Abstract
Congenital neutropenia and cyclic neutropenia are disorders of neutrophil production predisposing patients to recurrent bacterial infections. Recently the locus for autosomal dominant cyclic neutropenia was mapped to chromosome 19p13.3, and this disease is now attributable to mutations of the gene encoding neutrophil elastase (the ELA2 gene). The authors hypothesized that congenital neutropenia is also due to mutations of neutrophil elastase. Patients with congenital neutropenia, cyclic neutropenia, or Shwachman-Diamond syndrome were referred to the Severe Chronic Neutropenia International Registry. Referring physicians provided hematologic and clinical data. Mutational analysis was performed by sequencing polymerase chain reaction (PCR)-amplified genomic DNA for each of the 5 exons of the neutrophil ELA2 gene and 20 bases of the flanking regions. RNA from bone marrow mononuclear cells was used to determine if the affected patients expressed both the normal and the abnormal transcript. Twenty-two of 25 patients with congenital neutropenia had 18 different heterozygous mutations. Four of 4 patients with cyclic neutropenia and 0 of 3 patients with Shwachman-Diamond syndrome had mutations. For 5 patients with congenital neutropenia having mutations predicted to alter RNA splicing or transcript structure, reverse transcriptase-PCR showed expression of both normal and abnormal transcripts. In cyclic neutropenia, the mutations appeared to cluster near the active site of the molecule, whereas the opposite face was predominantly affected by the mutations found in congenital neutropenia. This study indicates that mutations of the gene encoding neutrophil elastase are probably the most common cause for severe congenital neutropenia as well as the cause for sporadic and autosomal dominant cyclic neutropenia.
Introduction
Cyclic and congenital neutropenia are severe disorders of neutrophil production that cause lifelong problems with recurrent infections. Cyclic neutropenia was first recognized as a distinct entity in 1910 because of the extremely regular recurrence of neutropenia, fever, and mouth ulcers in a 19-month-old boy.1 Its autosomal dominant pattern of inheritance was first suggested by de Berardinis and Reiman in 19492 and confirmed by Morley and colleagues3 in a study of 5 families in 1967. Characteristically in cyclic neutropenia, the blood neutrophil counts oscillate with a 21-day periodicity with extremely low circulating neutrophils at the nadir of the cycle and peak counts that are less than 2000/μL. The diagnosis of cyclic neutropenia depends on serial measurements of absolute neutrophil counts over a period of several weeks.4
Congenital neutropenia is a less well-defined entity and is generally regarded as a heterogeneous disorder. In 1954, Kostmann described autosomal recessive congenital neutropenia in a large Swedish family.5 Subsequently many other cases of severe congenital neutropenia have been described,6 usually sporadic cases as well as cases with autosomal dominant inheritance.7 Currently, the diagnosis of severe congenital neutropenia is made if the onset is recognized at birth or soon thereafter, blood neutrophil counts are less than 200/μL, the hematocrit or hemoglobin and platelet levels are normal or near normal, and the marrow shows a selective defect in neutrophil formation with many promyelocytes but relatively few myelocytes, metamyelocytes, and neutrophils or “promyelocytic maturation arrest.”
We recently reported positional cloning studies that map the locus for autosomal dominant cyclic neutropenia to chromosome 19p13.3, the locus for several serine proteases.8 Mutational analysis in 13 families and a sporadic case of cyclic neutropenia showed 7 different mutations of the gene for neutrophil elastase (ELA2), a serine protease synthesized chiefly at the promyelocytic stage in neutrophil development. The same mutations were found in several families and clustered around the active site of this enzyme. Because patients with congenital neutropenia have many hematologic features that are similar to patients with cyclic neutropenia, we hypothesized that they might result from mutations in the same gene. We found that most patients with congenital neutropenia also have neutrophil elastase mutations, results suggesting that mutations of the gene encoding neutrophil elastase cause both cyclic and severe congenital neutropenia.
Patients, materials, and methods
Patients
Patients were referred to the Severe Chronic Neutropenia International Registry (SCNIR). They or their parents or guardians gave consent for these studies under the auspices of the Human Subjects Committee of the University of Washington. Diagnoses were assigned at enrollment by established criteria of the SCNIR. The diagnosis of severe congenital neutropenia was based on at least 3 blood neutrophil counts less than 500/μL obtained at least 3 months after birth; a typical pattern of recurrent fevers, chronic gingivitis, and infections at irregular intervals; a bone marrow aspirate showing “maturation arrest” at the promyelocyte or myelocyte stage; and a normal cytogenetic analysis. The diagnosis of cyclic neutropenia required serial neutrophil counts for 3 to 6 weeks showing obvious oscillations at approximately 21-day intervals. The diagnosis of Shwachman-Diamond syndrome was made based on findings of steatorrhea, short stature, and pancytopenia in children with severe neutropenia. Patients with myelodysplasia, autoimmune disorders, or previous cytotoxic chemotherapy were excluded. Healthy individuals with normal blood counts served as controls.
Blood counts and DNA analyses
Routine blood cell counts were performed in laboratories of the patients' referring physicians. DNA was extracted from peripheral blood or bone marrow cells by standard techniques. Mutational analysis was performed by directly sequencing polymerase chain reaction (PCR)-amplified genomic DNA with an ABI/PE Biosystems PRISM Big Dye terminator chemistry on an ABI/PE Biosystems 310 Analyzer (Applied Biosystems, Foster City, CA). Each exon of neutrophil elastase and 20 bases of the flanking regions were sequenced from both directions in each individual.
Reverse transcriptase-PCR (RT-PCR)
Total RNA was isolated from fresh Ficoll-Hypaque–separated bone marrow mononuclear cells using a RNeasy kit (Qiagen, Valencia, CA). RT-PCR used Qiagen Omniscript RT with subsequent PCR and direct DNA sequencing. The primers amplified a 728-bp complementary DNA (cDNA) fragment between exons 2 and 5, which was subsequently digested with the restriction enzyme Taq I (New England Biolabs, Bethesda, MD) to produce a 411-bp fragment of interest in normal controls.
Statistical analysis
Differences between means were compared with the Studentt test using Prism Graph Pad Software (San Diego, CA).
Protein structure modeling
Coordinates of neutrophil elastase were obtained from the Protein Data Bank9 and mutations were made using the program of Jones and colleagues.10 Homology modeling was carried out with tools available in the suite of programs in the “Molecular Operating Environment” available from the Chemical Computing Group, Montreal, Quebec, (http://www.chemcomp.com) with the assistance of E. Adman at the University of Washington.
Results
Clinical characteristics
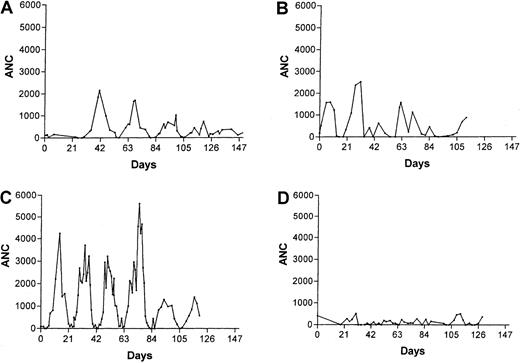
Hematologic data for 25 unrelated patients with congenital neutropenia are summarized in Table 1. The blood neutrophil counts were severely reduced (mean, 112 ± 21/μL) and monocyte counts increased (mean, 1489 ± 227/μL). Platelet counts were increased and hematocrit values mildly decreased in most patients. Marrow examinations showed scarce or absent neutrophils, bands, and metamyelocytes, with a typical pattern of “maturation arrest” of the neutrophil series; exceptions may be attributable to infection at the time of marrow sampling, which may transiently increase neutrophil production in these patients. In 3 patients with a diagnosis of congenital neutropenia who had long series of frequent blood counts (Table 1, patients 9, 20, and 22), there were periods with regular oscillations in blood neutrophil levels and other periods when oscillations were not apparent (Figure1).
Characteristics of patients with congenital neutropenia
| Subject . | Age (y) . | Gender . | ANC1-1/ mm1-3 . | AMC1-2/ mm1-3 . | Platelets1-3× 101-3/mm1-3 . | HCT1-4% . | Bone marrow evaluations1-5 . | Elastase mutations . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mblast % . | Promyelo % . | Myelo % . | Meta % . | B/Poly % . | Exon/intron . | Nucleotide1-6 . | Effect on protein1-7 . | |||||||
| 1 | 4 | M | 0 | 2943 | 558 | 36 | Severe maturation arrest | Exon 2 | G34010A | A28T | ||||
| 2 | 13 | M | 0 | 1891 | 348 | 32 | 2 | 5 | 23 | 7 | 2 | Exon 2 | T34020C | I31T |
| 3 | 15 | F | 106 | 1484 | 349 | 36 | 0 | 6 | 0 | 0 | 1 | Exon 2 | T34052A | C42S |
| 4 | 3 | F | 97 | 2928 | 547 | 30 | Arrest at myelocyte, very few mature neutrophils | Exon 3 | G34371A | V72M | ||||
| 5 | 8 | F | 126 | 1071 | 513 | 27 | 0 | 5 | 7 | 0 | 0 | Exon 3 | G34371A | V72M |
| 6 | 11 | M | 0 | 5372 | 576 | 29 | 0 | 7 | 1 | 0 | 1 | Exon 4 | C15823T | S97L |
| 7 | 3 | F | 79 | 2196 | 546 | 30 | 0 | 2 | 7 | 2 | 0 | Exon 4 | C15862T | P110L |
| 8 | 15 | F | 159 | 599 | 403 | 37 | 0 | 21 | 19 | 0 | 0 | Exon 4 | C15862T | P110L |
| 9 | 25 | M | 215 | 320 | 214 | 39 | 3 | 8 | 21 | 11 | 4 | Exon 4 | C15862T | P110L |
| 10 | 33 | F | 220 | 740 | 227 | 32 | 0 | 4 | 6 | 2 | 4 | Exon 4 | C15862T | P110L |
| 11 | 14 | M | 96 | 1088 | 320 | 25 | 3 | 11 | 2 | 0 | 0 | Exon 4 | ins5T16063 | delV161-F170 +2fs |
| 12 | 18 | F | 33 | 990 | 483 | 36 | 0 | 10 | 1 | 2 | 0 | Exon 4 | del15995-16018 | delV145-C152 |
| 13 | 4 | F | 0 | 3943 | 639 | 43 | Not done | Exon 5 | G16268T | G181V | ||||
| 14 | 10 | F | 0 | 1610 | 650 | 34 | 0 | 9 | 0 | 0 | 0 | Exon 5 | G16279A | G185R |
| 15 | 0.4 | M | 34 | 3041 | 272 | 39 | 3 | 1 | 1 | 0 | 1 | Exon 5 | G16300T | G192Stop |
| 16 | 4 | M | 318 | 504 | 534 | 35 | 1 | 10 | 26 | 15 | 6 | Exon 5 | C16313A | S196Stop |
| 17 | 3 | F | 200 | 4300 | 722 | 26 | Not done | Exon 5 | C16323A | Y199Stop | ||||
| 18 | 5 | M | 0 | 1797 | 345 | 34 | Not done | Exon 5 | delC16324 | Minus 1fs | ||||
| 19 | 26 | F | 58 | 1218 | 427 | 37 | 3 | 6 | 1 | 0 | 2 | Exon 5 | delC16340 | Minus 1fs |
| 20 | 21 | M | 272 | 318 | 418 | 37 | 1 | 4 | 16 | 1 | 1 | Intron 3 | C15805A | (ivs3 −8sa) insPQ94 |
| 21 | 1 | F | 0 | 1243 | 501 | 29 | 3 | 2 | 15 | 9 | 12 | Intron 4 | G16073A | (ivs4 +1sd)del V161-F170 |
| 22 | 33 | M | 124 | 990 | 235 | 45 | 6 | 6 | 4 | 0 | 0 | Intron 4 | G16077A | (ivs4 +5sd)del V161-F170 |
| 23 | 20 | M | 0 | 1449 | 413 | 34 | 3 | 9 | 13 | 3 | 0 | no mutation | ||
| 24 | 50 | M | 199 | 39 | 150 | 44 | 0 | 2 | 2 | 0 | 1 | no mutation | ||
| 25 | 14 | M | 330 | 465 | 281 | 37 | Not done | no mutation | ||||||
| Mean | 14 | 112 | 1489 | 415 | 35 | 1 | 7 | 9 | 3 | 2 | ||||
| SE | 2 | 21 | 227 | 31 | 1 | 0 | 1 | 2 | 1 | 1 | ||||
| 1-aNormal data | ||||||||||||||
| Mean | 3563 | 571 | 242 | 43 | 1 | 3 | 11 | 11 | 25 | |||||
| SE | 171 | 49 | 9 | 1 | 0 | 0 | 1 | 0 | 1 | |||||
| Subject . | Age (y) . | Gender . | ANC1-1/ mm1-3 . | AMC1-2/ mm1-3 . | Platelets1-3× 101-3/mm1-3 . | HCT1-4% . | Bone marrow evaluations1-5 . | Elastase mutations . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mblast % . | Promyelo % . | Myelo % . | Meta % . | B/Poly % . | Exon/intron . | Nucleotide1-6 . | Effect on protein1-7 . | |||||||
| 1 | 4 | M | 0 | 2943 | 558 | 36 | Severe maturation arrest | Exon 2 | G34010A | A28T | ||||
| 2 | 13 | M | 0 | 1891 | 348 | 32 | 2 | 5 | 23 | 7 | 2 | Exon 2 | T34020C | I31T |
| 3 | 15 | F | 106 | 1484 | 349 | 36 | 0 | 6 | 0 | 0 | 1 | Exon 2 | T34052A | C42S |
| 4 | 3 | F | 97 | 2928 | 547 | 30 | Arrest at myelocyte, very few mature neutrophils | Exon 3 | G34371A | V72M | ||||
| 5 | 8 | F | 126 | 1071 | 513 | 27 | 0 | 5 | 7 | 0 | 0 | Exon 3 | G34371A | V72M |
| 6 | 11 | M | 0 | 5372 | 576 | 29 | 0 | 7 | 1 | 0 | 1 | Exon 4 | C15823T | S97L |
| 7 | 3 | F | 79 | 2196 | 546 | 30 | 0 | 2 | 7 | 2 | 0 | Exon 4 | C15862T | P110L |
| 8 | 15 | F | 159 | 599 | 403 | 37 | 0 | 21 | 19 | 0 | 0 | Exon 4 | C15862T | P110L |
| 9 | 25 | M | 215 | 320 | 214 | 39 | 3 | 8 | 21 | 11 | 4 | Exon 4 | C15862T | P110L |
| 10 | 33 | F | 220 | 740 | 227 | 32 | 0 | 4 | 6 | 2 | 4 | Exon 4 | C15862T | P110L |
| 11 | 14 | M | 96 | 1088 | 320 | 25 | 3 | 11 | 2 | 0 | 0 | Exon 4 | ins5T16063 | delV161-F170 +2fs |
| 12 | 18 | F | 33 | 990 | 483 | 36 | 0 | 10 | 1 | 2 | 0 | Exon 4 | del15995-16018 | delV145-C152 |
| 13 | 4 | F | 0 | 3943 | 639 | 43 | Not done | Exon 5 | G16268T | G181V | ||||
| 14 | 10 | F | 0 | 1610 | 650 | 34 | 0 | 9 | 0 | 0 | 0 | Exon 5 | G16279A | G185R |
| 15 | 0.4 | M | 34 | 3041 | 272 | 39 | 3 | 1 | 1 | 0 | 1 | Exon 5 | G16300T | G192Stop |
| 16 | 4 | M | 318 | 504 | 534 | 35 | 1 | 10 | 26 | 15 | 6 | Exon 5 | C16313A | S196Stop |
| 17 | 3 | F | 200 | 4300 | 722 | 26 | Not done | Exon 5 | C16323A | Y199Stop | ||||
| 18 | 5 | M | 0 | 1797 | 345 | 34 | Not done | Exon 5 | delC16324 | Minus 1fs | ||||
| 19 | 26 | F | 58 | 1218 | 427 | 37 | 3 | 6 | 1 | 0 | 2 | Exon 5 | delC16340 | Minus 1fs |
| 20 | 21 | M | 272 | 318 | 418 | 37 | 1 | 4 | 16 | 1 | 1 | Intron 3 | C15805A | (ivs3 −8sa) insPQ94 |
| 21 | 1 | F | 0 | 1243 | 501 | 29 | 3 | 2 | 15 | 9 | 12 | Intron 4 | G16073A | (ivs4 +1sd)del V161-F170 |
| 22 | 33 | M | 124 | 990 | 235 | 45 | 6 | 6 | 4 | 0 | 0 | Intron 4 | G16077A | (ivs4 +5sd)del V161-F170 |
| 23 | 20 | M | 0 | 1449 | 413 | 34 | 3 | 9 | 13 | 3 | 0 | no mutation | ||
| 24 | 50 | M | 199 | 39 | 150 | 44 | 0 | 2 | 2 | 0 | 1 | no mutation | ||
| 25 | 14 | M | 330 | 465 | 281 | 37 | Not done | no mutation | ||||||
| Mean | 14 | 112 | 1489 | 415 | 35 | 1 | 7 | 9 | 3 | 2 | ||||
| SE | 2 | 21 | 227 | 31 | 1 | 0 | 1 | 2 | 1 | 1 | ||||
| 1-aNormal data | ||||||||||||||
| Mean | 3563 | 571 | 242 | 43 | 1 | 3 | 11 | 11 | 25 | |||||
| SE | 171 | 49 | 9 | 1 | 0 | 0 | 1 | 0 | 1 | |||||
Healthy volunteers = 39 subjects—21 males and 18 females.
ANC = Absolute neutrophil count. Measurement is the median of baseline counts.
AMC = Absolute monocyte count. Measurement is the median of baseline counts.
Measurement is the median of baseline counts.
Measurement is the median of baseline counts.
Mblast indicates myeloblast; Promyelo, promyelocyte; Myelo, Myelocyte; Meta, metamyelocyte; B/Polys, bands + polymorphonuclear cells.
Position of nucleotides for subjects 1-5 (mutations in exon 1-3) corresponds to GenBank AC004799 and for subjects 6-21 (mutations in intron 3-exon 5) to GenBank AC010648.
Numbering of splice site mutations is relative to the intron (negative)/exon (positive) boundary. Amino acid numbering begins from the first amino acid after the presignal peptide cleavage.
ins indicates insertion; del, deletion; ivs, intervening sequence; sa, splice acceptor; sd, splice donor; fs = frame shift.
Serial neutrophil counts for 3 patients with a diagnosis of congenital neutropenia, on no treatments, showing periods with regular oscillations of neutrophil counts.
Panels C and D are for the same patient observed for 2 prolonged periods 2 years apart.
Serial neutrophil counts for 3 patients with a diagnosis of congenital neutropenia, on no treatments, showing periods with regular oscillations of neutrophil counts.
Panels C and D are for the same patient observed for 2 prolonged periods 2 years apart.
Two groups of neutropenic patients were studied for comparison with the patients with congenital neutropenia (Table2). The 4 patients with cyclic neutropenia were sporadic cases with typical 21-day cycles of fever, mouth ulcers, and severe neutropenia. The 3 children with Shwachman-Diamond syndrome had neutropenia that was less severe than for the patients with congenital neutropenia.
Characteristics of patients with cyclic neutropenia and Shwachman-Diamond syndrome
| Subject . | Age (y) . | Gender . | ANC2-1/mm2-3 . | AMC2-2/mm2-3 . | Platelets2-3× 102-3/mm2-3 . | HCT2-4% . | Elastase mutations . | ||
|---|---|---|---|---|---|---|---|---|---|
| Exon/intron . | Nucleotide2-5 . | Effect on protein2-6 . | |||||||
| Cyclic neutropenia (sporadic cases) | |||||||||
| 1 | 23 | M | 246 | 928 | 330 | 41 | Intron 4 | G16073A | (ivs4 +1sd) delV161-F170 |
| 2 | 26 | M | 122 | 740 | 255 | 35 | Intron 4 | G16073A | (ivs4 +1sd) delV161-F170 |
| 3 | 4 | F | 178 | 816 | 416 | 33 | Exon 4 | C15862T | P110L |
| 4 | 10 | F | 109 | 53 | 421 | 34 | Intron 4 | G16073A | (ivs4 +1sd) del V161-F170 |
| Mean | 16 | 164 | 634 | 356 | 36 | ||||
| SE | 6 | 36 | 228 | 46 | 2 | ||||
| Shwachman-Diamond syndrome | |||||||||
| 1 | 4 | M | 201 | 206 | 191 | 22 | No mutation | ||
| 2 | 8 | F | 324 | 215 | 120 | 36 | No mutation | ||
| 3 | 4 | F | 452 | 339 | 299 | 32 | No mutation | ||
| Mean | 5 | 326 | 253 | 203 | 30 | ||||
| SE | 2 | 89 | 53 | 64 | 5 | ||||
| Subject . | Age (y) . | Gender . | ANC2-1/mm2-3 . | AMC2-2/mm2-3 . | Platelets2-3× 102-3/mm2-3 . | HCT2-4% . | Elastase mutations . | ||
|---|---|---|---|---|---|---|---|---|---|
| Exon/intron . | Nucleotide2-5 . | Effect on protein2-6 . | |||||||
| Cyclic neutropenia (sporadic cases) | |||||||||
| 1 | 23 | M | 246 | 928 | 330 | 41 | Intron 4 | G16073A | (ivs4 +1sd) delV161-F170 |
| 2 | 26 | M | 122 | 740 | 255 | 35 | Intron 4 | G16073A | (ivs4 +1sd) delV161-F170 |
| 3 | 4 | F | 178 | 816 | 416 | 33 | Exon 4 | C15862T | P110L |
| 4 | 10 | F | 109 | 53 | 421 | 34 | Intron 4 | G16073A | (ivs4 +1sd) del V161-F170 |
| Mean | 16 | 164 | 634 | 356 | 36 | ||||
| SE | 6 | 36 | 228 | 46 | 2 | ||||
| Shwachman-Diamond syndrome | |||||||||
| 1 | 4 | M | 201 | 206 | 191 | 22 | No mutation | ||
| 2 | 8 | F | 324 | 215 | 120 | 36 | No mutation | ||
| 3 | 4 | F | 452 | 339 | 299 | 32 | No mutation | ||
| Mean | 5 | 326 | 253 | 203 | 30 | ||||
| SE | 2 | 89 | 53 | 64 | 5 | ||||
ANC indicates absolute neutrophil count. Measurement is the median of baseline counts.
AMC indicates absolute monocyte count. Measurement is the median of baseline counts.
Measurement is the median of baseline counts.
Measurement is the median of baseline counts.
Positon of nucleotides for subjects 1-4 (mutations in intron 3-exon 5) corresponds to GenBank AC010648.
ns indicates insertion; del, deletion; ivs, intervening sequence; sa, splice acceptor; sd, splice donor; fs, frame shift.
Numbering is relative to the intron (negative)/exon (positive) boundary.
Amino acid numbering begins from the first amino acid after the presignal peptide cleavage.
Mutations of the neutrophil ELA2 gene
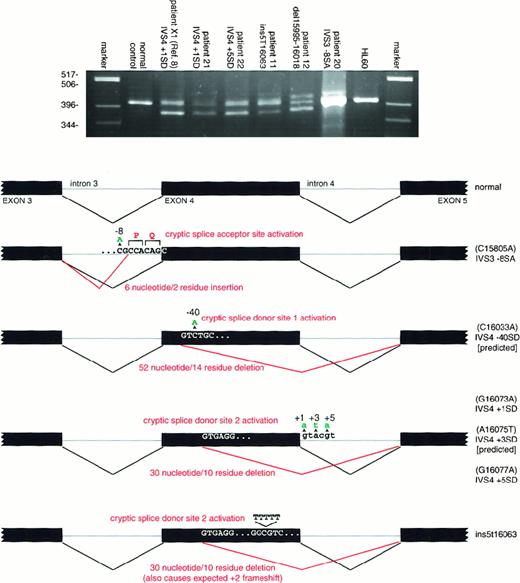
Twenty-two of 25 patients diagnosed as having congenital neutropenia were found to have neutrophil elastase mutations. Eighteen different heterozygous mutations were detected (Table 1). Four unrelated patients had the identical mutations in exon 4 (patients 7-10). Two other unrelated patients had the same mutation in exon 3 (patients 4 and 5). Five mutations occurred in families with 2 or more affected members—3 fathers and 3 daughters, (patients 5, 7, and 12), a mother and a son (patient 9), and a mother (patient 10) with 2 affected sons with different fathers, suggesting autosomal dominant inheritance. The identical mutation has been found in the affected parent and child (or children) for all 5 of these families. Hematologic comparisons of the relatively large group of patients with mutations (22 patients) with the smaller group without mutations (3 patients) showed that the neutrophil counts were somewhat lower and the monocyte counts higher in patients with the mutant neutrophilELA2 gene, but the differences were not statistically significant. Seven of these mutations were predicted to alter RNA splicing or transcript structure. RT-PCR analysis using cells from the 5 available marrow samples demonstrated the predicted effect in each of these individuals (Figure 2).
RT-PCR analysis of mutations affecting splicing and transcript structure.
Each aberrant splicing product appearing on the agarose gel was isolated and sequenced to confirm the interpretation shown in the schematic. The genomic DNA mutations are depicted in green, the resulting cDNA aberration in red. Because no bone marrow was available from patients with the IVS4 + 3SD and IVS4-40SD mutations, the result shown is the predicted result. Note that patient 11 has a 5T insertion and patient 12 a 24-bp deletion, neither of which would be expected to alter splicing. Nevertheless, the 5T insertion produces at least 2 transcripts, one containing the 5T insertion and another unanticipatedly forcing utilization of the 2nd cryptic upstream splice donor site. For patient 12, whereas only 2 transcripts were identified (the normal and one containing the 24-bp deletion) following subcloning and sequencing, uncharacterized higher molecular weight products that presumably represent misprocessed primary transcripts are apparent.
RT-PCR analysis of mutations affecting splicing and transcript structure.
Each aberrant splicing product appearing on the agarose gel was isolated and sequenced to confirm the interpretation shown in the schematic. The genomic DNA mutations are depicted in green, the resulting cDNA aberration in red. Because no bone marrow was available from patients with the IVS4 + 3SD and IVS4-40SD mutations, the result shown is the predicted result. Note that patient 11 has a 5T insertion and patient 12 a 24-bp deletion, neither of which would be expected to alter splicing. Nevertheless, the 5T insertion produces at least 2 transcripts, one containing the 5T insertion and another unanticipatedly forcing utilization of the 2nd cryptic upstream splice donor site. For patient 12, whereas only 2 transcripts were identified (the normal and one containing the 24-bp deletion) following subcloning and sequencing, uncharacterized higher molecular weight products that presumably represent misprocessed primary transcripts are apparent.
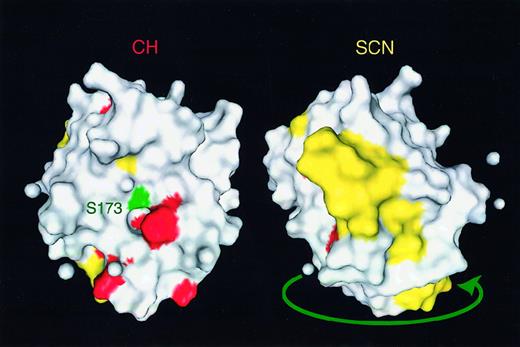
The diversity in the mutations in congenital neutropenia was substantially greater than in patients with cyclic neutropenia. The mutations in the 4 sporadic cases of cyclic neutropenia shown in Table2 and the 13 families with autosomal dominant cyclic neutropenia were predominantly in intron 4, creating splice donor mutations, or base substitutions near the junction of exon 4 and exon 5.8 One of the patients with congenital neutropenia (patient 22) with a mutation in intron 4 was at one time thought to have cyclic neutropenia based on a period of regularly recurring symptoms. The other child with a similar mutation (patient 21) was diagnosed in the first year of life as having congenital neutropenia without a long series of counts and then started on treatment. Based on examination of the tertiary structure of neutrophil elastase, the mutations in cyclic neutropenia appeared to cluster around the active site of the enzyme, whereas the mutations in congenital neutropenia were predominantly on the molecule's opposite face (Figure 3), suggesting a correlation between the genotype and the phenotype.
Mutations in severe congenital neutropenia.
Tertiary distribution of the severe congenital neutropenia (SCN) mutations (yellow) are compared to cyclic neutropenia/cyclic hematopoiesis (CH) mutations (red). Catalytic serine residue 173 is indicated in green near the active site pocket. The 2 figures are rotated 180° with respect to each other. The image was prepared from x-ray crystallographic coordinates taken from the Molecular Modeling DataBase (http://www.ncbi.nlm.nih.gov/Structure/MMDB/mmdb.html).
Mutations in severe congenital neutropenia.
Tertiary distribution of the severe congenital neutropenia (SCN) mutations (yellow) are compared to cyclic neutropenia/cyclic hematopoiesis (CH) mutations (red). Catalytic serine residue 173 is indicated in green near the active site pocket. The 2 figures are rotated 180° with respect to each other. The image was prepared from x-ray crystallographic coordinates taken from the Molecular Modeling DataBase (http://www.ncbi.nlm.nih.gov/Structure/MMDB/mmdb.html).
With the diversity of mutations in congenital neutropenia, there was no clear correlation of the sites of the mutations and the patient's blood neutrophil counts or clinical courses. Patients 2, 5, and 20 from the group with mutations (3 of 21, or 14%) have developed acute myeloid leukemia (AML), a recognized complication in about 10% of patients with congenital neutropenia.11 The neutropenic father of another patient (patient 12) has myelodysplasia. One of the 4 patients (25%) without a mutation (patient 24) has developed AML.
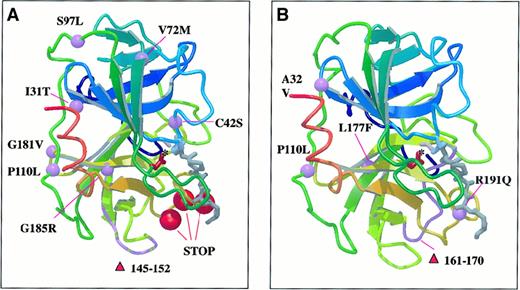
Schematic representation of neutrophil elastase structure with the positions of missense and deletion mutations is shown in Figure4. Preliminary structural analysis suggests that all of the substitutions led to conformational changes. Substitution of glycine 185 with positively charged arginine in the immediate proximity to the active site, the C42S missense mutation, and the deletion of 8 amino acid fragments (▿145-152) all probably alter biologic function. The 5 truncation and frame shift mutations in patients 15 through 19 remove the C-terminal portion of the molecule probably resulting in destabilization of the protein structure near the active site.
Ribbon diagram of neutrophil elastase with schematic representation of beta strands in complex with a peptide methoxysuccinyl-Ala-Ala-Pro-Ala chloromethyl ketone inhibitor.
Purple balls and chain fragment represent missense and deletion mutations. Red balls represent positions of truncation mutations in congenital neutropenia (A) and cyclic neutropenia (B). See text for explanations of effects of mutations noted. Drawing by E. Adman, University of Washington, using MOLSCRIPT (P. Kraulis. Molscript: a program to produce both detailed and schematic plots of protein structure. J Appl Crystallogr. 1991;24:946-950) and Raster3D (Merritt EA, Murphy MEP. Raster3D Version 2.0 a program for photorealistic molecular graphics. Acta Cyrstallogr. 1994;D50:869-873).
Ribbon diagram of neutrophil elastase with schematic representation of beta strands in complex with a peptide methoxysuccinyl-Ala-Ala-Pro-Ala chloromethyl ketone inhibitor.
Purple balls and chain fragment represent missense and deletion mutations. Red balls represent positions of truncation mutations in congenital neutropenia (A) and cyclic neutropenia (B). See text for explanations of effects of mutations noted. Drawing by E. Adman, University of Washington, using MOLSCRIPT (P. Kraulis. Molscript: a program to produce both detailed and schematic plots of protein structure. J Appl Crystallogr. 1991;24:946-950) and Raster3D (Merritt EA, Murphy MEP. Raster3D Version 2.0 a program for photorealistic molecular graphics. Acta Cyrstallogr. 1994;D50:869-873).
None of the patients with Shwachman-Diamond syndrome had mutations. Additionally, only one coding sequence change in the gene for neutrophil elastase was observed in 230 control chromosomes from normal individuals, and this change is presumed to represent a polymorphism, because of its appearance also in a family with cyclic neutropenia in which a mutation found in other pedigrees segregates with disease.
Discussion
This report describes the occurrence of mutations of the gene encoding neutrophil elastase in 22 patients with severe congenital neutropenia and extends our previous observation of mutations of this gene in cyclic neutropenia.8 Previous research has suggested that the pathophysiology of congenital neutropenia may relate to mutations of the granulocyte colony-stimulating factor (G-CSF) receptor, based on finding the mutation before any evidence suggests the patient has myelodysplasia or evolving AML.12 However, in almost all patients, G-CSF receptor mutations have been reported as acquired abnormalities detected in the process of evolution to AML.13 Current prevalence data suggest that a minority of patients manifest this mutation,14,15 and it now seems much more likely that mutations of the gene for neutrophil elastase lead to compromised myeloid differentiation and create the risk for development of AML. The familial cases reported here and the finding that heterozygous mutations are so consistently found in affected individuals suggest that both cyclic and most cases of congenital neutropenia have an autosomal dominant inheritance or are the result of a new dominant-acting germline mutation. The results also suggest that these are closely related disorders with overlapping molecular and clinical phenotypes. Visually and mathematically discernable oscillations in neutrophil counts are at times features of cases called “congenital neutropenia.”16 Similarly, affected family members with autosomal dominant cyclic neutropenia may be neutropenic without showing oscillations of their neutrophil counts.17Oscillations in both of these disorders are usually accentuated by treatment with G-CSF.18 19
The findings reported here suggest a primary role for mutations of neutrophil elastase in causing most, but not all, cases of severe congenital neutropenia. This protease is normally synthesized and packaged in promyelocytes at an early stage in neutrophil development.20 Neutrophil elastase has many recognized and possible substrates, including coagulation proteins, growth factors, and intracellular signaling molecules.21 It is normally synthesized in the developing neutrophil as a proenzyme but stored in the primary granules in its active form, ready with full enzymatic activity when released from the granules, normally at sites of inflammation.22,23 The pathogenic role of mutant elastase in causing neutropenia may be directly linked to the poor survival characteristics of early myeloid precursor cells in congenital and cyclic neutropenia.24 25 Accelerated apoptosis of promyelocytes and their progeny in vivo may explain the classic marrow finding of “promyelocytic arrest” in these conditions, but the specific protein-protein interactions causing these cellular abnormalities and causes for congenital neutropenia in patients without elastase mutations are yet to be discovered. The structural analysis presented here suggests that improper folding and destabilization of neutrophil elastase may be the critical effects of these mutations.
Oscillations in the hematopoietic system and the phenomenon of cyclic neutropenia have attracted the attention of clinicians and mathematicians for many years.26 Although numerous mechanisms have been proposed, the data in this report fit best with the hypothesis that oscillations of peripheral neutrophil counts will occur when there is reduced survival of early hematopoietic precursor cells and an intact feedback control system governing blood cell levels.27 This model assumes that neutrophil production is normally efficient, as indicated in careful quantitative studies.28 When precursor cell survival is poor, that is, the apoptotic rate is high, production of cells is very inefficient, and the output of cells is low. This state matches the high rate of apoptosis observed in severe congenital neutropenia and in patients with Shwachman-Diamond syndrome.29 Treatment with G-CSF can improve but does not completely reverse this defect in cell survival, probably through its antiapoptotic effect on myeloid cells.24,25 In cyclic neutropenia, the defect in cell survival is less severe, presumably because the mutant elastase less severely shortens precursor survival. In some individuals identified through family studies, either the cell survival may be better or the strength of the feedback loop weaker, resulting in no or substantially blunted oscillations. Because neither heterozygous nor homozygous neutrophil elastase knockout mice are neutropenic,30 it is quite likely the mutations in congenital and cyclic neutropenia lead to a gain of function or aberrant function of the enzyme.
Congenital and cyclic neutropenia are rare disorders, but the mechanisms governing the regulation and deployment of neutrophils are of broad importance. Treatment with G-CSF has greatly reduced the problems of recurrent and severe infections for patients with severe chronic neutropenia31; better understanding of the genetic and molecular mechanisms of these disorders should lead to even better therapies.
Acknowledgments
We are grateful for the participation of the many physicians worldwide who contribute data to the Severe Chronic Neutropenia International Registry. We appreciate the assistance of Tammy Cottle and staff at the Severe Chronic Neutropenia International Registry, Carol Fier at Amgen, Inc, and Elin Rodger for her technical assistance.
Supported by grants from Amgen, Inc, Thousand Oaks, CA; the National Institutes of Health (NIH) DK RO1 18951 and AI20065; the Markey Trust and the Doris Duke Charitable Foundation (T98006); the Leukemia Society of America; the American Cancer Society; the Leukemia Research Foundation; the National Leukemia Research Association; and UW Center grant number P30 ES07033 from the National Institute of Environmental Health Sciences, NIH.
Paid for in part by Amgen, Thousand Oaks, CA. D.C.D., M.A.B., L.A.B., G.K., and K.W. are consultants to Amgen, and D.C.D. and K.W. receive research support from Amgen. Amgen paid part or all of the salaries of R.E.P., A.A.B., A.G.A., and C.Z. while they were engaged in this study.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
David C. Dale, Severe Chronic Neutropenia International Registry, Department of Medicine, University of Washington School of Medicine, Box 356422, Seattle, WA, 98195-6422; e-mail: dcdale@u.washington.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal