Abstract
Sequencing the complete factor IX gene of 2 sisters with hemophilia B with different phenotypes and no family history of hemorrhagic diathesis revealed a common 5′ splice site mutation in intron 3 (T6704C) in both and an additional missense mutation (I344T) in one. The presence of dysfunctional antigen in the latter strongly suggested that these mutations are in trans. Neither mutation was found in leukocyte DNA from the asymptomatic parents, but the mother was in somatic mosaicism for the shared splice site mutation. This case illustrates the importance of defining the phenotype and considering somatic mosaicism in sporadic cases. It underlines the limitations of complete gene sequencing for the detection of mosaicism and has implication for genetic counseling.
Introduction
Hemophilia B is an X-linked bleeding disorder resulting from factor IX (F.IX) deficiency,1-3 caused by a wide range of mutations on the F.IX gene.4Hemophilia B in girls is extremely rare and results from different mechanisms, the most common of which is skewed inactivation of the normal X chromosome in heterozygous girls.5-10 In some cases, the inactivation process does not seem to be random and occurs by either selection in favor of the activity of an X chromosome involved in a balanced X:autosome translocation11 or as a result of genetic differences affecting the X chromosome inactivation itself.12 More rarely, the phenotypic expression of the disease can be related to compound heterozygosity for hemophilic mutations13 or Turner syndrome.
We present a family (Figure 1) with no prior bleeding history in whom moderate hemophilia B was diagnosed in 2 sisters having different (II3) and identical (II2) levels of F.IX activity (F.IXC) and antigen (F.IXAg). This discrepancy in the coagulation results implies a different molecular basis for hemophilia. Analysis of the F.IX gene nucleotide sequence confirmed this hypothesis: II2 is heterozygous for a null mutation with skewed inactivation of the normal X chromosome; II3 has an additional mutation in trans and is therefore a compound heterozygote. Neither mutation was detected in DNA from the parents. However, maternal buccal and uroepithelial studies showed somatic mosaicism for the mutation shared by the daughters. The second mutation must have resulted from a de novo mutation in the father's gametes.
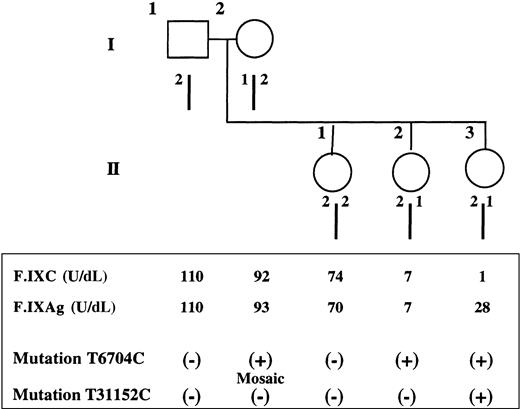
Pedigree, haplotypes, and coagulation and mutation studies.
(Top) Pedigree and haplotype results of the family with female hemophilia B. Haplotype analysis by restriction fragment length polymorphism using the enzyme Dde I demonstrates that the 2 affected sisters, II2 and II3, share a common allele (2/1) with the mother (I2), whereas the normal sister (II1) inherited the mother's other allele (2/2). (Bottom) Coagulation and mutation studies. For each member of the family, F.IXC and F.IXAg are indicated as well as the presence (+) or the absence (−) of each characterized mutation.
Pedigree, haplotypes, and coagulation and mutation studies.
(Top) Pedigree and haplotype results of the family with female hemophilia B. Haplotype analysis by restriction fragment length polymorphism using the enzyme Dde I demonstrates that the 2 affected sisters, II2 and II3, share a common allele (2/1) with the mother (I2), whereas the normal sister (II1) inherited the mother's other allele (2/2). (Bottom) Coagulation and mutation studies. For each member of the family, F.IXC and F.IXAg are indicated as well as the presence (+) or the absence (−) of each characterized mutation.
Study design
Patients and coagulation studies
The proband, a 14-year-old girl with moderate hemophilia B, suffered from hematomas, hemarthrosis, and epistaxis. There was no family history of a bleeding disorder and family studies revealed a milder form of the disease in a sister who suffered only from rare hematomas. The eldest sister and parents were asymptomatic. After informed consent, F.IXC was determined by a modified one-stage assay14 and F.IXAg by an immunoenzymatic method using a commercial kit (Asserachrom F.IXAg, Diagnostica Stago, Franconville, France).
Polymerase chain reaction (PCR) amplification and sequencing
Genomic DNA was prepared from leukocytes using standard procedures.15 All 8 exons, intron-exon boundaries, and 5′ and 3′ ends of the F.IX gene were amplified by PCR and, after purification on Sephacryl S400 columns (Pharmacia, Orsay, France), fragments were directly sequenced using the ABI PRISM Dye Terminator Cycle Sequencing Reaction Kit (PE Biosystems, Courtaboeuf, France) on an ABI PRISM 377 DNA sequencer according to the manufacturer's specifications.
Competitive oligo priming (COP) PCR
We performed COP PCR according to Tada.16 Leukocyte DNA as well as DNA obtained from uroepithelial and buccal cells were studied by COP with allele-specific primers conjugated to different fluorescent dyes, 6-carboxy-fluorescein (FAM), or tetrachloro-6-carboxy-fluorescein (TET), in conjunction with a common primer. The fluorescence of each dye with respect to its amplified DNA locus is scored on a 373 DNA sequencer as previously described.17 Primer sequences for intron 3 splice mutation are as follows:
F.IX-FOR1: 5′ (TET) TTGGAAGCAGTATGTTGGC 3′
F.IX-REV2: 5′ (FAM) TTGGAAGCAGTATGTTGGT 3′
F.IX-REV: 5′ TCAGGTTGGGAGATTGGGTAT 3′
Chromosome X inactivation
Results and discussion
The female proband (II3) had levels of F.IXC of 1 U/dL and F.IXAg of 28 U/dL, indicating moderately severe hemophilia B, and a karyotype of 46,XX. Her sister (II2) had mild hemophilia B with F.IXC and F.IXAg levels of 7 U/dL. The parents (I1 and I2) and elder sister (II1) showed normal coagulation results.
The F.IX gene analysis demonstrated heterozygosity for a unique splice site alteration (T6704C) in the affected sister (II2) as already described in severe hemophiliacs.20 The proband is therefore a carrier for hemophilia B and her low F.IXC and F.IXAg most probably result from a skewed X inactivation.6-8Inactivation patterns using the HUMARA gene showed that both II1 and II3 had a normal pattern. Unfortunately, X inactivation analysis in the third daughter (II2) was inconclusive because she was not polymorphic for the HUMARA gene (CAG) repeat. But her F.IX level demonstrates that at least a skewed inactivation occurred in the liver.
Families have been identified in which unbalanced X inactivation is apparently inherited as an X-linked dominant trait. A mutation involving a single nucleotide in the promoter region of theXIST gene has recently been shown to underlie a skewed pattern of X inactivation in multiple members of 2 unrelated families.19 Sequencing of the minimal promoter showed a normal nucleotide sequence especially the absence of the C43G mutation previously described by Plenge et al.19
The F.IX gene of the proband (II3) showed the same T6704C mutation as her sister's (II2). However, a second alteration, T31152C, was found in exon 8. This missense mutation, I344T, has not been reported to date but 2 mutations involving the same amino acid residue, I344F and I344S, have been found associated with moderate hemophilia (9th edition of the hemophilia B database;http://www.umds.ac.uk/molgen/haemBdatabase.htm).
The different levels of F.IXC and F.IXAg in II3 are only compatible with the presence of each mutation on different alleles and II3 is therefore a compound heterozygote.
Analysis of 2 intragenic polymorphic markers within the F.IXgene revealed that the 2 sisters, II2 and II3, inherited the same maternal haplotype, different from the normal eldest sister (II1). Therefore, the T6704C is of maternal origin. To investigate the possibility of somatic mosaicism in the mother (I2), DNA studies were performed on peripheral blood leukocytes and uroepithelial and buccal cells. Sequence analysis showed the unequivocal presence of a normal and mutant sequence at nucleotide 6704 in various amounts in these cells. COP PCR analysis confirmed the sequencing results with less than 5% leukocytes but 10% and 30% of uroepithelial or buccal cells, respectively, carrying the mutation. The second mutation of the proband (II3) probably originated in the father's gametes. Using the same approach, search for a mosaicism in the father's buccal and uroepithelial cells for the mutation in exon 8 was negative. Unfortunately, sperm cells were not available and germline mosaicism could not be excluded.
Mosaicism has been documented for chromosomal abnormalities, mitochondrial mutations, triplet repeats, and mutations in a growing number of dominant and X-linked single gene disorders.21For X-linked disorders, the detection of somatic mosaicism implies prior knowledge of the deleterious mutation. Actually, the method of choice for identification of the deleterious mutation relies on DNA sequencing, but the ability of this method to detect somatic mosaicism is poor because the ratio of the mutant to the wild-type allele can be quite small.
Somatic mosaicism may be more common than previously thought.22 23 In sporadic cases of hemophilia one must always consider the possibility of maternal somatic/germline mosaicism and therefore systematically examine buccal and uroepithelial DNA. In this family, the mother (I2) must be considered as a carrier even if the germinal mosaicism cannot be documented.
Moreover, the implication of the compound heterozygosity in II3 is that any male child has F.IX deficiency. Altogether, these results confirm the initial hypothesis based on coagulation studies that the molecular basis of hemophilia B in these 2 girls is different. Without determination of both low F.IXC and F.IXAg, such a hypothesis could not have been verified and if the entire F.IX gene had not been studied, one of the 2 mutations could have been overlooked with important consequences in genetic counseling.
Acknowledgment
We wish to thank Dr John F. Davidson for helpful advice.
Supported by a grant from Baxter Laboratories.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
J. M. Lavergne, INSERM U143, Hôpital Bicêtre, 94275 Bicêtre Cedex, France; e-mail: lavergne@infobiogen.fr.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal