Abstract
Interleukin-12 (IL-12) is a key immunoregulatory cytokine that promotes Th1 differentiation and cell-mediated immune responses. The transcription factor STAT4 (signal transducer and activator of transcription 4) is an important element in mediating IL-12 signals, as evidenced by the fact that STAT4−/− mice display impaired responsiveness to IL-12 and deficient Th1 differentiation. STAT4 is inducibly phosphorylated on tyrosine and serine in response to IL-12, but the kinase(s) responsible for the latter event is unknown. Here we show that IL-12 induces STAT4 phosphorylation on serine 721 and that mutation of serine 721 interferes with STAT4 transcriptional activity. In addition, we show that mutation of tyrosine 693 abrogates IL-12–induced STAT4 tyrosine phosphorylation and transcriptional activity. Although the site surrounding serine 721 is an optimum consensus sequence for mitogen-activated family of protein kinases (MAPKs)-mediated phosphorylation, we demonstrate that IL-12 does not induce extracellular signal-regulated kinase (ERK) or c-Jun N-terminal kinase (JNK) activation in T and natural killer (NK) cells and that IL-12–induced STAT4 transcriptional activity is not affected by these kinases. Rather, we show that IL-12 induces p38 activation. Moreover, we demonstrate that p38α and its upstream activator, MKK6, phosphorylate STAT4 on serine 721, and are required for STAT4 full transcriptional activity induced by IL-12, establishing the MKK6/p38α/STAT4 pathway as an important mediator of IL-12 actions.
Introduction
Interleukin (IL)-12 is a critical immunoregulatory cytokine that plays a central role in cell-mediated immune responses, enhancing proliferation and cytotoxic activity of natural killer (NK) and T cells, inducing the production of proinflammatory cytokines, and promoting Th1 cell differentiation.1 Like other cytokine receptors, the IL-12 receptor (IL-12R) lacks kinase activity but is associated with 3 cytoplasmic Janus tyrosine kinases (JAKs), JAK2 and TYK2.2,3 On ligand binding, the JAKs are activated and phosphorylate tyrosine residues on IL-12Rβ2, providing docking sites for the SH2 domain of signal transducer and activator of transcription (STAT) 3 and 4.4,5 STAT4 is of particular importance as most IL-12 functions are disrupted in STAT4 deficient mice;6,7 indeed the phenotype of STAT4−/−mice is completely concordant with the defects observed in IL-12 and IL-12R knockout mice and in humans carrying mutations of the IL-12R.8-10
STATs are a family of latent cytosolic transcription factors that, on binding to receptors, are themselves phosphorylated on a conserved C-terminal tyrosine residue.11 This is critical for dimer formation and DNA binding.12 13
In addition to tyrosine phosphorylation, it has been shown that most of the STATs are also phosphorylated on serine residues in response to cytokine stimulation. Serine phosphorylation appears to be important for maximal transactivating potential,14-17 though its role in STAT DNA binding is still somewhat controversial.18,19 For STAT1 and STAT3, a conserved site of serine phosphorylation has been mapped within the C-terminal transcriptional activation domain.19 Although it is clear that this site is phosphorylated in response to cytokine stimulation, the identity of the kinase responsible for this event remains elusive.17 20-22 This site resides in a consensus sequence for proline-directed serine/threonine kinases-mediated phosphorylation.
Mitogen-activated family of protein kinases (MAPKs) comprises several subfamilies of proline-directed serine/threonine kinases, including extracellular signal-regulated kinases 1/2 (ERKs), c-Jun N-terminal kinases (JNKs), and p38.23-26 The MAPK subfamilies are regulated by different extracellular stimuli. For example, while ERKs are activated rapidly by growth factors, JNKs and p38 are typically activated by environmental stress and proinflammatory cytokines.27 However, it has been recently demonstrated that JNKs and p38 can also be activated by T-cell receptor-mediated signaling28 and by several cytokines, interleukins, and colony-stimulating factors that regulate hemopoietic cell growth and differentiation.29-36
Like STAT1 and STAT3, we have demonstrated that STAT4 is also phosphorylated on both tyrosine and serine residues in response to IL-12 stimulation.5,16 It too has a putative MAPK phosphorylation site in its transcriptional activation domain.37,38 However, the serine residue phosphorylated on STAT4 and the kinase responsible for this phosphorylation had not been identified. In this study, we show that, in response to IL-12 stimulation, STAT4 is phosphorylated on tyrosine 693 and serine 721 and mutations of each of these residues strongly impair its transcriptional activity. Although it had been argued that IL-12 can induce ERK activity in human T cells,39 suggesting that ERKs are responsible for STAT4 serine phosphorylation, here we show that IL-12 does not activate ERKs or JNKs in NK and T cells. On the contrary, we demonstrate that stimulation of p38α, through its upstream activator, MKK6, mediates IL-12–dependent STAT4 serine 721 phosphorylation and is required for its full transcriptional activity.
Materials and methods
Cytokines, antibodies, and reagents
The following reagents were purchased: recombinant human or mouse IL-12 (R&D Systems, Minneapolis, MN); anti-phosphotyrosine, anti-Shc and anti-Grb2 antibody (Ab) (Upstate Biotechnology, Lake Placid, NY); anti-STAT4 and anti-p38 Ab and GST-ATF2 fusion protein (Santa Cruz Biotechnology, Santa Cruz, CA); anti-ERK2 Ab (Trasduction Laboratories, Lexington, KY); antiphosphoserine 727 STAT3 Ab (Biosource International, Camarillo, CA); anti-JNK1 Ab (PharMingen, San Diego, CA, and Santa Cruz Biotechnology); anti-HA Ab and Concanavalin A (ConA) (Boehringer Mannheim, Mannheim, Germany); myelin basic protein (MBP), anisomycin and phytohemagglutinin (PHA) (Sigma, St Louis, MO); PD98059 (Research Biochemical International, Notick, MA); SB202190 (Calbiochem, La Jolla, CA). Recombinant human IL-2 was provided by Dr C. Reynolds.
Cell culture
Lymphocytes were purified from mouse spleens using nylon wool fiber columns and activated with 2 μg/mL ConA. After 48 hours, the cells were serum starved for 4 hours and restimulated with IL-12. Normal human T cells (more than or equal to 95% CD3+) were isolated from the peripheral blood of healthy donors as described.2 Jurkat T cells were cultured in RPMI 1640 + 10% fetal bovine serum (FBS). NK3.3 cells (from Dr J. Kornbluth) were cultured as described.2 Before stimulation with cytokine, both T and NK cells were rested in RPMI 1640 + 1% bovine serum albumin (BSA). The 293T and NIH3T3 cells were grown in Dulbecco's modified Eagle's medium plus 10% FBS.
Transfections
The pEFBOS-IL-12Rβ1, pEFBOS-IL-12Rβ2 (from Dr U. Gubler), and pCDNA3-STAT4 (from Dr J. N. Ihle) were used to transfect 293T cells, a system that provides high efficiency of transfectability and high levels of expression. Site-directed mutagenesis of STAT4 was performed with the Transformer Site-Directed Mutagenesis Kit (Clontech, Palo Alto, CA), according to manufacturer's instructions. Oligonucleotides were designed to change tyrosine to phenylalanine at codon 693 (5′-GACAAGGGTTTCGTCCCTTCTGTTTTTATCCC-3′), serine to alanine at codon 721 (5′-CAGACCTTCTCCCCATGGCTCCAAGTGCA-3′), and serine to alanine at codon 723 (5′-CC ATGTCTCCAGCTGCATATGCTGTGC-3′).
The 293T cells were transfected using a calcium phosphate transfection kit (5 Prime 3 Prime, Boulder, CO). Thirty-six hours later, cells were rested for at least 16 hours, then harvested, stimulated, and lysed in a buffer containing 20 mmol/L HEPES pH 7.5, 2.5 mmol/L MgCl2, 10 mmol/L EDTA, 1 mmol/L dithiothreitol, 40 mmol/L β-glycerophosphate, 2 mmol/L vanadate, 1% NP-40, 20 μg/mL aprotinin, and 20 μg/mL leupeptin.
Reporter gene assay
NIH3T3 and Jurkat T cells were transfected by LipofectAMINE (Life Technologies, Gaithersburg, MD) and by SuperFect (Qiagen, Chatsworth, CA), respectively, following the manufacturer's protocol. The modest levels of expression of the transfected vectors in the cells (compared with 293T cells) were advantageous in permitting detection of ligand-dependent activation of the reporter. Except where indicated, cells were transfected with 0.2 μg p3 × GAS-luc (from Dr R. Pine), 0.2 μg pCMV-β-galactosidase (Clontech), 0.2 μg pEFBOS-IL-12Rβ1, 0.6 μg pEFBOS-IL-12Rβ2, 0.2 μg pCDNA3-STAT4 wild-type or mutated. Cells were cotransfected, as indicated, with various MAPK family members, including pCDNA3-MEKEE, pCDNA3-MEKAA, pCDNA3-HA-ERK2, pCEV-MEKK1, pCDNA3-HA-JNK1, pCEFL-GST-MKK6, pCEFL-GST-MKK6KR, pCEFL-HA-p38α, pCEFL-HA-p38γ, pCEFL-MEK5DD, and pCEFL-HA-ERK5.40-44 As required, the total amount of DNA was adjusted with vector with no insert. The cells were rested overnight and, then, stimulated, where indicated, with 10 ng/mL IL-12 for 8 hours. Where required, the cells were pretreated with 20 μmol/L SB202190 or DMSO for 1 hour before the stimulation with IL-12 for 6 hours. Cell were lysed and luciferase and β-galactosidase activities were determined using the Dual-Light Kit (Tropix, Bedford, MA), following the manufacturer's instructions. Luciferase activity in each sample was normalized by the corresponding β-galactosidase activity.
Cloning and bacterial expression of the glutathione S-transferase -STAT4 fusion protein
The C-terminal portion of STAT4 (amino acids 538-749) was cloned in the vector pGEX-4T3 (Amersham Pharmacia Biotech, Buckinghamshire, England) in frame with the glutathione S-transferase (GST) gene. The vector was transformed in the BL 21 Lys strain of Escherichia coli. The transformed bacteria were grown until the optical density was 0.5, at which time isopropyl-β-thiogalactopyranoside (1 mmol/L final concentration) was added for 3 hours. The GST-STAT4 fusion protein was purified using the Bulk GST Purification Module (Amersham Pharmacia Biotech Inc), following the manufacturer's instructions.
Immunoprecipitation, immunoblotting, and kinase assays
Results
Phosphorylation of tyrosine 693 and serine 721 is required for IL-12-induced STAT4 activation
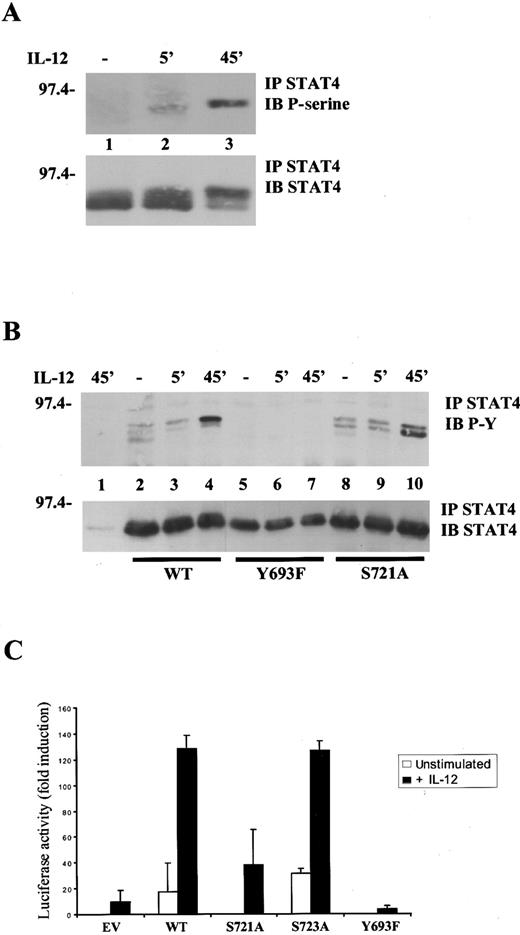
Serine 721 of STAT4 resides in a putative MAPK phosphorylation site and is analogous to serine 727 of STAT1 and STAT3.14,19 To assess whether IL-12 stimulation was able to induce STAT4 serine 721 phosphorylation in vivo, we took advantage of a phosphospecific antibody that recognizes STAT3 phosphorylated on serine 727 and cross-reacts with STAT4 phosphorylated on serine 721. This was not unexpected, as the core sequence surrounding these serine residues is very conserved. To this end, purified lymphocytes were activated for 48 hours to induce IL-12R expression and restimulated, after serum starvation, with IL-12 for various periods. As shown in Figure 1A, no basal phosphorylation was observed in unstimulated cells (lane 1), whereas STAT4 was phosphorylated on serine 721 beginning at 5 minutes (lane 2) but did not peak until 45 minutes of IL-12 stimulation (lane 3). The specificity of this antibody for serine 721 was confirmed using a STAT4 S721A mutant (see below, Figure6C). Of note, as we previously reported,16 the serine phosphorylated form of STAT4 had reduced electrophoretic mobility and phosphorylation on serine 721, correlated with the appearance of the shifted form of STAT4 (Figure 1A, compare upper and lower panels).
Importance of tyrosine 693 and serine 721 phosphorylation for IL-12-induced STAT4 activation.
(A), IL-12 induces STAT4 serine phosphorylation. Lymphocytes purified from mouse spleens were activated with ConA for 48 hours. After serum starvation, cells were left unstimulated (−) or were restimulated with IL-12 for the times indicated. Lysates were immunoprecipitated (IP) with anti-STAT4 Ab, resolved by SDS/polyacrylamide gel electrophoresis (SDS/PAGE), transferred to membrane and blotted (IB) with antiphosphoserine 727 STAT3 Ab that also detects serine phosphorylated STAT4 (upper panel). The blot was stripped and reprobed with anti-STAT4, demonstrating equal levels of proteins (lower panel). (B), Tyrosine 693 and serine 721 are important sites of phosphorylation in STAT4. 293T cells were transfected with 0.5 μg pEFBOS-IL-12Rβ1 and 1.5 μg pEFBOS-IL-12Rβ2 alone (lane 1) or together with 0.5 μg wild-type STAT4 (lanes 2-4), STAT4 Y693F (lanes 5-7) or STAT4 S721A (lanes 8-10). Cells were left unstimulated (−) or were stimulated with IL-12 for the times indicated. Lysates were immunoprecipitated with anti-STAT4 Ab, resolved by SDS/PAGE and blotted sequentially with antiphosphotyrosine Ab (upper panel) and anti-STAT4 (lower panel). (C), Tyrosine 693 and serine 721 are important for IL-12–mediated, STAT4-dependent transcriptional activity. Jurkat T cells were transiently transfected with the p3xGAS-luc and the pCMV-β-galactosidase reporters and expression vectors encoding the human IL-12R subunits. Where indicated, cells were cotransfected with control vector (EV), wild-type STAT4, or mutated STAT4 constructs. Transfected cells were left untreated or were stimulated with IL-12 before lysis. Luciferase and β-galactosidase activities were measured as described in “Materials and methods.” The data represent luciferase activity normalized by the β-galactosidase activity present in each sample, expressed as fold induction relative to the control, and are the averages ± SD of triplicate samples from a typical experiment. Unstimulated, ■; +IL-12, ▪.
Importance of tyrosine 693 and serine 721 phosphorylation for IL-12-induced STAT4 activation.
(A), IL-12 induces STAT4 serine phosphorylation. Lymphocytes purified from mouse spleens were activated with ConA for 48 hours. After serum starvation, cells were left unstimulated (−) or were restimulated with IL-12 for the times indicated. Lysates were immunoprecipitated (IP) with anti-STAT4 Ab, resolved by SDS/polyacrylamide gel electrophoresis (SDS/PAGE), transferred to membrane and blotted (IB) with antiphosphoserine 727 STAT3 Ab that also detects serine phosphorylated STAT4 (upper panel). The blot was stripped and reprobed with anti-STAT4, demonstrating equal levels of proteins (lower panel). (B), Tyrosine 693 and serine 721 are important sites of phosphorylation in STAT4. 293T cells were transfected with 0.5 μg pEFBOS-IL-12Rβ1 and 1.5 μg pEFBOS-IL-12Rβ2 alone (lane 1) or together with 0.5 μg wild-type STAT4 (lanes 2-4), STAT4 Y693F (lanes 5-7) or STAT4 S721A (lanes 8-10). Cells were left unstimulated (−) or were stimulated with IL-12 for the times indicated. Lysates were immunoprecipitated with anti-STAT4 Ab, resolved by SDS/PAGE and blotted sequentially with antiphosphotyrosine Ab (upper panel) and anti-STAT4 (lower panel). (C), Tyrosine 693 and serine 721 are important for IL-12–mediated, STAT4-dependent transcriptional activity. Jurkat T cells were transiently transfected with the p3xGAS-luc and the pCMV-β-galactosidase reporters and expression vectors encoding the human IL-12R subunits. Where indicated, cells were cotransfected with control vector (EV), wild-type STAT4, or mutated STAT4 constructs. Transfected cells were left untreated or were stimulated with IL-12 before lysis. Luciferase and β-galactosidase activities were measured as described in “Materials and methods.” The data represent luciferase activity normalized by the β-galactosidase activity present in each sample, expressed as fold induction relative to the control, and are the averages ± SD of triplicate samples from a typical experiment. Unstimulated, ■; +IL-12, ▪.
To ascertain whether serine 721 was a relevant site of phosphorylation, we next utilized site-directed mutagenesis and expression in a cell line, 293T cells, that lacks endogenous STAT4. We generated 2 STAT4 mutants in which the likely sites of phosphorylation, tyrosine 693 and serine 721, were mutated. We then transfected cells with constructs expressing the 2 chains of the human IL-12R, together with wild-type STAT4 or the STAT4 mutants. As shown in Figure 1B, wild-type STAT4 was tyrosine-phosphorylated after IL-12 stimulation (lane 4) but when tyrosine 693 of STAT4 was replaced by phenylalanine, no tyrosine phosphorylation was observed (lane 7). In addition, when serine 721 was replaced by alanine, tyrosine phosphorylation was unchanged, but serine phosphorylation of STAT4, assessed by its mobility shift, was significantly diminished (lane 10). As a control, we utilized another STAT4 mutant (S723A) in which the mutated serine does not reside in a proline-directed serine/threonine kinase consensus sequence. This mutant showed an identical phosphorylation and alteration of electrophoretic mobility compared with wild-type STAT4 (not shown).
To further establish the importance of tyrosine 693 and serine 721, we transfected a T-cell line, Jurkat T, with a STAT reporter construct (3xGAS-luc), IL-12R, and wild-type or mutated STAT4 constructs. As shown in Figure 1C, strong IL-12–induced transactivation of the reporter gene was detected when wild-type STAT4 was expressed but not in its absence. Interestingly, expression of the S721A mutant permitted reduced IL-12–inducibility of the reporter construct, whereas the S723A mutant had no effect on the functional activity of STAT4. As expected from the biochemical analysis, the Y693F mutant was completely unable to transactivate the reporter.
Thus, we conclude from these experiments that tyrosine 693 and serine 721 are important sites of phosphorylation in STAT4. Moreover, these results demonstrate that tyrosine and serine phosphorylation are required for maximal STAT4-mediated transcriptional activity in response to IL-12, and that serine 721 is likely to be the target of a proline-directed serine/threonine kinase.
IL-12 activates p38 but not ERKs or JNKs
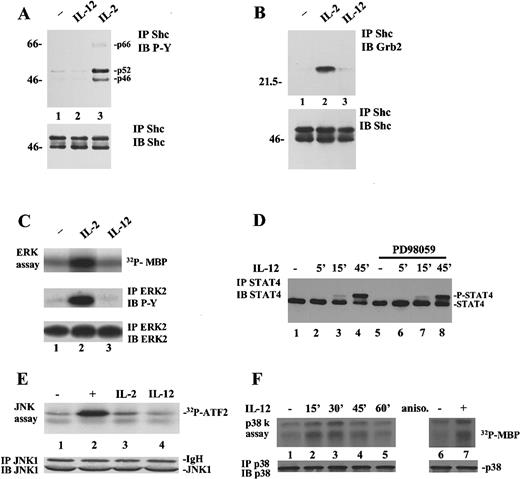
As STAT4 serine 721 resides within a putative MAPK phosphorylation site, we next investigated the possible contributions of ERKs, JNKs, and p38 in IL-12 signaling and STAT4 serine phosphorylation. It has been suggested that IL-12 induces ERK activity in T cells39; thus, we thought it was important to ascertain whether the ERKs were responsible for STAT4 serine 721 phosphorylation. Early events in the activation of the ERK pathway are the phosphorylation of the adapter molecule Shc, which, in turn, recruits Grb2 and the exchange factor SOS, thereby stimulating Ras activity.45 We first asked if IL-12 could induce tyrosine phosphorylation of Shc in human T cells. As shown in Figure2A, stimulation with IL-12 up to 1 hour resulted in no significant tyrosine phosphorylation of Shc (lane 2 and data not shown), whereas IL-2 was able to induce the phosphorylation of all the known isoforms of Shc (p46, p52, and p66) (lane 3).46 Consistent with the lack of Shc phosphorylation, Grb2 was not recruited into a complex with Shc after IL-12 stimulation of T cells (Figure 2B, lane 3), whereas strong Shc-Grb2 association was observed after treatment with IL-2 (lane 2). The same results were obtained using the NK3.3 cell line (not shown). In addition, we also directly assayed ERK activity on IL-12 stimulation of human T cells. As shown in Figure 2C, only IL-2, used as positive control,47was able to induce MBP phosphorylation (upper panel, lane 2), whereas IL-12 clearly failed to stimulate ERK activity (lane 3). Moreover, ERK2 immunoprecipitates were tyrosine phosphorylated in response to IL-2 but not IL-12 stimulation (Figure 2C, middle panel). Finally, consistent with the preceding results, a specific MEK inhibitor, PD98059,48 49 did not abrogate IL-12–induced STAT4 serine phosphorylation as measured by retardation in its electrophoretic gel migration (Figure 2D). As a control, PD98059 was able to inhibit IL-2–induced ERK phosphorylation, as assayed using anti-phosphoERK antibody (not shown).
IL-12 activates p38 but not ERKs or JNKs in T and NK3.3 cells.
(A), IL-2 but not IL-12 induces Shc phosphorylation. T cells were left unstimulated (lane 1) or were stimulated for 15 minutes with 10 ng/mL IL-12 (lane 2) or 2000 IU/mL IL-2 (lane 3). Lysates were immunoprecipitated with anti-Shc Ab, resolved by SDS/PAGE, transferred to membrane, and blotted with antiphosphotyrosine Ab (upper panel). The blot was stripped and reprobed with anti-Shc, demonstrating equal levels of proteins (lower panel). (B), IL-2 but not IL-12 induces Shc/Grb2 association. T cells were left unstimulated (lane 1) or were stimulated for 15 minutes with IL-2 (lane 2) or IL-12 (lane 3). Lysates were immunoprecipitated with anti-Shc Ab, resolved by SDS/PAGE, and blotted sequentially with anti-Grb2 (upper panel) and anti-Shc Ab (lower panel). (C), IL-2 but not IL-12 activates ERK2. T cells were left unstimulated (lane 1) or were stimulated for 15 minutes with IL-2 (lane 2) or IL-12 (lane 3). Lysates were immunoprecipitated with anti-ERK2 Ab and a kinase reaction was performed on the immunoprecipitates using MBP as a substrate (upper panel). Parallel samples were immunoprecipitated with anti-ERK2 and blotted with antiphosphotyrosine (middle panel) and anti-ERK2 Ab (lower panel). (D), A MEK inhibitor does not affect STAT4 serine phosphorylation. T cells were left unstimulated (lanes 1 and 5) or stimulated with IL-12 (lanes 2-4 and 6-8) for different periods. Cells in lanes 5 to 8 were pretreated with 100 μmol/L PD98059 for 30 minutes. Cell lysates were immunoprecipitated with anti-STAT4 Ab, resolved by SDS/PAGE, and blotted with anti-STAT4. (E), IL-12 does not activate JNK1. NK3.3 cells were left unstimulated (lane 1) or were stimulated with 10 μg/mL anisomycin (lane 2), IL-2 (lane 3), or IL-12 (lane 4). Lysates were immunoprecipitated with anti-JNK1 Ab. A kinase reaction was performed on the immunoprecipitates using GST-ATF2 as a substrate (upper panel). The filter was probed for JNK1 showing equal loading of the proteins (lower panel). (F), Activation of p38 by IL-12. NK3.3 cells were left unstimulated (lanes 1 and 6) or were stimulated with IL-12 for different periods (lanes 2-5) or with 10 μg/mL anisomycin for 15 minutes (lane 7). Lysates were immunoprecipitated with anti-p38 Ab. A kinase reaction was performed on the immunoprecipitates using MBP as a substrate (upper panel). The filter was probed for p38 showing equal loading of the proteins (lower panel).
IL-12 activates p38 but not ERKs or JNKs in T and NK3.3 cells.
(A), IL-2 but not IL-12 induces Shc phosphorylation. T cells were left unstimulated (lane 1) or were stimulated for 15 minutes with 10 ng/mL IL-12 (lane 2) or 2000 IU/mL IL-2 (lane 3). Lysates were immunoprecipitated with anti-Shc Ab, resolved by SDS/PAGE, transferred to membrane, and blotted with antiphosphotyrosine Ab (upper panel). The blot was stripped and reprobed with anti-Shc, demonstrating equal levels of proteins (lower panel). (B), IL-2 but not IL-12 induces Shc/Grb2 association. T cells were left unstimulated (lane 1) or were stimulated for 15 minutes with IL-2 (lane 2) or IL-12 (lane 3). Lysates were immunoprecipitated with anti-Shc Ab, resolved by SDS/PAGE, and blotted sequentially with anti-Grb2 (upper panel) and anti-Shc Ab (lower panel). (C), IL-2 but not IL-12 activates ERK2. T cells were left unstimulated (lane 1) or were stimulated for 15 minutes with IL-2 (lane 2) or IL-12 (lane 3). Lysates were immunoprecipitated with anti-ERK2 Ab and a kinase reaction was performed on the immunoprecipitates using MBP as a substrate (upper panel). Parallel samples were immunoprecipitated with anti-ERK2 and blotted with antiphosphotyrosine (middle panel) and anti-ERK2 Ab (lower panel). (D), A MEK inhibitor does not affect STAT4 serine phosphorylation. T cells were left unstimulated (lanes 1 and 5) or stimulated with IL-12 (lanes 2-4 and 6-8) for different periods. Cells in lanes 5 to 8 were pretreated with 100 μmol/L PD98059 for 30 minutes. Cell lysates were immunoprecipitated with anti-STAT4 Ab, resolved by SDS/PAGE, and blotted with anti-STAT4. (E), IL-12 does not activate JNK1. NK3.3 cells were left unstimulated (lane 1) or were stimulated with 10 μg/mL anisomycin (lane 2), IL-2 (lane 3), or IL-12 (lane 4). Lysates were immunoprecipitated with anti-JNK1 Ab. A kinase reaction was performed on the immunoprecipitates using GST-ATF2 as a substrate (upper panel). The filter was probed for JNK1 showing equal loading of the proteins (lower panel). (F), Activation of p38 by IL-12. NK3.3 cells were left unstimulated (lanes 1 and 6) or were stimulated with IL-12 for different periods (lanes 2-5) or with 10 μg/mL anisomycin for 15 minutes (lane 7). Lysates were immunoprecipitated with anti-p38 Ab. A kinase reaction was performed on the immunoprecipitates using MBP as a substrate (upper panel). The filter was probed for p38 showing equal loading of the proteins (lower panel).
We next tested the activity of another subfamily of MAPKs, the JNKs, in response to IL-12. As shown in Figure 2E, IL-12 did not induce JNK activity in NK3.3 cells (lane 4). Both anisomycin (lane 2) and a more physiologic stimulus, IL-2 (lane 3), used as positive controls, stimulated the activity of these kinases, even if, as expected,50 IL-2 did to a much lesser extent than anisomycin.
As IL-12 had no effect either on ERK or JNK activity, we next assayed p38 activation on IL-12 stimulation of NK3.3 cells. As shown in Figure2F, p38 was activated by 15 minutes of IL-12 stimulation (lane 2) at levels comparable with that induced by anisomycin, used as a positive control (lane 7). The activation was detected for up to 45 minutes (lane 4), returning at basal levels in 1 hour (lane 5).
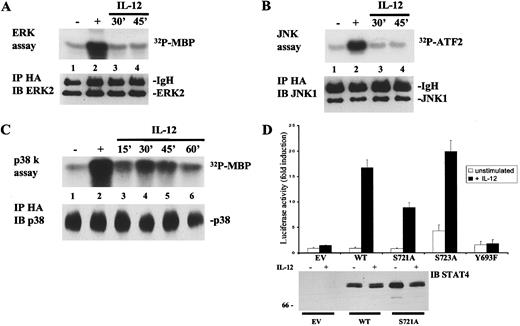
To further confirm the ability of IL-12 to activate p38 as opposed to ERKs and JNKs and to verify the utility of the cell line, we next evaluated the ability of IL-12 to activate transiently transfected, HA-tagged ERK2, JNK1, and p38α in NIH3T3 cells. An advantage of this approach was that the presence of the HA epitope allowed us to monitor the activity of the specific transfected MAPK. This avoided the possibility that the antibodies used to immunoprecipitate the endogenous MAPK were cross-reacting with close homologs of this family. As shown in Figure 3, the results obtained in NIH3T3 cells were perfectly comparable to those previously observed in T and NK3.3 cells. In fact, as shown in Figure 3, panels A through C, IL-12 was able to induce p38α, but not ERK2 or JNK1 activity in NIH3T3 cells, as we had found in T and NK3.3 cells (Figure2, panels C,E,F). Thus, the inability to couple to the ERKs appears to be an intrinsic propriety of the IL-12 receptor and not simply because of cell specific constituents. In addition, as shown in Figure 3D, in transfected NIH3T3 cells, IL-12 strongly induced transactivation of the STAT reporter construct when wild-type STAT4 was expressed. In agreement with the previous results in Jurkat T cells (Figure 1C), expression of the S721A mutant reduced IL-12 induction of the reporter, whereas mutation of serine 723 had no effect on STAT4-mediated transactivation. Mutation of tyrosine 693 completely abolished IL-12–induced STAT4 transcriptional activity. We confirmed that the various STAT4 constructs were equally expressed by blotting lysates from the transfected cells (lower panel). These data, demonstrating that NIH3T3 cells are a useful system to investigate IL-12 signaling pathways leading to STAT4 activation, allowed us to use this readily transfectable cell line for most of the experiments presented below.
IL-12 activates p38, but not ERKs or JNKs in non-lymphoid cells; serine 721 is important for IL-12-induced, STAT4-mediated transactivation in a heterologous system.
(A), IL-12 does not activate ERK2. NIH3T3 were transiently transfected with expression vectors encoding the human IL-12R subunits and pCDNA3-HA-ERK2. Cells were left unstimulated (lane 1) or were stimulated with serum (lane 2) or IL-12 for the times indicated (lanes 3 and 4). Lysates were immunoprecipitated with anti-HA Ab and a kinase reaction was performed on the immunoprecipitates using MBP as a substrate (upper panel). The filter was probed for ERK2 showing equal loading of the proteins (lower panel). (B), IL-12 does not activate JNK1. NIH3T3 were transiently transfected with expression vectors encoding the human IL-12R subunits and pCDNA3-HA-JNK1. Cells were left unstimulated (lane 1) or were stimulated with 10 μg/mL anisomycin (lane 2) or with IL-12 for the times indicated (lanes 3 and 4). Lysates were immunoprecipitated with anti-HA Ab. A kinase reaction was performed on the immunoprecipitates using GST-ATF2 as a substrate (upper panel). The filter was probed for JNK1 showing equal loading of the proteins (lower panel). (C), Activation of p38 by IL-12. NIH3T3 were transiently transfected with expression vectors encoding the human IL-12R subunits and pCEFL-HA-p38α. Cells were left unstimulated (lane 1) or were stimulated with 10 μg/mL anisomycin (lane 2) or with IL-12 for the times indicated (lanes 3-6). Lysates were immunoprecipitated with anti-HA Ab. A kinase reaction was performed on the immunoprecipitates using MBP as a substrate (upper panel). The filter was probed for p38 showing equal loading of the proteins (lower panel). (D), Importance of tyrosine 693 and serine 721 for IL-12–induced STAT4 transcriptional activity. NIH3T3 cells were transiently transfected with the p3xGAS-luc and the pCMV-β-galactosidase reporters together with expression vectors encoding the human IL-12R subunits. Where indicated, cells were cotransfected with control vector (EV), wild-type STAT4, or the mutated STAT4 constructs. The experiment was performed as described in Figure 1C. Unstimulated, ■; +IL-12, ▪. In the lower panel, 100 μg of the cell lysates, analyzed in the luciferase assay, were resolved by SDS/PAGE, transferred to membrane, and blotted with anti-STAT4 Ab.
IL-12 activates p38, but not ERKs or JNKs in non-lymphoid cells; serine 721 is important for IL-12-induced, STAT4-mediated transactivation in a heterologous system.
(A), IL-12 does not activate ERK2. NIH3T3 were transiently transfected with expression vectors encoding the human IL-12R subunits and pCDNA3-HA-ERK2. Cells were left unstimulated (lane 1) or were stimulated with serum (lane 2) or IL-12 for the times indicated (lanes 3 and 4). Lysates were immunoprecipitated with anti-HA Ab and a kinase reaction was performed on the immunoprecipitates using MBP as a substrate (upper panel). The filter was probed for ERK2 showing equal loading of the proteins (lower panel). (B), IL-12 does not activate JNK1. NIH3T3 were transiently transfected with expression vectors encoding the human IL-12R subunits and pCDNA3-HA-JNK1. Cells were left unstimulated (lane 1) or were stimulated with 10 μg/mL anisomycin (lane 2) or with IL-12 for the times indicated (lanes 3 and 4). Lysates were immunoprecipitated with anti-HA Ab. A kinase reaction was performed on the immunoprecipitates using GST-ATF2 as a substrate (upper panel). The filter was probed for JNK1 showing equal loading of the proteins (lower panel). (C), Activation of p38 by IL-12. NIH3T3 were transiently transfected with expression vectors encoding the human IL-12R subunits and pCEFL-HA-p38α. Cells were left unstimulated (lane 1) or were stimulated with 10 μg/mL anisomycin (lane 2) or with IL-12 for the times indicated (lanes 3-6). Lysates were immunoprecipitated with anti-HA Ab. A kinase reaction was performed on the immunoprecipitates using MBP as a substrate (upper panel). The filter was probed for p38 showing equal loading of the proteins (lower panel). (D), Importance of tyrosine 693 and serine 721 for IL-12–induced STAT4 transcriptional activity. NIH3T3 cells were transiently transfected with the p3xGAS-luc and the pCMV-β-galactosidase reporters together with expression vectors encoding the human IL-12R subunits. Where indicated, cells were cotransfected with control vector (EV), wild-type STAT4, or the mutated STAT4 constructs. The experiment was performed as described in Figure 1C. Unstimulated, ■; +IL-12, ▪. In the lower panel, 100 μg of the cell lysates, analyzed in the luciferase assay, were resolved by SDS/PAGE, transferred to membrane, and blotted with anti-STAT4 Ab.
Thus, our results indicated that ERKs and JNKs were not activated in response to IL-12, however, p38 was. Thus, this latter member of the MAPK family is likely to participate in the IL-12 signaling pathway and could be responsible for STAT4 serine 721 phosphorylation.
MKK6/p38α, but not other MAPKs, enhance STAT4 transcriptional activity
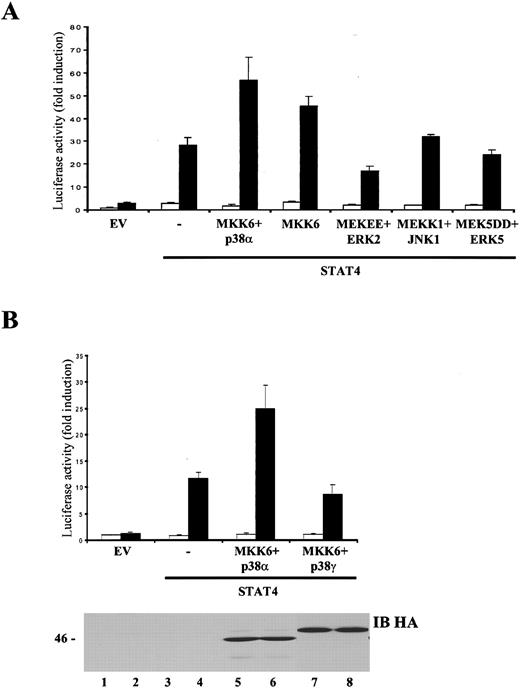
We next sought to establish the importance of the p38 signaling pathway in STAT4-mediated transcriptional activity. As shown in Figure 4A, on expression of p38α and its upstream activator, MKK6,51 IL-12 signaling was markedly enhanced, as STAT4-dependent transactivation of the 3xGAS-luc reporter was increased 20-fold compared with cells transfected with STAT4 alone. Interestingly, overexpression of MKK6 alone enhanced IL-12–mediated STAT4 transcriptional activity, without altering the basal level of luciferase activity. To confirm the specificity of this result, we next tested the other MAPK stimulators, namely, MEK1/2, MEKK1, and MEK5, which activate ERKs, JNKs, and the recently cloned MAPK ERK5, respectively. As shown in Figure 4A, a constitutively active form of MEK, MEKEE,40 failed to enhance STAT4 transactivation. Moreover, coexpression in NIH3T3 of a truncated JNK kinase, MEKK1, a potent activator of JNKs,52,53 and 2 different JNK isoforms, JNK1 or JNK2 (Figure 4A and data not shown), did not affect STAT4 transcriptional activity. In addition, coexpression of a constitutively active form of MEK5, MEK5DD, a strong stimulator of ERK5, had no effect on IL-12–induced STAT4 transcriptional activity. As a control, these kinases were tested for the ability to transactivate an Elk-1 (for MEKEE and MEKK1)- or a MEF2C (for MEK5DD)-dependent reporter construct43 and did so at the same concentration that failed to transactivate STAT4 (not shown). These data therefore argue that activation of the MKK6/p38α pathway is an important step in IL-12 signal trasduction, leading to STAT4-regulated gene expression.
p38α, but not other p38 isoforms or other kinases in the MAPK family, enhances STAT4 transcriptional activity.
(A), MKK6 and p38α selectively augment IL-12–dependent, STAT4-mediated transactivation. NIH3T3 cells were transiently transfected with the p3xGAS-luc and the pCMV-β-galactosidase reporters together with expression vectors encoding the human IL-12R subunits. Where indicated, cells were cotransfected with control vector (EV), wild-type STAT4, MKK6, p38α, MEKEE, ERK2, MEKK1, JNK1, MEK5DD, or ERK5. The experiment was performed as described in Figure 1C. Unstimulated, ■; +IL-12, ▪. (B), p38α, but not p38γ, enhances IL-12–dependent, STAT4-mediated transactivation. NIH3T3 cells were transiently transfected with the p3xGAS-luc and the pCMV-β-galactosidase reporters, together with expression vectors encoding the human IL-12R subunits. Where indicated, cells were cotransfected with control vector (EV), wild-type STAT4, MKK6, p38α, or p38γ. The experiment was performed as described in Figure 1C. In the lower panel, 100 μg of cell lysates, analyzed in the luciferase assay, were resolved by SDS/PAGE, transferred to membrane and blotted with anti-HA Ab to assure that the kinases were expressed at equal levels. Unstimulated, ■; +IL-12, ▪.
p38α, but not other p38 isoforms or other kinases in the MAPK family, enhances STAT4 transcriptional activity.
(A), MKK6 and p38α selectively augment IL-12–dependent, STAT4-mediated transactivation. NIH3T3 cells were transiently transfected with the p3xGAS-luc and the pCMV-β-galactosidase reporters together with expression vectors encoding the human IL-12R subunits. Where indicated, cells were cotransfected with control vector (EV), wild-type STAT4, MKK6, p38α, MEKEE, ERK2, MEKK1, JNK1, MEK5DD, or ERK5. The experiment was performed as described in Figure 1C. Unstimulated, ■; +IL-12, ▪. (B), p38α, but not p38γ, enhances IL-12–dependent, STAT4-mediated transactivation. NIH3T3 cells were transiently transfected with the p3xGAS-luc and the pCMV-β-galactosidase reporters, together with expression vectors encoding the human IL-12R subunits. Where indicated, cells were cotransfected with control vector (EV), wild-type STAT4, MKK6, p38α, or p38γ. The experiment was performed as described in Figure 1C. In the lower panel, 100 μg of cell lysates, analyzed in the luciferase assay, were resolved by SDS/PAGE, transferred to membrane and blotted with anti-HA Ab to assure that the kinases were expressed at equal levels. Unstimulated, ■; +IL-12, ▪.
Four mammalian p38 isoforms have been described: p38α, β, γ, and δ, the first 2 having the highest level of homology with each other. Although these different isoforms share both some upstream stimulators (such as MKK6) and downstream targets (ie, ATF2), recent reports have clearly indicated that each isoform can recognize a specific spectrum of substrates.54-58 To clarify which p38 family members may participate in IL-12 stimulation of STAT4 activity through an MKK6-dependent pathway, we cotransfected NIH3T3 with IL-12R subunits, MKK6, wild-type STAT4 and different HA-tagged p38 isoforms. Only the p38α isoform was able to enhance IL-12–dependent, STAT4-mediated transactivation (Figure 4B and data not shown). Comparable levels of expression of these kinases were confirmed by probing the lysates with an anti-HA antibody (Figure 4B, lower panel and data not shown).
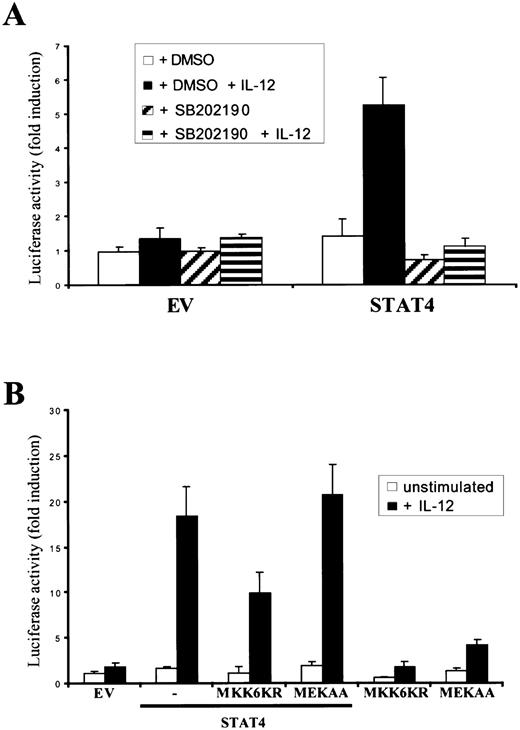
To further confirm that the MKK6/p38 pathway is responsible for enhancing STAT4 activity, we assessed the effect of a specific p38 inhibitor, SB202190. As shown in Figure5A, IL-12–induced, STAT4-mediated transactivation was completely abrogated on pretreatment of the cells with SB202190. As shown in Figure 5B, the expression of a dominant negative MKK6 construct, MKK6KR, was also able to significantly reduce IL-12–induced STAT4 transcriptional activity. In contrast, a dominant negative MEK construct, MEKAA, had no effect on STAT4 activity induced by IL-12, confirming the specific importance of MKK6 for STAT4 function on IL-12 stimulation.
A specific p38 inhibitor and a dominant negative MKK6 allele inhibit IL-12–induced STAT4 transcriptional activity.
(A), SB202190 inhibits IL-12–dependent, STAT4-mediated transactivation. NIH3T3 cells were transiently transfected with the p3xGAS-luc and the pCMV-β-galactosidase reporters, together with expression vectors encoding the human IL-12R subunits. Where indicated, cells were cotransfected with control vector (EV) or wild-type STAT4. As indicated, the cells were pretreated with DMSO or SB202190 for 1 hour before IL-12 stimulation. The experiment was performed as described in Figure 1C. (B), MKK6KR inhibits IL-12–dependent, STAT4-mediated transactivation. NIH3T3 cells were transiently transfected with 0.1 μg p3xGAS-luc reporter, along with 0.1 μg pCMV-β-galactosidase reporter, 0.1 μg pEFBOS-IL-12Rβ1, and 0.3 μg pEFBOS-IL-12Rβ2. Where indicated, cells were cotransfected with control vector (EV), 0.1 μg wild-type STAT4, 1.1 μg MKK6KR, or 0.5 μg MEKAA. The experiment was performed as described in Figure 1C.
A specific p38 inhibitor and a dominant negative MKK6 allele inhibit IL-12–induced STAT4 transcriptional activity.
(A), SB202190 inhibits IL-12–dependent, STAT4-mediated transactivation. NIH3T3 cells were transiently transfected with the p3xGAS-luc and the pCMV-β-galactosidase reporters, together with expression vectors encoding the human IL-12R subunits. Where indicated, cells were cotransfected with control vector (EV) or wild-type STAT4. As indicated, the cells were pretreated with DMSO or SB202190 for 1 hour before IL-12 stimulation. The experiment was performed as described in Figure 1C. (B), MKK6KR inhibits IL-12–dependent, STAT4-mediated transactivation. NIH3T3 cells were transiently transfected with 0.1 μg p3xGAS-luc reporter, along with 0.1 μg pCMV-β-galactosidase reporter, 0.1 μg pEFBOS-IL-12Rβ1, and 0.3 μg pEFBOS-IL-12Rβ2. Where indicated, cells were cotransfected with control vector (EV), 0.1 μg wild-type STAT4, 1.1 μg MKK6KR, or 0.5 μg MEKAA. The experiment was performed as described in Figure 1C.
Together, these results argue that MKK6/p38 are important elements that affect IL-12 activation of STAT4. That is, overexpression of p38α and MKK6 greatly enhances the IL-12–dependent STAT4 activation, expression of dominant negative MKK6 inhibits STAT4 signaling and the effect is specific as none of the other MAPK family members affected STAT4 transactivation.
Phosphorylation of STAT4 by p38
To confirm that p38α has the capacity to phosphorylate STAT4, we next tested whether STAT4 was an in vitro substrate of p38. We cloned the C-terminal portion of STAT4 in a vector for the expression of GST fusion protein in bacteria. The GST-STAT4 fusion protein was used as a substrate for an in vitro kinase assay on lysates from NK3.3 cells stimulated with IL-12. As shown in Figure 6A, IL-12–activated p38 was able to phosphorylate the STAT4-containing fusion protein (lane 3).
p38α phosphorylates STAT4 on serine 721.
(A), IL-12–activated p38 phosphorylates the C-terminus of STAT4 in vitro. NK3.3 cells were left unstimulated (lane 1) or were stimulated with 10 μg/mL anisomycin (lane 2) or with IL-12 (lane 3). Lysates were immunoprecipitated with anti-p38 Ab. A kinase reaction was performed on the immunoprecipitates using GST-STAT4 as a substrate (upper panel). The filter was probed for p38 showing equal loading of the proteins (lower panel). (B), Mutation of serine 721 blunts the enhancement seen by overexpression of p38α/ MKK6. NIH3T3 cells were transiently transfected with the p3xGAS-luc and the pCMV-β-galactosidase reporters, together with expression vectors encoding the human IL-12R subunits. Where indicated, cells were cotransfected with control vector (EV), wild-type STAT4, STAT4 S721A, MKK6, or p38α. The experiment was performed as described in Figure1C. Unstimulated, ■; +IL-12, ▪. (C), Overexpression of MKK6 and p38α is sufficient to mediate STAT4 serine 721 phosphorylation. 293T cells were transiently transfected with control vector (lane 1) or as indicated. Lysates were immunoprecipitated with anti-STAT4 Ab, resolved by SDS/PAGE, transferred to membrane and blotted with antiphosphoserine 727 STAT3 Ab that also detects serine phosphorylated STAT4 (upper panel). The blot was stripped and reprobed with anti-STAT4, demonstrating equal levels of proteins (lower panel).
p38α phosphorylates STAT4 on serine 721.
(A), IL-12–activated p38 phosphorylates the C-terminus of STAT4 in vitro. NK3.3 cells were left unstimulated (lane 1) or were stimulated with 10 μg/mL anisomycin (lane 2) or with IL-12 (lane 3). Lysates were immunoprecipitated with anti-p38 Ab. A kinase reaction was performed on the immunoprecipitates using GST-STAT4 as a substrate (upper panel). The filter was probed for p38 showing equal loading of the proteins (lower panel). (B), Mutation of serine 721 blunts the enhancement seen by overexpression of p38α/ MKK6. NIH3T3 cells were transiently transfected with the p3xGAS-luc and the pCMV-β-galactosidase reporters, together with expression vectors encoding the human IL-12R subunits. Where indicated, cells were cotransfected with control vector (EV), wild-type STAT4, STAT4 S721A, MKK6, or p38α. The experiment was performed as described in Figure1C. Unstimulated, ■; +IL-12, ▪. (C), Overexpression of MKK6 and p38α is sufficient to mediate STAT4 serine 721 phosphorylation. 293T cells were transiently transfected with control vector (lane 1) or as indicated. Lysates were immunoprecipitated with anti-STAT4 Ab, resolved by SDS/PAGE, transferred to membrane and blotted with antiphosphoserine 727 STAT3 Ab that also detects serine phosphorylated STAT4 (upper panel). The blot was stripped and reprobed with anti-STAT4, demonstrating equal levels of proteins (lower panel).
Given this result, we next tested whether serine 721 of STAT4 was a target for p38α in intact cells. We therefore transfected NIH3T3 cells with IL-12R subunits and wild-type STAT4 or the S721A mutant. As shown in Figure 6B, the enhanced activation seen with MKK6 and p38α overexpression was blunted when the S721A mutant was expressed, indicating that serine 721 contributes to the effects of p38α on STAT4 activity.
To confirm that p38 has the capacity to phosphorylate STAT4 on serine 721, we transfected cells with wild-type STAT4, alone or with MKK6 and p38α. STAT4 immunoprecipitates were immunoblotted with the antibody against phosphoserine 727 of STAT3 that cross-reacts with STAT4. As shown in Figure 6C, MKK6 alone (lane 3), or in combination with p38α (lane 4), readily induced STAT4 phosphorylation. In contrast, no STAT4 phosphorylation was detected on expression of the S721A mutant demonstrating the specificity of the antibody. Although IL-12 does not activate ERKs or JNKs, we cannot exclude the possibility that these MAPK family members may have the capacity to phosphorylate STAT4 when stimulated by other stimuli. Indeed, on overexpression in 293T cells, a constitutively active MEK construct did induce STAT4 serine 721 phosphorylation. Thus, it is possible that the ERKs may phosphorylate STAT4 under some circumstances.
Nonetheless, taken together, our data argue strongly that STAT4 appears an in vivo substrate for p38α serine kinase activity and that other MAPK family members are not relevant.
Discussion
In this report, we have shown that STAT4 is phosphorylated on tyrosine 693 and serine 721 in response to IL-12 stimulation. Moreover, we have demonstrated that phosphorylation of serine 721, a consensus site for MAPK-mediated phosphorylation, is required for full IL-12–induced STAT4 transcriptional activity. In addition, we have shown that p38, but not ERKs or JNKs, is activated in response to IL-12. Finally, we also provide evidence that the MKK6/p38α pathway mediates STAT4 serine 721 phosphorylation and is required for STAT4 activation.
It has been previously reported that IL-12 can induce tyrosine phosphorylation and activation of a 44 kd kinase in human T cells.39 In that report, Pignata et al39concluded that this protein was one of the ERK isoforms based on (1) the detected molecular weight, (2) the use of an anti-ERK antibody, and (3) the observation that this kinase recognizes MBP as a substrate. In contrast, here we show that IL-12 does not induce ERK tyrosine phosphorylation and activation. These seemingly disparate results could be in agreement. Since the Pignata study was published, many new members of the MAPK family, including p38, have been described. Most of them share characteristics very similar to ERKs: molecular weight in the 40 to 50 kd range, activation by tyrosine/threonine phosphorylation, and very high sequence homology. In addition, MBP has been recognized as a substrate for many MAPK family members, including p38α, p38γ, and ERK5.59-61 It is therefore reasonable to speculate that the kinase isolated by Pignata et al could have been p38α, and specific reagents were simply not available at the time of their report.
If IL-12 does not activate ERKs but does activate p38, the question arises, how? Unfortunately, the molecular mechanisms that might serve to couple IL-12R to MKK6 and p38 are still very unclear. Although many molecules have been identified that link cell surface receptors to ERKs45 and, to a lesser extent, to JNKs,62 the mechanism of activation of p38 is still poorly understood. It has been suggested that p38 activity might be regulated in T cells by kinases of the src family, through tyrosine phosphorylation of the Vav exchange factor and activation of the Rac small GTP-binding protein.63 In this regard, one such src family member, Lck, is induced on IL-12 stimulation of NK cells.64 Indeed, the possibility of Lck bridging the IL-12R to Vav/Rac and, in turn, p38 activation, as well additional mechanisms, will warrant further investigation.
We have shown that STAT4 serine phosphorylation peaks at 45 minutes after IL-12 stimulation (Cho et al16 and Figure 1A); on the other hand, p38 kinase activity begins at 15 minutes (Figure 2F). This discrepancy in kinetics is interesting and suggests, perhaps, that STAT4 and p38 both need to be translocated to the same intracellular compartment. It is unlikely that STAT4 serine phosphorylation is mediated by a de novo synthesized kinase induced by the p38 pathway, as pretreatment with cycloheximide does not affect STAT4 mobility shift (data not shown). Rather, it is appealing to speculate that STAT4 is phosphorylated by p38 in the nucleus in which both are described translocating after stimulation.11 25
The physiologic relevance of our data, showing that the MKK6/p38α pathway is necessary for IL-12–induced STAT4 transcriptional activity, is further supported by data obtained from transgenic mice expressing a dominant negative form of p38 in both peripheral T cells and thymocytes. Among these cells, the Th1 T-cell subset, the most specific cellular target for IL-12 activity, showed significant impairment in IFN-γ production.65 This defect has been attributed to the fact that the IFN-γ promoter contains 2 TREs66 that are considered a putative binding site for ATF2, which, in turn, is a p38 substrate. Nevertheless, it has been shown that STAT4 is able to induce expression of the IFN-γ gene.67 Our evidence that p38α activates STAT4 transcriptional potential allows us to postulate that, in mice carrying a p38 dominant negative, IL-12–dependent serine 721 phosphorylation of STAT4 and hence its transcriptional activity are impaired. This also suggests that p38-mediated serine 721 phosphorylation of STAT4 is physiologically required for production of IFN-γ by Th1 cells. The physiologic relevance of STAT4 serine 721 phosphorylation will warrant further investigation. However, an important caveat is that the function of STAT4 serine 721 for that matter, or of STATs serine phosphorylation in general, has not been finally established. Like our study, other studies have analyzed the function of STAT serine phosphorylation using reporter constructs. Exactly how important it is in physiologic gene regulation remains to be clarified.
Importantly, p38 may not be the sole MAPK family member that phosphorylates serine 721 in STAT4. It is entirely possible that the perfect consensus surrounding serine 721 in STAT4 may be recognized by other MAPKs. Nonetheless, we have proved that, among the different MAPKs, p38 is the only one activated by IL-12 and able to participate in IL-12–induced STAT4 transcriptional activity. It is therefore reasonable to speculate that other stimuli, such as IL-2, might activate other MAPKs, which in turn may induce STAT4 serine 721 phosphorylation and augment its transcriptional activity. In this scenario, STAT4 could represent a target for various signals, its final transcriptional response resulting from the integration of different biochemical pathways.
Despite a considerable effort of this and other laboratories in unveiling IL-12 signaling pathways, very little is known about the nature of molecules, other than JAKs and STATs, mediating its biologic functions. The finding that a specific MAPK, p38, participates to the signal transduction pathways elicited by IL-12 is therefore very important and is expected to shed new light on the complexity of the mechanisms by which IL-12 exerts its biologic responses.
In conclusion, our data suggests that MKK6 and p38 play an important role in regulating STAT4 serine phosphorylation in response to IL-12. Furthermore, our findings indicate that the optimal expression of the genes induced by IL-12, including IFN-γ, may depend on the activation of both JAKs and at least one member of the MAPK family, p38α.
Note added in proof. While our manuscript was under review, multiple studies, linking STAT serine phosphorylation to the activation of various MAPKs have been published. Significantly, they report apparently divergent results, probably because of the differences in the STAT proteins investigated and in the systems used.68-73 Thus, in agreement with our data, it has been reported that p38, activated in response to IFNs, is indispensable for STAT1 serine 727 phosphorylation and transcriptional activity.71 In contrast, other findings indicate that JNKs, but not p38, mediate STAT3 serine 727 phosphorylation in response to various stress treatments and that this event results in the inhibition of STAT3 activity.72 The most plausible hypothesis at the moment is that the effect of serine phosphorylation of STAT proteins depends on the cell type and on the class of serine kinases activated in response to different extracellular stimuli.
Acknowledgments
We thank U. Gubler, J. N. Ihle, J. Kornbluth, R. Pine, C. Reynolds for kindly providing reagents; M. J. Marinissen, A. Mazzoni, S. Pece, R. Sanchez-Prieto, M. Santoro for useful discussions and critical experimental advice.
R.V. and M.G. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Roberta Visconti, NIH-NIAMS, Building 10, Room 9N252, 10 Center Dr, MSC-1820, Bethesda, MD, 20892-1820; e-mail: viscontr@exchange.nih.gov.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal