 14.1promoter or duplex oligonucleotides spanning potential EBF-binding sites in the 14.1 promoter, as indicated. A duplex oligonucleotide spanning the EBF-binding site from the humanmb-1 promoter was used as positive control, and a BSAP-binding site from the human CD19 promoter as negative control. The gels have been cut.
EBF interacts with 5 independent binding sites in the promoter of the human surrogate light chain 14.1.
(A) Sequence alignment between the human 14.1 and mouseλ5 promoters. Conserved sequence is indicated by − and insertions/deletions by *. Potential EBF sites in the14.1 promoter as well as the experimentally confirmed EBF-binding sites in the λ5 promoter42 are aligned to a consensus EBF-binding site. Overlining indicates the oligonucleotides used in the EMSA analysis in panel B. E-boxes (CANNTG) in either the 14.1 or λ5 promoter are indicated by text and underlining. (B) EMSA obtained by competition for binding by hEBF to an excess of mb-1 promoter EBF site by the addition of increasing amounts of PCR-amplified 14.1promoter or duplex oligonucleotides spanning potential EBF-binding sites in the 14.1 promoter, as indicated. A duplex oligonucleotide spanning the EBF-binding site from the humanmb-1 promoter was used as positive control, and a BSAP-binding site from the human CD19 promoter as negative control. The gels have been cut.
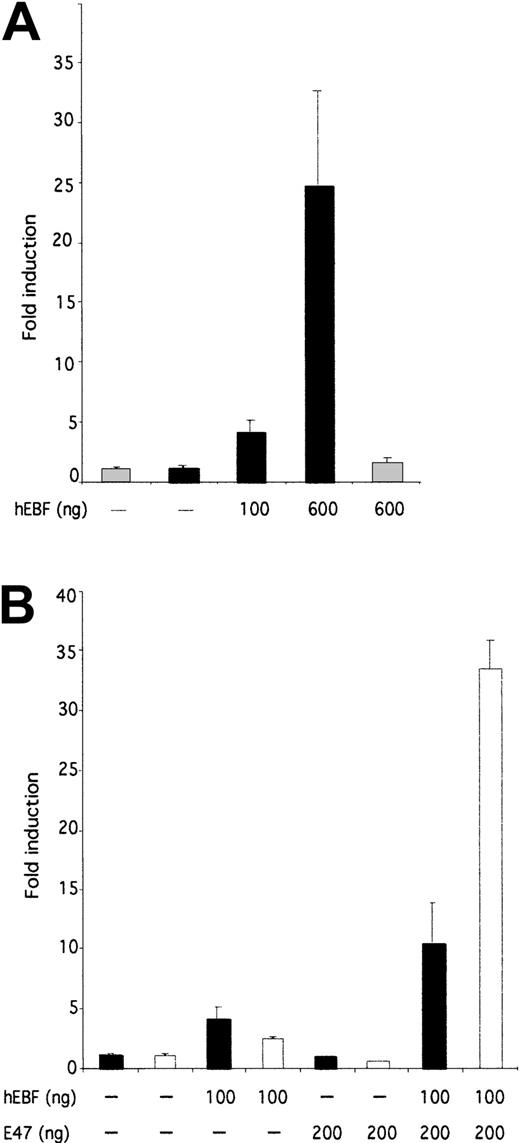
To investigate the ability of EBF to induce transcription from the 14.1 promoter, we cloned the PCR-amplified14.1 promoter in a luciferase reporter vector and made transient transfections of this construct and hEBF-encoding expression plasmid in HeLa cells (Figure 7A). Addition of 100 or 600 ng of hEBF-encoding expression vector resulted in a 4-fold or 24.5-fold induction of the 14.1 promoter activity, respectively, whereas no significant effect on the function of a fos promoter could be detected. This suggests that hEBF has the ability to interact functionally with the 14.1 promoter.
hEBF and E47 collaborate to induce functional activity of the 14.1 promoter.
(A) The relative luciferase activity after transient transfection of 250 ng of a 14.1 promoter-controlled reporter gene into epitheloid HeLa cells together with increasing amounts (100 or 600 ng) of hEBF expression plasmid (black bars). The gray bars indicate the activity of the fos promoter-controlled reporter construct. (B) Induction of either a 14.1 black bars or λ5white bars promoter-controlled luciferase reporter gene after transfection with either empty, hEBF-, E47-, or the combination of EBF- and E47-encoding expression plasmids, as indicated. The data in both panels were collected from 4 transfections in which the amount of DNA was normalized by the addition of empty expression plasmid (cDNA3). Error bars indicate standard deviation.
hEBF and E47 collaborate to induce functional activity of the 14.1 promoter.
(A) The relative luciferase activity after transient transfection of 250 ng of a 14.1 promoter-controlled reporter gene into epitheloid HeLa cells together with increasing amounts (100 or 600 ng) of hEBF expression plasmid (black bars). The gray bars indicate the activity of the fos promoter-controlled reporter construct. (B) Induction of either a 14.1 black bars or λ5white bars promoter-controlled luciferase reporter gene after transfection with either empty, hEBF-, E47-, or the combination of EBF- and E47-encoding expression plasmids, as indicated. The data in both panels were collected from 4 transfections in which the amount of DNA was normalized by the addition of empty expression plasmid (cDNA3). Error bars indicate standard deviation.
The function of the mouse λ5 promoter is dependent on template-specific interactions of EBF and E47 with their cognate sites.42 A striking feature of the comparison of theλ5 and 14.1 promoters (Figure 6A) was that even though no conserved E-boxes (E47-binding sites) could be found, the14.1 promoter contained 2 potential E47-binding sites. This led us to investigate whether E47 was acting also on the14.1 promoter. To this end, we transiently transfected epitheloid HeLa cells with reporter constructs under the control of either the λ5 or the 14.1 promoter together with hEBF and/or human E4725 expression constructs (Figure7B). Co-transfection of the λ5 promoter with 100 ng of hEBF expression vector induced the promoter 2.4-fold, whereas the inclusion of 200 ng of E47 expression plasmid did not induce the promoter significantly. The combination of 100 ng of EBF and 200 ng of E47 resulted in a 33.4-fold induction of λ5 promoter function, suggesting that the human homologues of EBF and E47 have the ability to act in synergy to activate the mouse λ5promoter. A similar experiment using the 14.1-controlled reporter construct resulted in a 4-fold induction of activity by the addition of EBF, whereas no significant induction could be detected by the inclusion of E47-encoding expression plasmid. The combination of E47 and EBF resulted in a 10.4-fold induction of 14.1promoter activity. This indicates that E47 acts in concert with EBF also in the activation of the 14.1 promoter, even though this cooperation is not as pronounced as on the λ5promoter.
Discussion
We here report the cloning of the human homologue of mouse EBF. Mouse and human EBFs have a high homology at the protein and RNA levels and also appear to share expression patterns in the hematopoietic system. We also show data supporting the idea that these proteins share a number of B-lineage–restricted target genes. Thus, we suggest that the role of EBF in human B-cell development may be highly analogous to that in mice, positioning EBF as a key transcriptional regulator in human B lymphopoiesis.
In the mouse, it has been suggested that EBF is an activator of several genes encoding components of the pre–B-cell and B-cell receptors.39,40,42 The data we present suggest not only that EBF is conserved as a protein, but also that the functional role of EBF in B-cell development may be conserved between man and mouse. This is based on the ability of hEBF to interact functionally with promoters of target genes defined in mouse B-cell development. In the case of the B2951 andmb-148 genes, this seems to be a consequence of conserved 5′ regions. As a result, the structural features of the promoter elements as well as the EBF-binding sites appear to be conserved between man and mouse. It is notable that mutations have occurred also within the EBF-binding sites, but that most of these are positioned so that they should not impair binding of EBF (Figure 5A). This may reflect a selection for functional ability to interact with EBF rather than a precise base-pair composition of the promoters. Functional conservation of EBF binding is even more striking when comparing the promoter of 14.1 with that of the mouse homologue λ5 (Figure 6A). The homology between these promoters is rather low, with sequence conservation only in certain regions (Figure 6A).53 As a result, the structural features of the whole element and in particular the EBF-binding sites are rather poorly conserved. Instead, the 14.1 promoter contains 5 EBF-binding sites not found in the mouse λ5promoter. A similar observation can be made when comparing the presence of E-boxes in the promoters. Even though only 1 of 4 E-boxes appears to be conserved, an additional element is present uniquely in the14.1 promoter. This appears to be sufficient to allow cooperation between hEBF and E47 in the regulation of the14.1 promoter, similar to what has been reported for theλ5 promoter.42 This could indicate that an essential feature of a surrogate light chain promoter is interaction with EBF and E47 to be able to mediate pre–B-cell–specific expression.
When studying the role of transcription factors in an evolutionary perspective, it may be interesting to compare functional and genetic conservation. One striking example of functional conservation is the role of NFκB homologues in stimulating inflammatory responses in both mouse and Drosophila.54-57 This occurs even though the inflammatory systems in these species are highly divergent. An example of genetic conservation could be the bh-l-h proteins encoded by theE2A gene, E12 and E47.24 Their Drosophila homologue daughterless is involved in neurogenesis and sex determination in flies,58 whereas targeted mutation of the E2A gene in mice results in a B-cell developmental block and less obvious neurologic or embryonal symptoms.29,31,59 EBF homologues have now been cloned from Drosophila melanogaster,60Caenorhabditis elegans,61Xenopus laevis,62and Zebrafish.63 However, the functional role of these homologues appears to reside mainly in the formation of the nervous system and, in Drosophila, in the formation of head structures. Even though EBF-deficient mice display a neurologic phenotype,64 the embryonal development appears rather normal with the exception of the generation of B lymphocytes, which do not develop either in the fetus or in the adult mouse.26This may depend on the expression of EBF homologues65 that might well compensate for EBF in neuronal development while they are unable to support the development of B lymphocytes. Thus, it appears as if the genetic conservation of EBF extends beyond the functional conservation in the immune system. This may be due to gene duplications allowing for the development of homologues with an ability to compensate for functional loss of one of the family members in certain tissues.
The presence of a helix–loop–helix dimerization domain in EBF has led to the suggestion that EBF belongs to the helix–loop–helix family of transcription factors. This family includes transcription factors such as myc, Tal, and E47, all known to play important roles in the development of leukemia.66-68 EBF has not yet been implicated in the development of human cancer. However, abnormalities in the p34 region of human chromosome 5, where hEBF is located,34 have been associated with a number of myeloid leukemias.69 A role for EBF in the development of myeloid leukemia could occur because EBF appears to be a key factor in the determination of the B-cell as opposed to the myeloid lineage. Ectopic expression of EBF in macrophage-converted 70Z/3 pre–B cells results in a partial restoration of the B-cell phenotype.45 Such a function of EBF may be of interest in human malignancies in which the conversion of B lymphomas into mixed or myeloid-like cells is associated with lower remission rates and poor prognosis.70-72
The data we present suggest that hEBF is involved in the regulation of several genes defining the B lineage. However, our understanding of the precise role of this factor in normal and malignant B-cell development remains elusive and demands further studies of genetic targets and of direct involvement in the initiation or progression of human cancer.
Acknowledgments
We thank Y. Zhuang and R. Grosschedl for the kind gifts of plasmids, K. Riesbeck for the gift of human peripheral blood cell RNA, Ingbritt Åstrand-Grundström for help with cell sorting, and D. Liberg and R. Carlsson for helpful comments and critical reading of the manuscript.
Supported by the Swedish Medical Research Council, The Swedish Cancer Foundation, Magnus Bergwall-Åke Wibergs-Österlunds Foundation, The Crafoord Foundation, and The American Cancer Society.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Mikael Sigvardsson, Immunology Group, CMB, Lund University, Sölvegatan 21, S-223 62 Lund, Sweden; e-mail:mikael.sigvardsson@immuno.lu.se.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal