Abstract
Deficiency of functional deoxycytidine kinase (dCK) is a common characteristic for in vitro resistance to cytarabine (AraC). To investigate whether dCK is also a target for induction of AraC resistance in patients with acute myeloid leukemia (AML), we determined dCK messenger RNA (mRNA) expression in (purified) leukemic blasts and phytohemagglutinin-stimulated T cells (PHA T cells) from patients with chemotherapy-sensitive and chemotherapy-resistant AML. In control samples from healthy donors (PHA T cells and bone marrow), only wild-type dCK complementary DNA (cDNA) was amplified. Also, in (purified) leukemic blasts from patients with sensitive AML, only wild-type dCK cDNAs were observed. These cDNAs coded for active dCK proteins in vitro. However, in 7 of 12 (purified) leukemic blast samples from patients with resistant AML, additional polymerase chain reaction fragments with a deletion of exon 5, exons 3 to 4, exons 3 to 6, or exons 2 to 6 were detected in coexpression with wild-type dCK. Deletion of exons 3 to 6 was also identified in 6 of 12 PHA T cells generated from the patients with resistant AML. The deleted dCK mRNAs were formed by alternative splicing and did code for inactive dCK proteins in vitro. These findings suggest that the presence of inactive, alternatively spliced dCK mRNA transcripts in resistant AML blasts may contribute to the process of AraC resistance in patients with AML.
Introduction
Remission induction treatment of patients with acute myeloid leukemia (AML) usually consists of a combination of 1-β-arabinofuranosylcytosine (AraC) and an anthracycline, occasionally supplemented by a third drug.1 The clinical outcome is still unsatisfactory, because 15% of the patients do not respond to initial induction chemotherapy (refractory AML) and 30% to 80% of the patients who achieve a complete remission (CR) relapse eventually.2,3 Treatment of the patients with relapsed or refractory AML using standard- or high-dose chemotherapy including AraC is unable to produce a prolonged leukemia-free survival in most patients.4 5 Resistance to the cytostatic drugs AraC and anthracyclines is thought to be the reason for the lack of long-term leukemia-free survival of patients with AML.
AraC is a deoxycytidine (dC) analogue that differs from dC by a substitution of the hydrogen atom at the 2′ position in the pentose ring by a hydroxyl group. When administered at high doses, AraC is taken up by the cell by passive diffusion. In the cytoplasm, AraC has to be phosphorylated to a triphosphate form (Ara-CTP) to become toxic. Ara-CTP can compete with dCTP for incorporation into DNA, where it subsequently inhibits the function of DNA polymerase, resulting in a block of DNA synthesis.6-9 The phosphorylation of AraC is catalyzed by 3 different kinases, in which the first phosporylation is performed by deoxycytidine kinase (dCK).
Deoxycytidine kinase (EC 2.7.1.74) catalyzes the transfer of the γ-phosphate of adenosine triphosphate (ATP) or uridine triphosphate (UTP) to dC to give rise to dC 5′-monophosphate.10-13 It can also phosphorylate antimetabolites such as AraC. The dCK protein functions as a homodimer of 60 kd, consisting of 2 subunits of 30.5 kd. Also, dCK is present in a variety of tissues, with the highest expression in thymus and in T-lymphocytic lineages.10,14The enzyme is located in the cytoplasm; however, when overexpressed in vitro, the dCK protein translocates to the nucleus. Its expression is not cell-cycle regulated,15 and the promoter contains several binding sites of ubiquitously expressed transcription factors such as Sp1 and E2F.16 Enzyme activity of dCK is increased during the S-phase of the cell cycle.17
Regarding resistance to AraC, many mechanisms have been postulated to be involved, such as an increased dCTP pool inducing the negative feedback inhibition of dCK, increased cytidine deaminase function leading to an increase in the deamination of AraC to AraU,18-21 decreased AraC transport over the cell membrane into the cytoplasm,22 defects in the cell death pathways (apoptosis),23-27 and inactivation of dCK.28-34 We and others have shown that when resistance to AraC is induced in vitro, inactivation of dCK was the predominant mechanism of AraC resistance.28-34 We have generated 2 different AraC-resistant rat leukemic cell lines by in vitro exposure to AraC. These cell lines expressed inactive dCK proteins as a result of either a point mutation or a genomic rearrangement of the dCK gene.34 Also, in a rat leukemic cell line resistant to AraC, which was generated ex vivo, dCK was inactivated by a deletion of the dCK locus.34
To investigate the possible involvement of dCK in patients with chemotherapy-resistant AML, we have analyzed dCK messenger RNA (mRNA) expression in purified leukemic blasts from patients with sensitive and resistant AML. In parallel, we also analyzed the dCK expression in phytohemagglutinin-stimulated T cells (PHA T cells) cultured in vitro from the same individuals. In this study we report the presence of alternatively spliced forms of dCK mRNA only detectable in leukemic blasts and normal PHA T cells from patients with clinically resistant AML but not in leukemic blasts from patients with sensitive AML or in healthy donors. These alternatively spliced forms of dCK mRNA could be translated into proteins in vitro, which were shown to be inactive in an in vitro dCK activity assay. Furthermore, alternatively spliced variants of dCK were also detected in rat leukemic cells during induction of AraC resistance in vitro. We propose that the presence of dCK splice products may contribute to the occurrence of AraC resistance in patients with AML.
Materials and methods
Chemicals
We purchased 2-chloro-2′-deoxyadenosine, adenosine 5′-triphosphate magnesium salt, uridine 5′-triphosphate sodium salt, and bovine serum albumin from Sigma Chemical Co (St Louis, MO). Interleukin-2 was purchased from Roussel-Uclaf (Paris, France). Creatine kinase and creatine phosphate were obtained from Boehringer (Mannheim, Germany) and NaF from Merck (Darmstadt, Germany). [3H] 2-chloro-2′-deoxyadenosine (8.14 × 1011Bq/mmol) was bought from Campro Scientific (Veenendaal, The Netherlands) and L-[4,5-3H] leucine (3.7 × 107 Bq/mL) (TRK 6830) from Amersham Pharmacia Biotech (Buckinghamshire, UK). TnT T7 Coupled Wheat Germ Extract System and RNasin Ribonuclease Inhibitor (40 U/μL) were obtained from Promega (Madison, WI). TRIzol Reagent for total RNA isolation was from GIBCO BRL, Life Technologies (Gaithersburg, MD). The Random Primed DNA labeling kit was obtained from Boehringer Mannheim. QIAquick PCR Purification Kit was from QIAGEN GmbH (Hilden, Germany). Taq polymerase was purchased from PE Biosystems (Foster City, CA).
Rat leukemic cell lines
The AraC- and 5-aza-2′-deoxycytidine– (decitabine, or DAC) sensitive rat leukemic cell line RCL/0 was originally purchased from TNO (Rijswijk, The Netherlands). The AraC-sensitive cell line RO/1 was derived from the RCL/0 cell line by limiting dilution. From this RO/1 cell line, an AraC-resistant cell line RO/1-A was derived by in vitro exposure to increasing concentrations of AraC (up to 10−5 mol/L).35-37
Rat cell line culture conditions
All rat cell lines were cultured in HEPES-buffered RPMI-1640 medium supplemented with 10% fetal calf serum, 4-mmol/L L-glutamine, 50-μg/mL streptomycin, 50-U/mL penicillin, and amphotericin B. Resistant clones were frequently tested for their resistant phenotype by culturing them in the presence of 10−5-mol/L AraC or DAC. Cells were retained in exponential growth.
Isolation of leukemic blasts from bone marrow or peripheral blood of patients with AML
Mononuclear cells were isolated by sedimentation on Ficoll-Isopaque (Pharmacia, Uppsala, Sweden) density gradient (density 1.077 g/cm3) centrifugation and cryopreserved in liquid nitrogen until use. Leukemic blasts were purified by a fluorescence-activated cell sorter (FACS) using at least 2 different patient-specific leukemic cell markers (Table1). Therefore, post-Ficoll (PF) samples from peripheral blood (PB-PF) or bone marrow (BM-PF) were thawed and immediately used for FACS. Cell pellets were resuspended in 0.5 mL of RPMI-1640 medium and incubated with phycoerythrin- or fluorescein isothiocyanate–labeled monoclonal antibodies for 60 minutes at 4°C. Labeled leukemic blasts were washed 3 times, resuspended in RPMI-1640 plus 2% fetal calf serum, and sorted on a FACSVantage (Becton Dickinson, Mountain View, CA). The purity of the leukemic blasts was confirmed by FACS analysis and was always more than 95%. All specimens were collected as part of protocols that had been approved by the medical ethical committee of the Leiden University Medical Center.
Characteristics of patients with chemotherapy-resistant AML (rAML) and chemotherapy-sensitive AML (sAML)
| Patient . | FAB . | Karyotype . | Age . | Disease state of blasts obtained* . | Chemotherapy, including AraC . | Material . | Blasts before FACS, % . | Leukemic-specific markers used for FACS . | Blasts after FACS, % . |
|---|---|---|---|---|---|---|---|---|---|
| rAML1 | M0 | t(3;6), del(11), del(12) | 38 | Relapse | 3 | BM | 95 | CD33/34 | >95 |
| rAML2 | M0 | Normal | 60 | Relapse | 2 | BM | 90 | CD13/34 | >99.5 |
| rAML3 | M2 | t(8;21), t(1;10)(minor) | 42 | Relapse | 3 | BM | 75 | CD33/34 | 99 |
| rAML4 | M5A | t(9;11) | 22 | Relapse | 2 | BM | 80 | CD33/HLA-DR | 98 |
| rAML5 | M2 | t(1;9), del(6) | 21 | Relapse | 3 | PB | 35 | CD13/14/33/45 | >95 |
| rAML6 | M6 | Unknown | 34 | Refractory | 2 | BM | 35 | CD15/33 | 95 |
| rAML7 | M1 | t(9;22), inv(3), −7 | 41 | Refractory | 2 | PB | >90 | CD34/54 | >99.9 |
| rAML8 | M5 | Unknown | 50 | Refractory | 3 | BM | 80 | CD33/HLA-DR | >95 |
| rAML9 | M3 | +(13) | 56 | Refractory | 3 | BM | 97 | No sort | 97 |
| rAML10 | M2 | Unknown | 53 | Refractory | 3 | BM | 94 | CD33/34 | >95 |
| rAML11 | M2 | Normal | 68 | Diagnosis | 0 | BM | 50 | CD13/14/33 | >95 |
| rAML12 | M1 | Del(7) | 57 | Diagnosis | 0 | BM | 95 | No sort | 95 |
| sAML1 | M1 | Normal | 44 | Diagnosis | 0 | BM | 95 | No sort | 95 |
| sAML2 | M5A | Normal | 48 | Diagnosis | 0 | BM | 95 | No sort | 95 |
| sAML3 | M5A | t(9;11), +8 | 55 | Diagnosis | 0 | BM | 93 | CD33/HLA-DR | 98.5 |
| sAML4 | M4 | Unknown | 46 | Diagnosis | 0 | BM | 60 | CD33/HLA-DR | >95 |
| sAML5 | M5 | t(8;21) | 55 | Diagnosis | 0 | BM | 96 | No sort | 96 |
| sAML6 | M4 | Inv(16), +8, +14, +21 | 48 | Diagnosis | 0 | BM | 60 | CD13/CD65 | >98 |
| sAML7 | M2 | t(8;21) | 17 | Diagnosis | 0 | PB | 75 | CD34/CD54 | 99.5 |
| sAML8 | M5A | Normal | 45 | Diagnosis | 0 | BM | 87 | CD11c/CD33 | 99.5 |
| sAML9 | M4 | Inv(16), del(7) | 49 | Diagnosis | 0 | BM | 90 | CD13/HLA-DR | 99 |
| sAML10 | M2 | t(8;21), −Y, +8 | 53 | Diagnosis | 0 | BM | 79 | CD13/HLA-DR | 97 |
| Patient . | FAB . | Karyotype . | Age . | Disease state of blasts obtained* . | Chemotherapy, including AraC . | Material . | Blasts before FACS, % . | Leukemic-specific markers used for FACS . | Blasts after FACS, % . |
|---|---|---|---|---|---|---|---|---|---|
| rAML1 | M0 | t(3;6), del(11), del(12) | 38 | Relapse | 3 | BM | 95 | CD33/34 | >95 |
| rAML2 | M0 | Normal | 60 | Relapse | 2 | BM | 90 | CD13/34 | >99.5 |
| rAML3 | M2 | t(8;21), t(1;10)(minor) | 42 | Relapse | 3 | BM | 75 | CD33/34 | 99 |
| rAML4 | M5A | t(9;11) | 22 | Relapse | 2 | BM | 80 | CD33/HLA-DR | 98 |
| rAML5 | M2 | t(1;9), del(6) | 21 | Relapse | 3 | PB | 35 | CD13/14/33/45 | >95 |
| rAML6 | M6 | Unknown | 34 | Refractory | 2 | BM | 35 | CD15/33 | 95 |
| rAML7 | M1 | t(9;22), inv(3), −7 | 41 | Refractory | 2 | PB | >90 | CD34/54 | >99.9 |
| rAML8 | M5 | Unknown | 50 | Refractory | 3 | BM | 80 | CD33/HLA-DR | >95 |
| rAML9 | M3 | +(13) | 56 | Refractory | 3 | BM | 97 | No sort | 97 |
| rAML10 | M2 | Unknown | 53 | Refractory | 3 | BM | 94 | CD33/34 | >95 |
| rAML11 | M2 | Normal | 68 | Diagnosis | 0 | BM | 50 | CD13/14/33 | >95 |
| rAML12 | M1 | Del(7) | 57 | Diagnosis | 0 | BM | 95 | No sort | 95 |
| sAML1 | M1 | Normal | 44 | Diagnosis | 0 | BM | 95 | No sort | 95 |
| sAML2 | M5A | Normal | 48 | Diagnosis | 0 | BM | 95 | No sort | 95 |
| sAML3 | M5A | t(9;11), +8 | 55 | Diagnosis | 0 | BM | 93 | CD33/HLA-DR | 98.5 |
| sAML4 | M4 | Unknown | 46 | Diagnosis | 0 | BM | 60 | CD33/HLA-DR | >95 |
| sAML5 | M5 | t(8;21) | 55 | Diagnosis | 0 | BM | 96 | No sort | 96 |
| sAML6 | M4 | Inv(16), +8, +14, +21 | 48 | Diagnosis | 0 | BM | 60 | CD13/CD65 | >98 |
| sAML7 | M2 | t(8;21) | 17 | Diagnosis | 0 | PB | 75 | CD34/CD54 | 99.5 |
| sAML8 | M5A | Normal | 45 | Diagnosis | 0 | BM | 87 | CD11c/CD33 | 99.5 |
| sAML9 | M4 | Inv(16), del(7) | 49 | Diagnosis | 0 | BM | 90 | CD13/HLA-DR | 99 |
| sAML10 | M2 | t(8;21), −Y, +8 | 53 | Diagnosis | 0 | BM | 79 | CD13/HLA-DR | 97 |
FAB, French-American-British AML subclassification.
Indicates the moments of sampling of the leukemic blasts.
Generation of PHA T cells of healthy donors and patients with AML
BM-PF or PB-PF samples were thawed and cultured in the presence of 120-U/mL interleukin-2 and 0.8-μg/mL PHA in HEPES-buffered RPMI-1640 medium supplemented with 10% human AB negative serum, 4-mmol/L L-glutamine, 50-μg/mL streptomycin, 50-U/mL penicillin, and amphotericin B. After 3 days of PHA stimulation, PHA was washed away and stimulated T cells were maintained in medium with fresh interleukin-2 (120 U/mL).38 Cells were analyzed by FACS scan before RNA isolation.
RNA isolation, cDNA synthesis, and genomic DNA isolation
Total cellular RNA was isolated from a minimum of 105 cells by using TRIzol (GIBCO BRL Life Technologies) according to the manufacturer's protocol. After precipitation with isopropanol, the RNA pellet was dissolved in 50 μL of RNase-free milliQ water. Two micrograms of total RNA was reverse transcribed into single-strand complementary DNA (cDNA) as previously described.39 The cDNA yield was determined by performing a polymerase chain reaction (PCR) on cDNA derived from the hypoxanthine-guanine phosphoribosyltransferase (HPRT) housekeeping gene for human samples and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) for rat samples. The HPRT-PCR generated a 351–base pair (bp) fragment and the GAPDH-PCR a 450-bp construct, as analyzed on a 1% agarose gel.
Genomic DNA was isolated using the Puregene DNA Isolation Kit (Gentra Systems, Minneapolis, MN) according to the manufacturer's protocol.
Polymerase chain reaction
The amount of cDNA for dCK reverse transcriptase (RT)-PCR amplifications was standardized to the HPRT-PCR yield from one healthy donor (dCO). Full-length dCK was amplified in triplicate experiments from rat and human cDNA samples using 2 dCK-specific primers, A7 and B5 (Table 2), generating a 857-bp PCR fragment, representing the complete coding region of dCK. Complementary DNA from a healthy donor (dCO) was used as a positive control and milliQ water as a negative control. A nested PCR was subsequently performed to introduce a T7 RNA polymerase promoter upstream of the start codon of the dCK cDNA, using 50 pmol of the T7 and B6 dCK primers (Table 2). Wild-type dCK fragments of 823 bp for human and 819 bp for rat were formed and analyzed on a 1% agarose gel.
Nucleotide sequences of the primers used in RT-PCR and exon-intron PCR analysis subdivided in human dCK-specific primers, rat dCK-specific primers, and primers for housekeeping genes
| . | 5′-3′ primer sequence . | Annealings temperature . |
|---|---|---|
| Human dCK | ||
| Forward | ||
| A7-human | TCT TTG CCG GAC GAG CTC TG- | 65°C |
| T7-human | GGA TCC TAA TAC GAC TCA CTA | 55°C |
| TAG GAA CAG ACC ACC ATG GCC | ||
| ACC CCG CCC AAG AGA A– — | ||
| Exon 1 | CCG CCA GTG TCC TCA GCT GC- | 65°C |
| Exon 2 | CCT GAC AAC TTT TCT TCC TC- | 55°C |
| Exon 3 | CCT ATT GAC CAT TAA TTT GC- | 55°C |
| Exon 4 | GAT ACA TGT GTT GAT GAA GA- | 55°C |
| Exon 5-6 | CCC TGC CTT TTT CTT CCA TC- | 55°C |
| Exon 7 | GAT ACC TCA ATT AGT CAA GG- | 55°C |
| Reverse | ||
| B5-human | TGG AAC CAT TTG GCT GCC TG- | 65°C |
| B6-human | CAA GAT CAC AAA GTA CTC AA- | 55°C |
| Exon 1 | CTT GCG TCC CAC ATT TCC GG- | 65°C |
| Exon 2 | CAT ACC TCA AAT TCA TCT TG- | 55°C |
| Exon 3 | TTC AAA TGG CCA CGT ACA AG- | 55°C |
| Exon 4 | ATC CAC TGA AAA GAG TTC TA- | 55°C |
| Exons 5-6 | TTC TCA CCA CTC AGT ACA CT- | 55°C |
| Exon 7 | CAC CAT TAG GCT CAA TTC TA- | 55°C |
| Rat dCK | ||
| Forward | ||
| A7-rat | CCT GAG GTC CCG CGT CCT TA- | 65°C |
| T7-rat | GGA TCC TAA TAC GAC TCA CTA | 55°C |
| TAG GAA CAG ACC ACC ATG GCC | ||
| ACC CCA CCT AAG AGG — — | ||
| Reverse | ||
| B5-rat | TTG CCT GTT GTC TCC TGT GC- | 65°C |
| B6-rat | TGC AAT CAC AAA GTA CTC AA- | 55°C |
| Housekeeping genes | ||
| Forward | ||
| HPRT A | TGA CCA GTC AAC AGG GGA CA- | 55°C |
| GAPDH 5′ | ACC ACA GTC CAT GCC ATC AC- | 55°C |
| Reverse | ||
| HPRT B | CTT GAA CTC TGA TCT TAG GC- | 55°C |
| GAPDH 3′ | TCC ACC ACC CTG TTG CTG TA- | 55°C |
| . | 5′-3′ primer sequence . | Annealings temperature . |
|---|---|---|
| Human dCK | ||
| Forward | ||
| A7-human | TCT TTG CCG GAC GAG CTC TG- | 65°C |
| T7-human | GGA TCC TAA TAC GAC TCA CTA | 55°C |
| TAG GAA CAG ACC ACC ATG GCC | ||
| ACC CCG CCC AAG AGA A– — | ||
| Exon 1 | CCG CCA GTG TCC TCA GCT GC- | 65°C |
| Exon 2 | CCT GAC AAC TTT TCT TCC TC- | 55°C |
| Exon 3 | CCT ATT GAC CAT TAA TTT GC- | 55°C |
| Exon 4 | GAT ACA TGT GTT GAT GAA GA- | 55°C |
| Exon 5-6 | CCC TGC CTT TTT CTT CCA TC- | 55°C |
| Exon 7 | GAT ACC TCA ATT AGT CAA GG- | 55°C |
| Reverse | ||
| B5-human | TGG AAC CAT TTG GCT GCC TG- | 65°C |
| B6-human | CAA GAT CAC AAA GTA CTC AA- | 55°C |
| Exon 1 | CTT GCG TCC CAC ATT TCC GG- | 65°C |
| Exon 2 | CAT ACC TCA AAT TCA TCT TG- | 55°C |
| Exon 3 | TTC AAA TGG CCA CGT ACA AG- | 55°C |
| Exon 4 | ATC CAC TGA AAA GAG TTC TA- | 55°C |
| Exons 5-6 | TTC TCA CCA CTC AGT ACA CT- | 55°C |
| Exon 7 | CAC CAT TAG GCT CAA TTC TA- | 55°C |
| Rat dCK | ||
| Forward | ||
| A7-rat | CCT GAG GTC CCG CGT CCT TA- | 65°C |
| T7-rat | GGA TCC TAA TAC GAC TCA CTA | 55°C |
| TAG GAA CAG ACC ACC ATG GCC | ||
| ACC CCA CCT AAG AGG — — | ||
| Reverse | ||
| B5-rat | TTG CCT GTT GTC TCC TGT GC- | 65°C |
| B6-rat | TGC AAT CAC AAA GTA CTC AA- | 55°C |
| Housekeeping genes | ||
| Forward | ||
| HPRT A | TGA CCA GTC AAC AGG GGA CA- | 55°C |
| GAPDH 5′ | ACC ACA GTC CAT GCC ATC AC- | 55°C |
| Reverse | ||
| HPRT B | CTT GAA CTC TGA TCT TAG GC- | 55°C |
| GAPDH 3′ | TCC ACC ACC CTG TTG CTG TA- | 55°C |
The PCR reactions were performed in a reaction mixture containing 1.5-mmol/L MgCl2, 50-mmol/L KCl, 10-mmol/L Tris-HCl (pH 8.4), 0.2 mg of bovine serum albumin, 0.25-mmol/L of each deoxyribonucleoside triphosphate, 50 pmol of each primer, and 1 unit of Taq polymerase. The PCR was started after denaturation for 5 minutes at 95°C, followed by 30 to 33 cycles consisting of 48 seconds at 95°C, 48-seconds primer-specific annealing at 55 or 65°C and 48 seconds at 72°C, and a final elongation at 72°C for 5 to 10 minutes.
The exon-intron boundaries of the dCK gene were analyzed following PCR amplifications of genomic DNA. Six different primer sets were used, annealing in the introns flanking exons 1, 2, 3, 4, 5 to 6, and 7 (Table 2). Primers were chosen complementary to intron sequences approximately 15 bp from the splice sites, generating PCR fragments from exons 1, 2, 3, 4, 5 to 6, and 7 (nucleotide data from the intron sequences flanking the exons were kindly provided by Dr Beverly S. Mitchell, Department of Medicine, University of North Carolina, Chapel Hill, NC). PCR amplification of exon 1 generated a fragment of 316 bp, exon 2 of 164 bp, exon 3 of 278 bp, exon 4 of 246 bp, exon 5 to 6 of approximately 1000 bp, and exon 7 of 1721 bp.40
Southern blot analysis
Southern blot analysis was performed on 7.5-μg genomic DNA from PHA T cells from healthy donors and from unpurified BM cells from patients with resistant AML. Genomic DNA was digested withEcoRI, HindIII, and a combination ofEcoRI and HindIII. Digested DNA was separated on an 0.8% agarose gel, transferred to nylon membranes (Hybond N+, Amersham, Buchler, Germany) in 10 × SSC, and hybridized at 65°C with A7B5 dCK-PCR construct from a healthy donor as probe, labeled with 32P by random priming. Blots were washed twice with 2 × SSC plus 0.1% sodium dodecyl sulfate (SDS) at 65°C, with increasing stringent conditions when necessary, followed by exposure to Kodak films overnight at −80°C.
Sequence analysis
Sequence analysis was performed using an ABI 310 automatic sequencer; 15 to 30 ng of PCR product was cycle sequenced using the ABI-prism BigDye sequence kit according to the manufacturer's protocol, making use of the GeneAmp PCR System 9700 (all from PE Applied Biosystems, Perkin-Elmer-Cetus). Briefly, PCR products were purified using QIAgen PCR purification kit, and PCR concentration was determined on a 1% agarose gel. A total of 15to 30 ng of purified PCR product was sequenced using a POP-6 polymer and 50-μm capillary.
Coupled in vitro transcription-translation for dCK protein synthesis
For the synthesis of the dCK protein, we made use of the TnT Coupled Wheat Germ Extract System from Promega. The coupled in vitro transcription-translation reaction was performed according to the manufacturer's protocol on 1 μg of the nested dCK-PCR construct, making use of the T7 RNA polymerase promoter introduced 5′ of the start codon. In vitro transcription-translation was performed at 30°C for 90 minutes, and dCK protein was stored at −20°C.
To a part of the reaction, 3H-leucine (1.11 × 105 Bq) was added to determine the yield and molecular weight of dCK protein as described in the TnT protocol, with minor modifications. Briefly, 3H-leucine–labeled protein was incubated in 1-mol/L NaOH and 25% trichloroacetic acid (TCA), respectively, spotted on glass fiber filters (Whatman GF/C glass fiber filters, Whatman, Maidstone, England), and were then washed thoroughly with 5% TCA and acetone. Filters were then incubated in Filtercount (Packard), and 3H incorporation was estimated by counting for 1 minute using a scintillation counter. The protein concentration (mg/mL of dCK) was calculated from the counts per minute (cpm) as follows: (cpm/cpm standard) × (mol3H-leucine) × (1/number of leucine amino acids in dCK) × (molecular weight dCK) × (106).
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis
Five microliters of 3H-leucine–labeled dCK protein (approximately 150 ng) from the TnT transcription-translation protocol was separated on a 12.5% polyacrylamide denaturing gel to check for dCK protein truncations. The gel was fixed for 30 minutes in 10% methanol plus 10% acetic acid and, after fixation, incubated for 30 minutes in Amplifier (Amersham, UK). The gel was dried on a vacuum slab gel dryer, and autoradiography was determined after exposure to Kodak film overnight at −80°C.
Deoxycytidine kinase activity assay
The phosphorylating properties of the in vitro–translated dCK proteins were analyzed using a dCK activity protocol as originally described by Cheng et al,41 with minor modifications. Briefly, dCK activity was estimated using 3H-labeled 2-CdA as substrate, at a concentration of 0.002 mmol/L. Duplicate experiments were performed in reaction mixture containing 20-mmol/L Tris-HCl (pH 7.4); 5-mmol/L MgUTP; 27-U/mL creatine phosphokinase; 7.5-mmol/L creatine phosphate; 7.03 × 103 Bq/mL [3H]-labeled 2-CdA (specific activity 1.48 × 1011 Bq/mmol); 10-mmol/L unlabeled 2-CdA; 7-mmol/L NaF; 0.2% bovine serum albumin; and 0.2-mmol/L tetrahydrouridine to block dC-deaminase activity. The reactions were initiated by the addition of 2.5 ng of dCK protein per reaction and incubated at 37°C for 0, 10, 20, and 40 minutes and, at each time point, 50-μL aliquots were spotted on DEAE-coated paper discs (Whatman DE-81, Whatman). Filters were dried and washed in 1-mmol/L ammonium formate 4 times. Phosphorylated substrates bound to the filters were eluted from the filters by 0.6-mol/L HCl/1.5-mol/L NaCl, and3H-labeled reaction products were determined by scintillation counting in Atomlight using an LKB Rackbeta liquid scintillation counter. Enzyme kinetic properties were calculated by linear regression analysis and given in nmol/min × mg.
Results
Expression of dCK mRNA in purified AML blasts
To investigate the involvement of dCK in the process of AraC resistance in patients with resistant AML, we performed dCK RT-PCR analysis on (purified) leukemic blasts from patients with AML. AML patients were divided into 2 groups (resistant and sensitive) according to their clinical response to the chemotherapy. Table 1 shows the characteristics of 12 patients with clinically resistant AML (and 10 patients with sensitive AML). Clinical resistance was defined as persistence of leukemic blasts after at least 2 courses of intensive chemotherapy containing AraC. Patients with an early relapse of the AML (within 6 months) were also included in the population of clinically resistant patients. The leukemic blasts were purified by FACS using at least 2 patient-specific leukemic markers. The purity of the AML blasts after FACS was more than 95% in all patient samples.
The dCK mRNA expression was analyzed by RT-PCR in purified leukemic blasts and PHA T cells from patients with resistant AML (12 patients) and sensitive AML (10 patients), PHA T cells from healthy donors (10 samples), and total BM from 16 healthy donors. In all samples from healthy donors (total BM and PHA T cells) only wild-type dCK of 823 bp was amplified. However, in 7 of 12 purified leukemic blast samples from patients with resistant AML, additional bands of different lengths were observed in coexpression with wild-type dCK (Figure1A). Aberrant PCR fragments of 707, 481, 275, and 158 bp were detected in at least 2 of 3 PCR amplifications per patient sample. The aberrant PCR fragment of 158 bp was not found in coexpression with wild-type dCK in 2 of 4 PCR reactions on leukemic blasts from patient rAML12. In the other 2 PCR reactions of patient rAML12, a different additional PCR fragment of 481 bp was observed in coexpression with wild-type dCK. The aberrant dCK-PCR transcripts of 707, 481, and 275 bp were observed in more than 1 patient. Also, within a single resistant patient, more than 1 aberrant PCR fragment could be observed. The altered dCK-PCR fragment of 275 bp was not only observed in the leukemic blasts from resistant patients, but also in 6 of 12 PHA T cells generated from the resistant AML patients. In contrast to the resistant patients, in 9 of 10 patients with sensitive AML, only wild-type dCK-PCR fragments were observed (Table3). In 1 patient, sAML2, 1 aberrant dCK-PCR fragment of 275 bp was observed in 1 of 4 PCR reactions (Table3).
A representation of the detection of dCK mRNA expression by RT-PCR analysis in leukemic blasts from patients with resistant AML.
For each sample, at least 3 PCR amplifications were performed. T7B6 dCK-PCR constructs were separated on a 1.5% agarose gel and visualized by ethidium bromide staining. Indicated are dCK-PCR constructs amplified from purified leukemic blasts from patients with resistant AML and PHA-stimulated T cells from the same individuals.
A representation of the detection of dCK mRNA expression by RT-PCR analysis in leukemic blasts from patients with resistant AML.
For each sample, at least 3 PCR amplifications were performed. T7B6 dCK-PCR constructs were separated on a 1.5% agarose gel and visualized by ethidium bromide staining. Indicated are dCK-PCR constructs amplified from purified leukemic blasts from patients with resistant AML and PHA-stimulated T cells from the same individuals.
RT-PCR analysis of dCK of leukemic blasts and corresponding PHA T cells in patients with resistant and sensitive AML in at least triplicate experiments
| Resistant AML . | dCK-PCR in leukemic blasts . | dCK-PCR in PHA T cells . | Sensitive AML . | dCK-PCR in leukemic blasts . | dCK-PCR in PHA T cells . |
|---|---|---|---|---|---|
| rAML1 | 823 + 481 + 275 | 823 | sAML1 | 823 | 823 |
| rAML2 | 823 | 823 | sAML2 | 823 + 275 | 823 |
| rAML3 | 823 + 481 | 823 + 275 | sAML3 | 823 | 823 |
| rAML4 | 823 + 275 | 823 + 275 | sAML4 | 823 | 823 |
| rAML5 | 823 | 823 | sAML5 | 823 | 823 |
| rAML6 | 823 + 707 | 823 + 275 | sAML6 | 823 | 823 |
| rAML7 | 823 + 707 | 823 + 275 | sAML7 | 823 | 823 |
| rAML8 | 823 + 481 + 275 | 823 + 275 | sAML8 | 823 | 823 |
| rAML9 | 823 | 823 | sAML9 | 823 | 823 |
| rAML10 | 823 | 823 + 275 | sAML10 | 823 | 823 |
| rAML11 | 823 | 823 | |||
| rAML12 | 823 + 481 + 158 | 823 |
| Resistant AML . | dCK-PCR in leukemic blasts . | dCK-PCR in PHA T cells . | Sensitive AML . | dCK-PCR in leukemic blasts . | dCK-PCR in PHA T cells . |
|---|---|---|---|---|---|
| rAML1 | 823 + 481 + 275 | 823 | sAML1 | 823 | 823 |
| rAML2 | 823 | 823 | sAML2 | 823 + 275 | 823 |
| rAML3 | 823 + 481 | 823 + 275 | sAML3 | 823 | 823 |
| rAML4 | 823 + 275 | 823 + 275 | sAML4 | 823 | 823 |
| rAML5 | 823 | 823 | sAML5 | 823 | 823 |
| rAML6 | 823 + 707 | 823 + 275 | sAML6 | 823 | 823 |
| rAML7 | 823 + 707 | 823 + 275 | sAML7 | 823 | 823 |
| rAML8 | 823 + 481 + 275 | 823 + 275 | sAML8 | 823 | 823 |
| rAML9 | 823 | 823 | sAML9 | 823 | 823 |
| rAML10 | 823 | 823 + 275 | sAML10 | 823 | 823 |
| rAML11 | 823 | 823 | |||
| rAML12 | 823 + 481 + 158 | 823 |
All dCK-PCR fragment sizes are presented in base pairs.
Nucleotide sequence analysis of the dCK-PCR fragments
Table 4 shows the results of the sequence analysis of the PCR fragments that were detected in the dCK RT-PCR amplifications. All aberrant dCK-PCR fragments appeared to be deletion mutants of wild-type dCK, in which complete exons were excised. Deletion of exon 5, exons 3 to 4, exons 3 to 6, and exons 2 to 6 were observed. In the wild-type dCK-PCR constructs, mutation was detected in only 1 patient (rAML3). This patient, with an additional PCR product with deletion of exons 3 to 4, did have a heterozygous mutation in exon 3 (C→T nucleotide 364 [Pro→Ser]).
Structure of the aberrant dCK-PCR transcripts and the crude genomic structure of the dCK gene in patients with resistant AML
| T7B6 dCK-PCR in base pairs . | cDNA sequence . | Southern blot analysis . | Exon-intron sequence . |
|---|---|---|---|
| 823 | Wild type | Normal | Wild type |
| 707 | Deletion exon 5 | Normal | Wild type |
| 481 | Deletion exons 3-4 | Normal | Wild type |
| 275 | Deletion exons 3-6 | Normal | Wild type |
| 158 | Deletion exons 2-6 | Normal | Wild type |
| T7B6 dCK-PCR in base pairs . | cDNA sequence . | Southern blot analysis . | Exon-intron sequence . |
|---|---|---|---|
| 823 | Wild type | Normal | Wild type |
| 707 | Deletion exon 5 | Normal | Wild type |
| 481 | Deletion exons 3-4 | Normal | Wild type |
| 275 | Deletion exons 3-6 | Normal | Wild type |
| 158 | Deletion exons 2-6 | Normal | Wild type |
Analysis of the genomic structure of the dCK gene
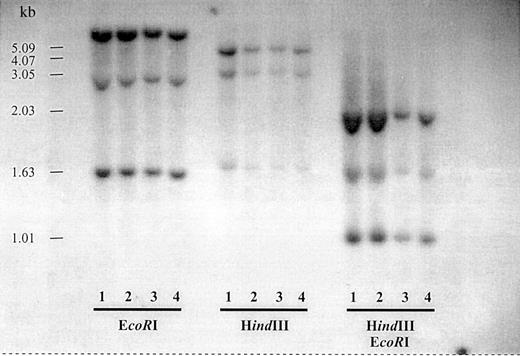
To check for possible genomic rearrangements of the dCK locus, Southern blot analysis was performed on genomic DNA from PHA stimulated T cells from healthy donors and unpurified blast samples from patients with resistant AML expressing an aberrant dCK transcript or only wild-type dCK. Figure 2 is representative for all patients with an aberrant dCK-PCR fragment. No genomic rearrangements were observed.
Southern blot analysis of the dCK genomic region of patients with resistant AML.
Genomic DNA from PHA T cells from a healthy donor and unpurified BM cells from 3 patients with resistant AML were digested withEcoRI, HindIII, and EcoRI plusHindIII. Lane 1, PHA T cells from a healthy donor; lane 2, unpurified leukemic blasts from patient rAML3; lane 3, unpurified leukemic blasts from rAML12; and lane 4, unpurified leukemic blasts from rAML2.
Southern blot analysis of the dCK genomic region of patients with resistant AML.
Genomic DNA from PHA T cells from a healthy donor and unpurified BM cells from 3 patients with resistant AML were digested withEcoRI, HindIII, and EcoRI plusHindIII. Lane 1, PHA T cells from a healthy donor; lane 2, unpurified leukemic blasts from patient rAML3; lane 3, unpurified leukemic blasts from rAML12; and lane 4, unpurified leukemic blasts from rAML2.
Because no genomic rearrangements seemed to be involved in the formation of the aberrant dCK-PCR fragments, we investigated whether mutations within the splice sites were responsible for the exon deletions in the aberrant dCK-PCR fragments. No mutations were detected in the 5′-splice donor or the intron 3′-splice acceptor nucleotide sites (Table 4).
Determination of dCK activity
To investigate whether the altered dCK cDNAs could be translated into functional proteins, we performed a coupled in vitro transcription-translation assay using the T7 RNA polymerase promoter sequence upstream of the coding region. The translated proteins were visualized on a 12.5% SDS–polyacrylamide gel electrophoresis (PAGE) gel. Figure 3 shows the translated proteins of the wild-type dCK and dCK-PCR fragments missing exon 5, exons 3 to 4, and exons 3 to 6. The molecular weights of the proteins were in agreement with the predicted molecular weight as calculated from the cDNA sequences (Table 5). The deletion of exon 5 did code for a protein of 23 kd, deletion of exons 3 to 4 of 17 kd, and deletion of exons 3 to 6 did result in a protein of only 8.5 kd. The cDNA missing exons 2 to 6 could not be translated into a detectable dCK protein in our in vitro transcription-translation system because of the absence of leucine amino acids as a result of a frame shift in the reading frame of the dCK gene caused by the exon deletions.
Coupled in vitro transcription-translation test.
3H-leucine–labeled dCK proteins were synthesized and analyzed on a 12.5% SDS-PAGE gel. The dCK protein generated from a dCK-PCR T7B6 construct from healthy donor PHA T cells (wt), from a T7B6 construct with a deletion of exon 5 (del 5; rAML6), from a T7B6 construct with a deletion of exons 3 to 4 (del 3-4; rAML3), and from a T7B6 construct with a deletion of exons 3 to 6 (del 3-6; rAML4).
Coupled in vitro transcription-translation test.
3H-leucine–labeled dCK proteins were synthesized and analyzed on a 12.5% SDS-PAGE gel. The dCK protein generated from a dCK-PCR T7B6 construct from healthy donor PHA T cells (wt), from a T7B6 construct with a deletion of exon 5 (del 5; rAML6), from a T7B6 construct with a deletion of exons 3 to 4 (del 3-4; rAML3), and from a T7B6 construct with a deletion of exons 3 to 6 (del 3-6; rAML4).
Predicted protein translations of the altered dCK cDNA constructs
| cDNA sequence . | T7B6 dCK-PCR, bp . | Coding region, bp . | Frame shift . | Predicted amino acids . | Predicted molecular weight . | dCK activity [nmol per min × mg] . |
|---|---|---|---|---|---|---|
| Wild type | 823 | 780 | No | 260 | 30.5 | 129.1 |
| Deletion exon 5 | 707 | 597 | Yes | 199 | 22.8 | 0 |
| Deletion exon 3-4 | 481 | 438 | No | 146 | 17.0 | 0 |
| Deletion exon 3-6 | 275 | 231 | No | 77 | 8.5 | 0 |
| Deletion exon 2-6 | 158 | 120 | Yes | — | — | — |
| cDNA sequence . | T7B6 dCK-PCR, bp . | Coding region, bp . | Frame shift . | Predicted amino acids . | Predicted molecular weight . | dCK activity [nmol per min × mg] . |
|---|---|---|---|---|---|---|
| Wild type | 823 | 780 | No | 260 | 30.5 | 129.1 |
| Deletion exon 5 | 707 | 597 | Yes | 199 | 22.8 | 0 |
| Deletion exon 3-4 | 481 | 438 | No | 146 | 17.0 | 0 |
| Deletion exon 3-6 | 275 | 231 | No | 77 | 8.5 | 0 |
| Deletion exon 2-6 | 158 | 120 | Yes | — | — | — |
Corresponding dCK activities as determined by the phosphorylation of 3H-labeled 2-CdA on 2.5 ng of dCK protein. Wild-type dCK was generated from a dCK T7B6-PCR construct from PHA T cells generated from healthy donors. The activity represents the mean activity in nmol/min × mg on 2.5-ng in vitro–translated dCK proteins from 5 individuals.
We performed dCK activity tests on 2.5 ng of in vitro–translated wild-type and aberrant dCK proteins. All altered dCK proteins had lost their activity by using 0.002-mmol/L 3H–2-CdA as substrate, whereas wild-type dCK showed a normal activity compared with the dCK activity in healthy donors (Table 5).
Detection of altered dCK forms in rat leukemic cell lines during induction of AraC resistance
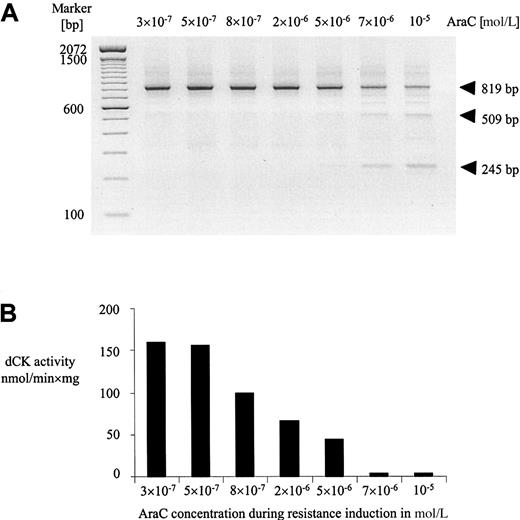
To investigate whether altered dCK mRNA transcripts could also be detected in AraC-resistant leukemic cell lines, we analyzed the dCK expression at different time intervals during the period of resistance induction in the rat leukemic cell line RO/1. Previously we described the generation of this AraC resistant cell line (RO/1-A), which was made resistant to AraC by in vitro exposure to increasing concentrations of AraC over a period of 160 days.37Aberrant dCK-PCR fragments were observed during the course of resistance induction at AraC concentrations of 7 × 10−6mol/L and 10−5 mol/L (Figure4A). Sequence analysis of these altered dCK-PCR fragments revealed deletion of exons 2 to 3 (509 bp) and exons 2 to 5 (245 bp). No mutations were detected in the wild-type dCK-PCR transcripts at all AraC concentrations during the period of resistance induction. In parallel, we determined the dCK activities of the cell lines during the period of resistance induction. Activity of dCK was markedly reduced at an AraC concentration of 5 × 10−6mol/L and lost at 7 × 10−6 mol/L and 10−5 mol/L of AraC (Figure 4B).
Resistance to AraC was introduced by in vitro exposure to increasing concentrations of AraC.
(A) Expression of dCK mRNA determined by RT-PCR analysis in rat leukemic cells during the course of induction of AraC resistance in vitro. Cell samples at different concentrations of AraC (in moles per liter) were obtained from exponentionally growing cells with more than 90% viability. The cells at 10−5-mol/L AraC were defined as the RO/1-A cell line. (B) Corresponding dCK activities in nmol/min × mg of protein during induction of AraC resistance in vitro as determined on in vitro–translated dCK proteins of the total PCR reaction.
Resistance to AraC was introduced by in vitro exposure to increasing concentrations of AraC.
(A) Expression of dCK mRNA determined by RT-PCR analysis in rat leukemic cells during the course of induction of AraC resistance in vitro. Cell samples at different concentrations of AraC (in moles per liter) were obtained from exponentionally growing cells with more than 90% viability. The cells at 10−5-mol/L AraC were defined as the RO/1-A cell line. (B) Corresponding dCK activities in nmol/min × mg of protein during induction of AraC resistance in vitro as determined on in vitro–translated dCK proteins of the total PCR reaction.
Discussion
Resistance toward chemotherapy is a major obstacle in the treatment of patients with AML. Resistance to an anthracycline is associated with increased expression and activity of P-glycoprotein (Pgp) in vivo and in vitro. However, resistance toward AraC, the most important component of the chemotherapeutic regimen for patients with AML, cannot be explained by increased activity of Pgp, because AraC is not a substrate of Pgp. Loss of dCK activity by mutational inactivation or genomic rearrangements is thought to play an important role in resistance to AraC in vitro. However, relatively few mutations have been reported in patients with chemotherapy-resistant AML. In the present study, we report the expression of alternatively spliced dCK mRNAs that may contribute to the occurrence of AraC resistance in patients with resistant AML. Different alternatively spliced forms of the dCK gene were detected in purified leukemic blasts from patients with resistant AML as well as in PHA-stimulated T cells generated from the same patients. These dCK splice products were not present in leukemic blasts or PHA T cells from patients with sensitive AML or in BM or PHA T cells from healthy donors. The alternatively spliced cDNA transcripts code for inactive dCK proteins in vitro.
No mutations within the splice sites of the dCK gene could be detected, and no gross genomic rearrangements were observed by Southern blot analysis, implying that the cDNA transcripts with exon deletions were generated by alternatively splicing dCK. Different splice variants of dCK were observed with deletion of one or more internal exons. In our patient material from resistant AML, splice variants missing exon 5, exons 3 to 4, exons 3 to 6, and exons 2 to 6 were detected. To our knowledge, this is the first report of the detection of alternatively spliced dCK in patients with resistant AML. Alternatively spliced forms of dCK were also detected in rat leukemic cells during resistance induction to AraC. A deletion of exons 2 to 3 and 2 to 5 were observed at the end point of AraC resistance induction in vitro. Deletion of exon 5 has also been described in human cell lines by others. A deletion of exon 5 was observed in a human T-lymphoblast cell line (Ara-C-8D) that had been made resistant to AraC by in vitro mutagenesis.28 More recently, an erythroleukemic cell line resistant to fludarabine and cross-resistant to AraC (K562-fluda) has been generated in vitro, expressing dCK transcripts with deletion of exon 5.31
The enrichment of the AML blasts by FACS has probably contributed to the detection of the aberrant PCR fragments. Nonpurified leukemic material contains a heterogeneous cell population with a significant amount of nonleukemic cells expressing wild-type dCK. In addition, leukemic blasts will probably consist of resistant blasts that express altered dCK and sensitive blasts expressing wild-type dCK. Previously, a low incidence of inactivation of dCK in patients with resistant AML was observed by Flasshove et al.42 These authors demonstrated point mutations within the dCK locus in 7 of 16 patients with resistant AML. However, only one point mutation resulted in a loss of dCK activity. In that study, an RT-PCR analysis was performed on unpurified material with a relatively low overall blast percentage. Contamination of the sample with nonleukemic cells and sensitive blasts with wild-type dCK may explain the absence of alternatively spliced forms of dCK in the study of Flasshove et al. The alternatively spliced forms of dCK that we observed were always found in coexpression with wild-type dCK. The wild-type dCK-PCR fragment in our RT-PCR are probably derived from the low percentage of nonleukemic cells still present in the purified sample or from leukemic blasts expressing wild-type dCK. In a titration experiment in which cells with no dCK expression were mixed with cells with wild-type dCK expression, dCK-PCR fragments were observed when 0.5% of the cells did express wild-type dCK. The presence of nonleukemic cells and leukemic blasts expressing wild-type dCK may also explain why no differences in dCK activities in cellular extract were observed between patients with alternatively spliced forms of dCK compared with patients without alternatively spliced dCK (data not shown).
No mutations were found in the wild-type cDNA transcripts of dCK or in the genomic DNA of the patients with resistant AML, except for one patient (rAML3) with an additional PCR fragment with deletion of exons 3 to 4, which had a heterozygous mutation in exon 3. However, because this mutation was not observed in any of the patients with the same alternatively spliced form of dCK, we assume that this mutation was introduced during the PCR amplification. In all cases, wild-type dCK mRNAs did code for dCK proteins of 30.5 kd and were shown to be able to phosphorylate 2-CdA as substrate. In addition, no point mutations were detected in the wild-type dCK-PCR fragments observed in the rat leukemic cell line during resistance induction to AraC in vitro.
All in vitro translatable splice proteins with lower molecular weights were inactive for their dCK activity in vitro. Loss of dCK activity is reflected by the inability of the enzyme to phosphorylate its substrates like AraC and 2-CdA. Deletion of the functional domains of dCK such as the ATP binding site (exon 2) or the tyrosine kinase domain (exon 6),43 replacement of the STOP codon, and major changes in the secondary structure of the protein will be the reason for loss of activity in the alternatively spliced dCK proteins. In our in vitro transcription-translation system, the transcript with deletion of exons 2 to 6 could not be detected because of the absence of the incorporation of 3H-labeled leucine amino acids as a result of a frame shift caused by the deletion.
The alternatively spliced dCK missing exons 3 to 6 was not only detected in leukemic blasts from patients with resistant AML but also in PHA T cells that were generated from the same patients with resistant AML, suggesting that alternative splicing of dCK is a general phenomenon in resistant patients. The presence of this alternatively spliced form in PHA T cells can probably not be explained by contamination of leukemic blasts still present at very low levels in the PHA–T-cell population, because only the splice variant with deletion exons 3 to 6 was detected in all samples. Alternative splicing may even be a common event in drug-resistant cancer in general, because alternatively spliced forms of the estrogen receptor have been detected in tamoxifen-resistant breast cancer cell lines.44,45Also, alternative splicing of the androgen receptor has been observed in some prostate cancers that exhibit resistance to androgen therapy.45 46
In conclusion, this study has demonstrated that inactivation of dCK by the formation of alternatively spliced dCK transcripts may mediate AraC resistance in patients with resistant AML. It is not clear whether these alternatively spliced forms indeed lead to resistance to AraC and whether the formation of alternatively spliced dCK transcripts are a result of AraC resistance. It remains to be investigated whether the alternatively spliced dCK proteins are expressed at a protein level in vivo and in vitro, because the alternatively spliced dCK mRNAs may be unstable and therefore not translated. More in vitro studies will be necessary to prove the biologic importance of alternatively spliced forms of dCK for the development of resistance to AraC in vitro and in vivo. Also, the mechanism by which the alternatively spliced forms of dCK are formed still needs to be elucidated. It is clear, however, that the alternatively spliced forms are only detectable in leukemic blasts and PHA T cells from patients with resistant AML and not in leukemic blasts or PHA T cells from patients with sensitive AML or in BM or PHA T cells from healthy donors. Therefore, we conclude that the presence of inactive, alternatively spliced dCK mRNA transcripts may contribute to the process of AraC resistance in patients with AML.
Acknowledgments
The authors thank Dr Beverly S. Mitchell (Department of Medicine, University of North Carolina, Chapel Hill, NC) for the genomic DNA sequence of human dCK and Dr Hans L. Vos (Department of Hematology, Leiden University Medical Center, Leiden, The Netherlands) for insightful discussions during the course of these studies and critical reading of the manuscript.
Supported by the Dutch Cancer Society (grant RUL96-1347).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
R. M. Y. Barge, Leiden University Medical Center, Department of Hematology, C2-R, PO Box 9600, 2300 RC Leiden, The Netherlands; e-mail: barge@hematology/lumc.nl.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal