Abstract
Epstein-Barr virus (EBV)–specific CD8 T lymphocytes are present at remarkably high frequencies in healthy EBV+individuals and provide protection from EBV-associated lymphoproliferative diseases. Allogeneic peripheral blood stem cell transplantation (allo-PBSCT) is a commonly used therapy in which T-cell surveillance for EBV is temporarily disrupted. Herein, human leukocyte antigen (HLA) class I tetramers were used to investigate the reestablishment of the EBV-specific CD8 T-cell repertoire in patients following allo-PBSCT. CD8+ T cells specific for lytic and latent cycle–derived EBV peptides rapidly repopulate the periphery of matched sibling allo-PBSCT patients. The relative frequencies of T cells specific for different EBV peptides in transplantation recipients closely reflect those of their respective donors. Investigation of patients at monthly intervals following unmanipulated allo-PBSCT demonstrated that the frequency of EBV-specific T cells correlates with the number of EBV genome copies in the peripheral blood and that expansion of EBV-specific T-cell populations occurs even in the setting of immunosuppressive therapy. In contrast, patients undergoing T-cell–depleted or unrelated cord blood transplantation have undetectable EBV-specific T cells, even in the presence of Epstein-Barr viremia. The protective shield provided by EBV-specific CD8 T cells is rapidly established following unmanipulated matched sibling allo-PBSCT and demonstrates that HLA class I tetramers complexed with viral peptides can provide direct and rapid assessment of pathogen-specific immunity in this and other vulnerable patient populations.
Introduction
Allogeneic peripheral blood stem cell transplantation (allo-PBSCT) is a lifesaving therapy for neoplastic and genetic hematologic diseases1 and is increasingly used to treat autoimmune diseases.2,3 Graft-versus-host disease (GVHD), an alloimmune attack on host tissues mediated by donor T cells, is a major complication of allo-PBSCT. Measures to limit or prevent GVHD include (1) pharmacologic immunosuppression with T-cell inhibitory drugs such as cyclosporin A or tacrolimus4,5; (2) in vivo depletion of T cells in the transplant recipient with antibodies that recognize the lymphocyte surface protein CD52 (CAMPATH-1) or other T-cell–specific surface proteins (antithymocyte globulin [ATG])6,7; and (3) ex vivo depletion of T lymphocytes from the allograft prior to infusion into the transplant recipient. Although these methods can be effective at modulating or ameliorating GVHD, they are associated with the second major complication of allografting, a heightened susceptibility to infection. Epstein-Barr virus (EBV) is one among a panoply of pathogens that can cause severe complications in immunosuppressed patients.8
EBV causes persistent infection of B lymphocytes in greater than 90% of healthy adults.9 Infection of human B lymphocytes with EBV results in their transformation, and EBV-specific T cells play a central role in preventing their uncontrolled growth. Compromise of cell-mediated immunity results in a spectrum of lymphoproliferative diseases ranging from polyclonal lymphoid hyperplasia to rapidly fatal malignant monoclonal lymphomas.10 These diseases are found in people with acquired and congenital immunodeficiencies and in solid organ or bone marrow transplantation patients. Major histocompatability complex (MHC) class I molecules on the surface of EBV-transformed B lymphocytes present virally derived peptides to CD8+cytotoxic T lymphocytes (CTLs), a process that appears to be vital for the control of latent EBV infection in healthy EBV+individuals.9 Recipients of T-cell–depleted stem cell or solid organ allografts or those who require immunosuppression to treat GVHD are at risk for EBV-associated posttransplantation lymphoproliferative disease (PTLD).8
Evidence of the role of CD8+ CTLs in controlling viral infections following allogeneic bone marrow transplantation (allo-BMT) comes from adoptive transfer studies. Riddell and colleagues11 generated cytomegalovirus (CMV)-specific CTL clones and infused them into allo-BMT recipients. Transferred T cells persisted for several months, and none of the recipients developed fatal CMV disease. Papadopoulos et al12 transferred unmanipulated donor lymphocytes into T-cell–depleted allo-BMT recipients and eradicated PTLD. Along similar lines, Rooney et al13 transferred EBV-specific polyclonal T cell lines into patients, which lead to regression of PTLD.
Recently, MHC class I tetramers have been developed as reagents to identify antigen-specific T lymphocytes. Tetramers consist of 4 identical MHC class I peptide complexes that are bound to a fluorochrome-conjugated streptavidin molecule. Our ability to measure and characterize virus- and bacteria-specific T-cell populations during the course of acute and chronic infections has been greatly advanced by this new technology.14-18 Quantitative studies using MHC class I tetramers complexed with peptides derived from latent and lytic cycle EBV proteins have demonstrated that primary EBV infection is accompanied by a massive expansion of virus-specific CD8 T-cell populations, with up to 40% of CD8+ T cells responding to one epitope.19 Remarkably, in healthy EBV+individuals, up to 5% of CD8 T cells are specific for EBV even years following primary infection.20 The impact of immunosuppressive therapy on EBV-specific T-cell populations and the kinetics of EBV-specific T-lymphocyte reconstitution in patients undergoing allo-PBSCT has not been determined with this methodology.
In this report we investigate PBSCT recipients, measuring the redevelopment of the EBV-specific T-cell repertoire within the CD8 T-cell compartment. Using a broad panel of human leukocyte antigen (HLA)–A2, HLA-B8, and HLA-B7 tetramers complexed with multiple latent and lytic EBV peptides, we show that EBV-specific T cells form a substantial percentage of CD8 T cells within one month of transplantation. The frequency and absolute number of T cells that are specific for lytic EBV peptides correlate directly with the in vivo viral load, even in the face of aggressive immunosuppression. Our studies demonstrate the feasibility of monitoring EBV-specific CTLs in allo-PBSCT recipients and suggest that tetramer analysis may enable rapid and direct identification of individuals at risk of PTLD.
Patients, materials, and methods
Human subjects
Healthy subjects were typed at the A, B, and C major histocompatibility antigen (HLA) loci and serologically tested for infection with EBV. We isolated peripheral blood mononuclear cells (PBMCs) from EBV+ donors for direct flow cytometric analysis and the generation of cell lines. Blood for isolation of PBMCs from individuals undergoing allo-PBSCT and renal transplantation was obtained at various time intervals following transplantation. All of these subjects signed an informed consent form in accordance with the Yale University School of Medicine Institutional Review Board guidelines. Additionally, blood was obtained from patients undergoing allogeneic matched unrelated donor (MUD) or umbilical cord blood (UCB) transplantation at various time intervals. All of these subjects signed an informed consent form in accordance with the University of Minnesota School of Medicine Institutional Review Board guidelines.
Patient conditioning
The details of conditioning for recipients of stem cell or umbilical cord allografts are listed in Table1. The solid organ allograft recipients were all treated with antithymocyte globulin induction, anti-CD25 monoclonal antibody (mAb) induction, and a prednisone taper per institutional protocol. In addition, cyclosporin A or tacrolimus was started at the time of transplantation, and mycophenolate was given to high-risk patients or to patients who had an episode of acute rejection.
Stem cell allograft recipient characteristics
| Patient . | HLA type . | Age/sex . | Diagnosis . | Conditioning . | |
|---|---|---|---|---|---|
| Donor . | Recipient . | ||||
| A | A3,32; B8,35 | Same | 50/M | NHL | BEAM |
| B | A1,2; B44,51 | Same | 32/F | NHL | TBI/CY |
| C | A1,2; B8,44 | Same | 31/M | NKL | TBI/CY |
| D | A2,11; B27,52 | Same | 29/F | AML | TBI/CY |
| E | A2,28; B37,44 | Same | 38/F | NHL | BU/CY |
| F | A11,23; B49,— | A2,23; B49,60 | 49/M | ALL | TBI/CY/ATG/TCD |
| M-1 | A2,74; B35,70 | A2,30; B35,70 | 47/F | CML | TBI/CY/ATG |
| M-2 | A3,—; B60,62 | A3,—; B7,40 | 9/F | ALL | TBI/CY/ATG |
| M-3 | A2,—; B56,57 | A2,—; B18,57 | 49/M | MDS | TBI/CY/ATG |
| M-4 | A24,—; B7,56 | A24,—; B7,56 | 52/M | AML | TBI/CY |
| M-5 | A1,2; B15,— | A2,32; B15,— | 19/F | ALL | TBI/CY/ATG |
| Patient . | HLA type . | Age/sex . | Diagnosis . | Conditioning . | |
|---|---|---|---|---|---|
| Donor . | Recipient . | ||||
| A | A3,32; B8,35 | Same | 50/M | NHL | BEAM |
| B | A1,2; B44,51 | Same | 32/F | NHL | TBI/CY |
| C | A1,2; B8,44 | Same | 31/M | NKL | TBI/CY |
| D | A2,11; B27,52 | Same | 29/F | AML | TBI/CY |
| E | A2,28; B37,44 | Same | 38/F | NHL | BU/CY |
| F | A11,23; B49,— | A2,23; B49,60 | 49/M | ALL | TBI/CY/ATG/TCD |
| M-1 | A2,74; B35,70 | A2,30; B35,70 | 47/F | CML | TBI/CY/ATG |
| M-2 | A3,—; B60,62 | A3,—; B7,40 | 9/F | ALL | TBI/CY/ATG |
| M-3 | A2,—; B56,57 | A2,—; B18,57 | 49/M | MDS | TBI/CY/ATG |
| M-4 | A24,—; B7,56 | A24,—; B7,56 | 52/M | AML | TBI/CY |
| M-5 | A1,2; B15,— | A2,32; B15,— | 19/F | ALL | TBI/CY/ATG |
M indicates male; F, female; NHL, non-Hodgkin lymphoma; AML, acute myeloid leukemia; NKL, natural killer cell lymphoma; —, no other allele detected; CML, chronic myeloid leukemia; ALL, acute lymphoblastic leukemia; MDS, myelodysplastic syndrome; BEAM, BCNU, etoposide, ara-C, and melphalan; TBI, total body irradiation; CY, cytoxan; ATG, antithymocyte globulin; and TCD, T-cell depletion.
Cell lines
Lymphoblastoid cell lines (B-LCLs) were established from healthy donors AD, EA, EP, and KK. PBMCs were isolated from heparinized blood on lymphocyte separation medium (ICN Biomedicals, Aurora, OH), and EBV-transformed B-LCLs were generated by incubation with the B95-8 EBV strain (Dr George Miller, Yale University) in the presence of 1 nmol/L tacrolimus. B-LCLs were maintained in Roswell Park Memorial Institute (RPMI) media supplemented with 10% fetal calf serum (FCS). EBV-specific T cell lines were generated by stimulating 1-2.5 × 107 Ficoll-purified PBMCs with 5-7.5 × 106 irradiated autologous B-LCLs. T cell lines were maintained by weekly restimulation with autologous B-LCLs. After the second restimulation T-cell cultures were supplemented with 5% human phytohemagglutinin (PHA) T stim (Becton Dickinson, San Jose, CA).
Generation of HLA-A2, HLA-B7, and HLA-B8 tetramers
Tetrameric HLA-A2, HLA-B7, and HLA-B8 complexes were generated with human β2-microglobulin and EBV peptides using previously described methods. Materials used included the plasmid construct expressing HLA-A2 with a biotinylation signal (Coulter, Miami, FL), the construct for HLA-B8 (John Altman, Emory University, Atlanta, GA), and complementary DNA (cDNA) for HLA-B7 (Peter Parham, Stanford University, Palo Alto, CA). The cDNA was used as a template for polymerase chain reaction (PCR) mutagenesis, which eliminated the signal sequence and replaced the transmembrane and cytoplasmic regions with a biotinylation sequence, as previously described,21 22 using the following primers: 5′-CATGCCATGGATACCATTCCATGAGGTATTTCTACACC-3′ and 5′-CTAGCTAGCGGACTGGGAAGACGGCTCCCA-3′. The PCR product was cloned into the PET 21d vector (Novagen, Madison, WI), and recombinant proteins were generated as insoluble proteins following induction with IPTG inEscherichia coli strain BL21 (DE3) LysS and purified as previously described. Insoluble HLA-A2, HLA-B7, and HLA-B8 and human β2-microglobulin were dissolved in 8 mol/L urea and refolded in the presence of 25 μg/mL of the appropriate peptide with the following protease inhibitors: 1 μg/mL pepstatin A, 1 μg/mL leupeptin, and 0.4 mmol/L phenylmethylsulfonyl fluoride (PMSF). The following peptides (Research Genetics, Huntsville, AL) were used in the generation of tetramers: GLCTLVAML, CLGGLLTMV, and LLDFVRFMGV (HLA-A2–restricted epitopes); RPPIFIRRL and VPAPAGPIV (HLA-B7–restricted epitopes); and RAKFKQLL, QAKWRLQTL, and FLRGRAYGL (HLA-B8–restricted epitopes). Soluble monomeric complexes were purified by gel filtration over a Superdex 200HR column (Amersham Pharmacia Biotech). Purified monomeric complexes were biotinylated at room temperature for 12 hours in the presence of 15 μg Bir A enzyme (Avidity, Boulder, CO), 80 μmol/L biotin, 10 mmol/L adenosine 5′-triphosphate (ATP), 10 mmol/L MgOAc, and 20 mmol/L bicine; purified by gel filtration to remove excess biotin; and then tetramerized with phycoerythrin (PE)-conjugated streptavidin (Molecular Probes, Sunnyvale, CA) at a 4:1 molar ratio. Finally, tetramers were purified by gel filtration and stored at 4°C in phosphate-buffered saline (PBS) (pH 8.0) with 0.02% sodium azide, 1 μmol pepstatin, 1 μg/mL leupeptin, and 0.5 mmol/L ethylenediamine tetraacetic acid (EDTA).
Cell staining
Fresh PBMCs or cell lines generated from healthy individuals, stem cell allograft recipients, or renal allograft recipients were incubated on ice for 1 hour in staining buffer (comprising PBS with 0.5% bovine serum albumin [BSA] and 0.02% sodium azide [pH 7.45]) containing (1) 0.25-0.5 mg/mL PE-labeled tetrameric complex; (2) saturating amounts of anti-CD3 and anti-CD8 mAbs conjugated to APC (Immunotech International, Marseilles, France) or Cychrome (PharMingen, San Diego, CA); and (3) either anti-CD44, anti-CD45RA, or anti-CD62L–FITC (fluorescein isothiocyanate) (Immunotech). The stained cells were fixed in 1% paraformaldehyde and analyzed by a fluorescence-activated cell sorter (FACS) using CellQuest software (Becton Dickinson). Lymphocytes were gated by forward and side angle light scatter.
EBV quantitative PCR
Blood was drawn into EDTA-containing tubes and processed on the same day. Mononuclear cells were purified on lymphocyte separation medium and the cells were treated with 60 μg/mL proteinase K (Roche Molecular Biochemicals, Indianapolis, IN). DNA from aliquots of 100 000 cells was amplified using primers 1100 and 1181 (Operon, Alameda, CA) for the EBV BMLF1 gene.23The 50 μL PCR reaction components (Roche Molecular Biochemicals) included 10 times PCR buffer; 1.5 mmol/L MgCl2; 0.025 μmol/L each of adenosine, cytadine, guanosine, and uridine 5′-triphosphate (dATP, dCTP, dGTP, and dUTP, respectively); and 1.0 U Taq DNA polymerase. The EBV-containing Raji cell line (50 EBV genomes per cell) was used to develop a standard curve. Thermal cycle parameters were completed for 35 cycles at 94°C for 30 seconds, 55°C for 30 seconds, and 72°C for 1 minute in a Perkin-Elmer 9600 thermal cycler (Perkin Elmer). The PCR product was hybridized to a BMLF 1 probe (10 pm) labeled with Tris (tris[hydroxymethyl] aminomethane; 2, 2′-bipyridine) rutherium II chelate (TBR) electrochemiluminescent label (Baron Biotech, Milford, CT).24 The hybridized PCR product was quantified using a Perkin Elmer QPCR 5000. All patient samples were amplified using β-globin primers to test for DNA integrity.
Results
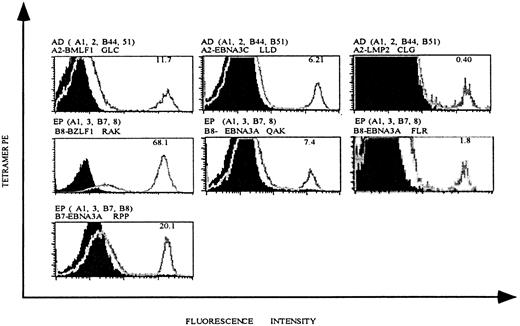
As a first step toward investigating EBV-specific T-cell populations in patients undergoing allo-PBSCT, we generated an HLA tetramer panel with 3 different HLA class I molecules, each complexed with multiple antigenic EBV-derived peptides (Table2). In addition to HLA-A2 and HLA-B8 tetramers, which have been used previously to measure EBV-specific T-cell populations in patients during and following acute EBV infection, our panel also included HLA-B7 tetramers complexed with 2 EBV peptides and HLA-A2 and HLA-B8 tetramers complexed with additional antigenic EBV peptides. To determine the specificity of our tetramer panel, we generated EBV-specific T cell lines from healthy EBV+ individuals by in vitro restimulation of fresh PBMCs with autologous B-LCLs. T cell lines were stained with mAbs specific for CD8 and CD62L and with HLA tetramers complexed with EBV peptides (Figure 1). Staining of a T cell line derived from individual AD (who expresses HLA-A2) demonstrates that 11.7% of CD8-gated T lymphocytes bound HLA-A2 tetramers complexed with a peptide derived from the lytic BMLF1 protein (Figure 1, upper left panel), and 6.21% and 0.4% of CD8 T cells bound HLA-A2 tetramers complexed with peptides derived from the latent EBNA3C and LMP2 proteins (Figure 1, upper middle and right panels).
EBV tetramers with HLA restriction
| Tetramer . | EBV epitope . | Epitope AA sequence . |
|---|---|---|
| A2-GLC-lytic | BMLF1 280-288 | GLCTLVAML |
| A2-LLD-latent | EBNA3C 284-293 | LLDFVRFMGV |
| A2-CLG-latent | LMP2 426-434 | CLGGLLTMV |
| B7-RPP-latent | EBNA3A 379-387 | RPPIFIRRL |
| B7-VPA-latent | EBNA3A 502-510 | VPAPAGPIV |
| B8-RAK-lytic | BZLF1 190-197 | RAKFKQLL |
| B8-QAK-latent | EBNA3A 158-166 | QAKWRLQTL |
| B8-FLR-latent | EBNA3A 325-333 | FLRGRAYGL |
| Tetramer . | EBV epitope . | Epitope AA sequence . |
|---|---|---|
| A2-GLC-lytic | BMLF1 280-288 | GLCTLVAML |
| A2-LLD-latent | EBNA3C 284-293 | LLDFVRFMGV |
| A2-CLG-latent | LMP2 426-434 | CLGGLLTMV |
| B7-RPP-latent | EBNA3A 379-387 | RPPIFIRRL |
| B7-VPA-latent | EBNA3A 502-510 | VPAPAGPIV |
| B8-RAK-lytic | BZLF1 190-197 | RAKFKQLL |
| B8-QAK-latent | EBNA3A 158-166 | QAKWRLQTL |
| B8-FLR-latent | EBNA3A 325-333 | FLRGRAYGL |
HLA tetramers complexed with EBV epitopes stain EBV-specific CD8 T cells.
EBV-specific T cell lines were generated from EBV+individuals AD and EP by in vitro restimulation with autologous B-LCLs, as described in “Materials and methods.” HLA-A andHLA-B alleles expressed in these individuals and the HLA tetramer used for staining are indicated above each histogram. Each plot is gated on live, CD8+ T lymphocytes, and the percentage of tetramer-positive T cells is indicated in the right upper corner of each histogram.
HLA tetramers complexed with EBV epitopes stain EBV-specific CD8 T cells.
EBV-specific T cell lines were generated from EBV+individuals AD and EP by in vitro restimulation with autologous B-LCLs, as described in “Materials and methods.” HLA-A andHLA-B alleles expressed in these individuals and the HLA tetramer used for staining are indicated above each histogram. Each plot is gated on live, CD8+ T lymphocytes, and the percentage of tetramer-positive T cells is indicated in the right upper corner of each histogram.
T cell lines generated from HLA-A2− individuals did not stain with HLA-A2 tetramers, confirming the specificity of these reagents (data not shown). Similar experiments were performed with a T cell line generated from individual EP, who expresses both HLA-B8 and HLA-B7. HLA-B8 tetramers complexed with peptide derived from the lytic protein BZLF1 or 2 different peptides derived from the latent protein EBNA3A labeled 68.1%, 7.4%, and 1.8% of CD8 T cells respectively, and HLA-B7 tetramers complexed with peptide-derived EBNA3A labeled 20.1% of CD8 T cells (Figure 1). As noted above for the HLA-A2 tetramers, cell lines derived from HLA-B8− or HLA-B7− individuals did not stain with these HLA-B8 or HLA-B7 tetramers (results not shown).
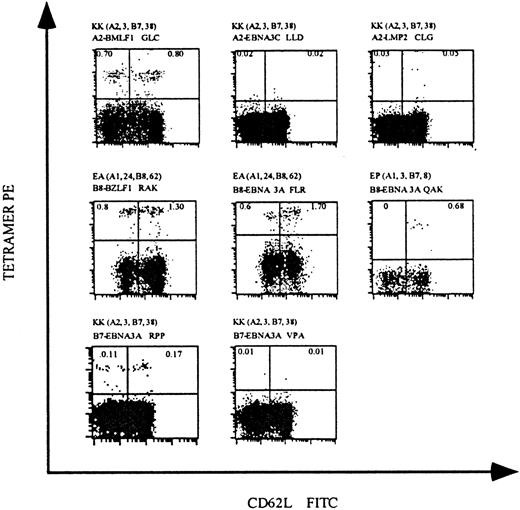
To confirm the feasibility of tetramer staining for the direct analysis of EBV-specific T cells in peripheral blood, we isolated PBMCs from 4 healthy EBV+ individuals and stained cells with our panel of MHC class I tetramers. In individual KK, who expresses HLA-A2, approximately 1.5% of the CD8 T cells stained with HLA-A2 tetramers complexed with the lytic protein BMLF1 peptide; tetramers complexed with the latent protein EBNA3C and LMP2 peptides did not stain detectable CD8 T-cell populations (Figure2, upper panels). Individuals EA and EP, who express HLA-B8, had readily detectable CD8 T-cell populations that stained HLA-B8 tetramers complexed with the lytic protein BZLF1 peptide (2.1%) and 2 latent protein EBNA3A peptides (2.3% and 0.68%). Staining of PBMCs from individual KK, who also expresses HLA-B7, demonstrated that approximately 0.28% of CD8 T cells bound HLA-B7 tetramers complexed with one of the latent protein EBNA3A peptides; the binding of tetramers complexed with a second EBNA3A peptide was undetectable (Figure 2, lower panels).
HLA tetramers complexed with EBV peptides detect virus-specific CD8 T cells in the peripheral blood of EBV+individuals.
PBMCs were isolated from 3 EBV+ individuals and stained with mAbs specific for CD8 and CD62L and with HLA tetramers complexed with EBV epitopes. The HLA-A and HLA-B alleles expressed by these individuals and the HLA/epitope complexes used for staining are indicated above each dot-plot. Each dot-plot is gated on live CD8+ T cells, and the percentage of CD8 T cells staining with HLA tetramers is indicated in the upper quadrants of each plot.
HLA tetramers complexed with EBV peptides detect virus-specific CD8 T cells in the peripheral blood of EBV+individuals.
PBMCs were isolated from 3 EBV+ individuals and stained with mAbs specific for CD8 and CD62L and with HLA tetramers complexed with EBV epitopes. The HLA-A and HLA-B alleles expressed by these individuals and the HLA/epitope complexes used for staining are indicated above each dot-plot. Each dot-plot is gated on live CD8+ T cells, and the percentage of CD8 T cells staining with HLA tetramers is indicated in the upper quadrants of each plot.
These analyses demonstrate that HLA tetramers complexed with EBV-derived peptides can directly identify EBV-specific CD8 T cells ex vivo from healthy individuals. All of our tetramers were tested for the staining of PBMCs isolated from individuals not expressing the particular MHC class I molecule. All were negative, confirming once again the specificity of these reagents (results not shown). Additionally, in these and subsequent analyses, staining with antibodies specific for CD3 (which is only expressed on T cells) and CD56 (a natural killer cell marker) revealed that tetramer-staining cells were CD3+ T cells and not natural killer cells. EBV-specific CD8 T cells identified by HLA tetramer staining generally fell into 2 subpopulations that expressed either high or low levels of CD62L, an adhesion molecule that is down-regulated on the cell surface upon T-cell activation. This pattern of expression of CD62L has been described previously for EBV-specific T cells in EBV+individuals.20
To determine whether allo-PBSCT recipients regenerate EBV-specific T-cell populations that are similar in magnitude to healthy individuals, we next evaluated 2 patients who had undergone matched sibling donor (MSD) allogeneic PBSCT. The clinical characteristics of the patients included in this study and the HLA typing of the patients and donors are summarized in Table 2. CD8 T cells from patient A, who expresses HLA-B8 and underwent allo-PBSCT 5 months prior to this analysis, are shown in Figure 3A. Approximately 0.9% of CD8 T cells stained with HLA-B8 tetramers complexed with the lytic BZLF1 peptide, and 0.3% and 0.62% of CD8 T cells were specific for the 2 latent EBNA3A-derived peptides (Figure3A). EBV-specific CD8 T cells from patient B, who expresses HLA-A2 and underwent transplantation 9 months prior to analysis, are shown in Figure 3B. Nearly 7% of CD8 T cells in this patient are specific for the HLA-A2 restricted lytic BMLF1-derived peptide. Phenotypic analysis of the tetramer-positive cells revealed that the population is split between CD45RA+ and CD45RA− as well as CD62L high and low, and that staining with another activation marker, CD44, was high. This pattern was seen for all patients examined. Staining with the HLA-B8 tetramer complexed with the lytic BZLF1 peptide was negligible, as expected, because the patient did not express HLA-B8 (Figure 3B, bottom panel).
EBV-specific CD8 T cells are present in the peripheral blood of allo-PBSCT recipients.
PBMCs were obtained from 2 allo-PBSCT recipients at 155 days (patient A) and 212 days (patient B) following transplantation. (A) Patient A was triple-stained with anti-CD8+, CD62L, and HLA tetramers. (B) Patient B was stained with tetramer and anti-CD8 with either anti-CD44, CD45RA, or CD62L. Finally, patient B was stained with an unrelated tetramer/peptide complex as a negative control. The HLA haplotype and the HLA tetramer/peptide complex used for staining are indicated above each plot. Each dot-plot is gated on live CD8+ T cells, and the percentage of CD8 T cells that stain with the respective tetramer is indicated in the upper quadrants of each plot.
EBV-specific CD8 T cells are present in the peripheral blood of allo-PBSCT recipients.
PBMCs were obtained from 2 allo-PBSCT recipients at 155 days (patient A) and 212 days (patient B) following transplantation. (A) Patient A was triple-stained with anti-CD8+, CD62L, and HLA tetramers. (B) Patient B was stained with tetramer and anti-CD8 with either anti-CD44, CD45RA, or CD62L. Finally, patient B was stained with an unrelated tetramer/peptide complex as a negative control. The HLA haplotype and the HLA tetramer/peptide complex used for staining are indicated above each plot. Each dot-plot is gated on live CD8+ T cells, and the percentage of CD8 T cells that stain with the respective tetramer is indicated in the upper quadrants of each plot.
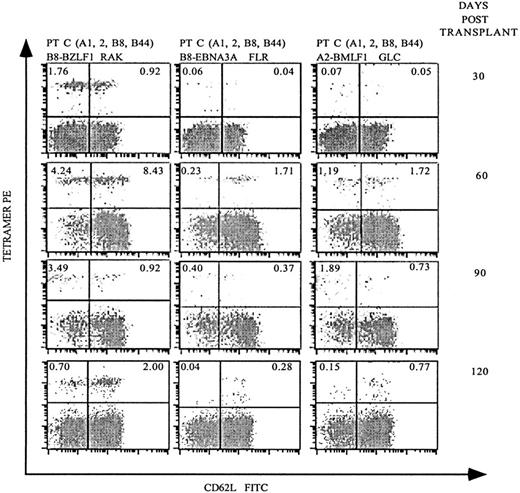
Although the previous analyses demonstrated that allo-PBSCT recipients generate EBV-specific T-cell populations, it was unclear how rapidly EBV-specific immunity redeveloped following transplantation. Therefore, we next investigated patient C, who expresses HLA-A2 and HLA-B8 at monthly intervals following allo-PBSCT. Remarkably, 30 days following transplantation, 2.68% of the CD8 T cells stained with HLA-B8 tetramers complexed with the lytic BZLF1 peptide (Figure4). In contrast, HLA-B8–restricted CD8 T cells specific for the EBNA3A peptide or HLA-A2–restricted T cells specific for a BMLF1 peptide were present at 20- to 30-fold lower frequencies. Analysis of patient C 60 days following transplantation demonstrated a dramatic expansion of EBV-specific CD8 T cells specific for HLA-B8/BZLF1 (12.67%), HLA-B8/EBNA3A (1.94%), and HLA-A2/BMLF1 (2.91%). Of note, patient C developed GVHD of the skin and gastrointestinal tract 30 days following transplantation, which required, in addition to tacrolimus, immunosuppression with steroids and eventually antithymocyte globulin. In spite of this immunosuppressive regimen, patient C had a substantial expansion of EBV-specific T cells, both in frequency (Figure 4) and absolute number in terms of EBV-specific T cells per μL of blood (Figure5). Patient C's GVHD stabilized in the following months, after which his immunosuppression was gradually tapered, and the frequency of EBV-specific T cells decreased 90 and 120 days following transplantation (Figure 4).
Serial analysis of PBMCs following allo-PBSCT reveals that EBV-specific T-cell populations reconstitute the periphery rapidly and fluctuate in frequency.
PBMCs were obtained 30, 60, 90, and 120 days after allo-PBSCT from patient C, who expresses both HLA-A2 and HLA-B8, and stained as described in Figures 2 and 3A. The percentage of live CD8-gated T cells that stained with the respective tetramers is indicated in the upper quadrants of each dot-plot.
Serial analysis of PBMCs following allo-PBSCT reveals that EBV-specific T-cell populations reconstitute the periphery rapidly and fluctuate in frequency.
PBMCs were obtained 30, 60, 90, and 120 days after allo-PBSCT from patient C, who expresses both HLA-A2 and HLA-B8, and stained as described in Figures 2 and 3A. The percentage of live CD8-gated T cells that stained with the respective tetramers is indicated in the upper quadrants of each dot-plot.
The frequency of EBV-specific T cells correlates with the EBV burden following allo-PBSCT.
PBMCs were obtained from allo-PBSCT patient C (HLA-A2/B8), patient D (HLA-A2), and patient E (HLA-A2) at monthly intervals following transplantation, and EBV-specific CD8 T cells, total CD8 T cells, and EBV genome copies were quantified as described in “Materials and methods.” The graphs indicate the number of EBV genome copies per 105 PBMCs, the number of EBV-specific CD8 T cells per μL (measured by MHC tetramer staining with A2-GLC-lytic, B8-RAK-lytic, and B8-FLR-latent in patient C and with A2-GLC-lytic, A2-LLD-latent, and A2-CLG-latent in patients D and E), and the number of CD8 T cells per μL. The duration of therapy with tacrolimus and corticosteroids is indicated above each graph, as is the administration of ATG to patient C. The days following allo-PBSCT are indicated on the x-axis of each graph.
The frequency of EBV-specific T cells correlates with the EBV burden following allo-PBSCT.
PBMCs were obtained from allo-PBSCT patient C (HLA-A2/B8), patient D (HLA-A2), and patient E (HLA-A2) at monthly intervals following transplantation, and EBV-specific CD8 T cells, total CD8 T cells, and EBV genome copies were quantified as described in “Materials and methods.” The graphs indicate the number of EBV genome copies per 105 PBMCs, the number of EBV-specific CD8 T cells per μL (measured by MHC tetramer staining with A2-GLC-lytic, B8-RAK-lytic, and B8-FLR-latent in patient C and with A2-GLC-lytic, A2-LLD-latent, and A2-CLG-latent in patients D and E), and the number of CD8 T cells per μL. The duration of therapy with tacrolimus and corticosteroids is indicated above each graph, as is the administration of ATG to patient C. The days following allo-PBSCT are indicated on the x-axis of each graph.
To correlate the frequency of EBV-specific T cells with the EBV viral load, we used HLA tetramers to measure the frequency of EBV-specific CD8 T cells and PCR to determine the number of EBV genomes per 105 PBMCs at monthly intervals in 3 patients following allo-PBSCT. This quantitative analysis revealed that patient C had a dramatic increase in the number of EBV genomes 60 days following transplantation, at the same time that the EBV-specific T-cell population underwent expansion (Figure 5, upper panel). Despite aggressive immunosuppression, the EBV load dropped precipitously 90 days following transplantation and was accompanied by a similar decrease in the number of EBV-specific CD8 T lymphocytes. Interestingly, although the number of EBV-specific CD8 T cells increased and then decreased following transplantation, the patient's CD8 T-cell count decreased between days 30 and 120 following treatment for GVHD. Although the decrease in CD8 T-cell numbers can be attributed to antithymocyte globulin and steroid administration, the relative insensitivity of EBV-specific T cells is remarkable. The EBV-specific T-cell number, CD8+ T-cell absolute number, and EBV genome titer for patients D and E, who are both HLA-A2, were less dramatic (Figure 5, middle and lower panels). Patients D and E, who received steroids for GVHD, did not have an increase in EBV genome numbers, and both the EBV-specific and total CD8+ T-cell numbers remained stable throughout the duration of follow-up.
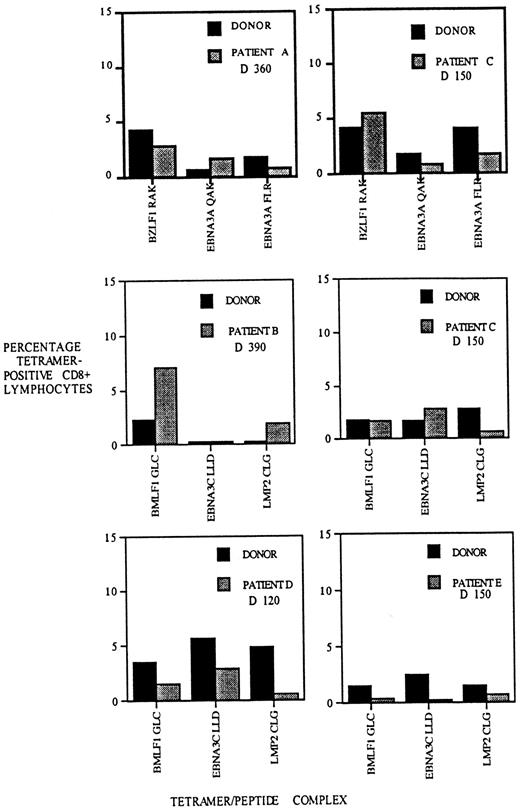
Different peptides derived from single pathogens are known to elicit T-cell responses that differ in size.25,26 The factors responsible for the T-cell hierarchies include antigen stability; T-cell repertoire; and to a lesser extent, antigen prevalence.27-30 We decided to compare the hierarchy of T-cell responses to EBV epitopes in recipients of allo-PBSCT and their donors following reconstitution of the T-cell compartment. We determined the frequency of EBV-specific T cells to 3 HLA-A2– and 3 HLA-B8–restricted EBV epitopes in patients A, B, C, D, and E and in their respective stem cell donors. This analysis revealed that although not identical, the relative frequencies of T cells specific for the different EBV epitopes are similar in the donor and recipient (Figure6). In some instances, such as the HLA-A2/BMLF1–specific T-cell populations in the patient B/donor pair, the frequencies appear to be disparate, but the overall patterns suggest a similarity between the recipients and their donors. In contrast, comparison of the T-cell hierarchies between different recipient/donor pairs indicates a higher degree of disparity (eg, compare hierarchies of HLA-A2 patients B and D). At a quantitative level, these results suggest that the homeostatic mechanisms determining the frequency and hierarchy of virus-specific T cells in the healthy donor have been recapitulated in the matched sibling stem cell allograft recipient.
The hierarchy of CD8 T-cell populations specific for different EBV epitopes is similar in allo-PBSCT recipients and their respective donors.
PBMCs were isolated from allograft recipients A-E at the indicated times following transplantation and stained with HLA-A2 or HLA-B8 tetramers, each complexed with 3 different EBV epitopes, as indicated below each bar graph. Donor PBMCs were stained with the same tetramers, and the frequency of EBV-specific T cells was determined. The percentage of CD8 T cells staining with HLA tetramers is indicated on the y-axis.
The hierarchy of CD8 T-cell populations specific for different EBV epitopes is similar in allo-PBSCT recipients and their respective donors.
PBMCs were isolated from allograft recipients A-E at the indicated times following transplantation and stained with HLA-A2 or HLA-B8 tetramers, each complexed with 3 different EBV epitopes, as indicated below each bar graph. Donor PBMCs were stained with the same tetramers, and the frequency of EBV-specific T cells was determined. The percentage of CD8 T cells staining with HLA tetramers is indicated on the y-axis.
Solid organ transplant recipients or patients with HLA disparity undergoing PBSCT are at higher risk for PTLD when compared to MSD stem cell allograft recipients. We performed a preliminary study of patients undergoing unrelated UCB, MUD, or haploidentical (HAPLO) stem cell transplantation or solid organ transplantation (Table3). Of the 5 solid organ allograft recipients, 4 recipients had detectable populations of EBV-specific T cells and a normal EBV genome titer. Patient YS-5 had no detectable EBV-specific T cells and a high EBV genome titer. We are currently following this patient to evaluate for the development of EBV-specific CD8 T cells. We studied 4 patients who received unrelated UCB transplantations, 1 patient who received a non–T-cell–depleted MUD transplantation, and 1 patient who received a HAPLO allograft. We did not detect EBV-specific T cells in any of these stem cell allograft recipients. Although EBV viral titers were not elevated in the UCB or HAPLO recipients, the patient who received a MUD transplantation (M-4) had an EBV genome titer of 176.
Correlation of EBV-specific T cells with EBV viral load in high-risk stem cell and solid organ allograft recipients
| Patient . | Recipient HLA type . | Allograft, type (day following transplant) . | EBV genomes, 105PBMCs . | EBV-specific T cells, % . | |||
|---|---|---|---|---|---|---|---|
| A2 . | B7 . | B8R . | B8F . | ||||
| YS-1 | A1,—; B8,37 | renal (D240) | <5 | — | — | 0 | 0.5 |
| YS-2 | A1,2; B8,44 | renal (D210) | <5 | 0.4 | — | 0.1 | 0.9 |
| YS-3 | A1,2; B8,18 | renal (D220) | <5 | 0.2 | — | 0.6 | 0.3 |
| YS-4 | A30,31; B8,35 | renal (D330) | <5 | — | — | 0.4 | 0.3 |
| YS-5 | A1,26; B8,35 | renal (D450)3-150 | 371 | — | — | 0 | 0 |
| M-1 | A2,30; B35,70 | UCB (D60) | <5 | 0 | — | — | — |
| M-2 | A3; B7,40 | UCB (D60) | <5 | — | 0 | — | — |
| M-3 | A2; B18,57 | UCB (D180) | <5 | 0 | — | — | — |
| M-4 | A24; B7,56 | MUD (D28) | 176 | — | 0 | — | — |
| M-5 | A2,32; B15 | UCB (D90) | <5 | 0 | — | — | — |
| F | A2,23; B49,60 | HAPLO (D180) | <5 | 0 | — | — | — |
| Patient . | Recipient HLA type . | Allograft, type (day following transplant) . | EBV genomes, 105PBMCs . | EBV-specific T cells, % . | |||
|---|---|---|---|---|---|---|---|
| A2 . | B7 . | B8R . | B8F . | ||||
| YS-1 | A1,—; B8,37 | renal (D240) | <5 | — | — | 0 | 0.5 |
| YS-2 | A1,2; B8,44 | renal (D210) | <5 | 0.4 | — | 0.1 | 0.9 |
| YS-3 | A1,2; B8,18 | renal (D220) | <5 | 0.2 | — | 0.6 | 0.3 |
| YS-4 | A30,31; B8,35 | renal (D330) | <5 | — | — | 0.4 | 0.3 |
| YS-5 | A1,26; B8,35 | renal (D450)3-150 | 371 | — | — | 0 | 0 |
| M-1 | A2,30; B35,70 | UCB (D60) | <5 | 0 | — | — | — |
| M-2 | A3; B7,40 | UCB (D60) | <5 | — | 0 | — | — |
| M-3 | A2; B18,57 | UCB (D180) | <5 | 0 | — | — | — |
| M-4 | A24; B7,56 | MUD (D28) | 176 | — | 0 | — | — |
| M-5 | A2,32; B15 | UCB (D90) | <5 | 0 | — | — | — |
| F | A2,23; B49,60 | HAPLO (D180) | <5 | 0 | — | — | — |
The following tetramers were used in this table: A indicates A2-GLC-lytic; B7, B7-RPP-latent; B8R, B8-RAK-lytic; and B8F, B8-FLR-latent.
Denotes that the patient underwent a renal and pancreas transplantation.
Discussion
The recent introduction of methods that directly identify antigen-specific T lymphocytes has provided unprecedented views of the interaction between the human immune system and infectious pathogens.14 18 In this report we have used MHC class I tetramers to characterize the dynamics of EBV-specific T lymphocytes in patients undergoing one of the most dramatic immunomodulatory therapies in current medical practice: immune ablation followed by allogeneic stem cell transplantation. In the face of this upheaval within the patient's immune system, we find that EBV-specific CD8 T-cell populations are readily detectable in patients following matched sibling allo-PBSCT and in solid organ transplant recipients, and their frequencies rapidly approximate those found in healthy individuals. Despite potent immunosuppressive therapy to prevent GVHD, the reconstitution of EBV-specific immunity is rapid and effective. In a patient analyzed at monthly intervals following transplantation, the frequency of EBV-specific T lymphocytes correlated with the amount of EBV detected by PCR in PBMCs. This suggests that EBV-specific T-cell populations expanded in response to the increased viral burden even while the number of CD8 T cells declined.
Our finding that the frequency of EBV-specific CD8 T cells correlates with the abundance of EBV genomes is reminiscent of human immunodeficiency virus (HIV)–specific CD8 T-cell populations in patients treated with antiretroviral therapy. In adults and children receiving highly active antiretroviral therapy (HAART), dramatic reductions in viral load were associated with decreased HIV-specific CTL frequencies,14,31 suggesting that the size of antigen-specific T-cell populations in the setting of chronic infections is determined by antigen prevalence. A similar correlation between the prevalence of viral antigen and the frequency of HIV-specific T lymphocytes has also been demonstrated in the CD4+ T-cell compartment.32 Thus, although there is abundant evidence that pathogen-specific T lymphocytes play a critical role in the suppression of chronic viral infections, our study and those performed in HIV-infected individuals demonstrate that the relative activity of viral infection dramatically influences the frequency of virus-specific T lymphocytes in the peripheral blood.
UCB allografts are EBV−, and they are generally given to an EBV+ recipient. The incidence of PTLD in this population has not been determined, but 2 cases have been reported in the literature.33 34 MUD and HAPLO allogeneic transplantation recipients are at risk for PTLD due to marked HLA disparity and the delay in T-cell reconstitution after transplantation. Our results indicate that these patients have far fewer EBV-specific CD8 T cells than recipients of fully matched related stem cell allografts. Despite the absence of EBV-specific T cells by tetramer analysis, only one of these patients had an elevated EBV titer in the peripheral blood (patient M-4, Table 3). Solid organ transplant patients are at risk for PTLD due to the immunosuppression necessary to prevent organ rejection. Of the renal transplant patients that we investigated in this report, 4 patients had readily detectable EBV-specific CD8 T cells in the peripheral blood. Interestingly, the renal transplantation patient who had an elevated EBV titer in the blood (YS-5, Table 3) did not have EBV-specific T cells detectable by tetramer staining. Longitudinal follow-up of these patients will provide an opportunity to correlate EBV titers and EBV T-cell frequencies with clinical outcomes and the risk for PTLD.
The reconstitution of EBV-specific T-cell populations was carefully investigated in the pretetramer era. Using limiting dilution analysis, which we now know substantially underestimates the true frequency of antigen-specific T lymphocytes, Lucas et al35 demonstrated that EBV-specific T cells were present 3 months following BMT in normal frequencies in 19% of the patients, in intermediate frequencies in 23%, and undetectable in 58%. That study, which measured the frequency of T cells responding to B-LCLs, did not differentiate between T cells specific for lytic or latent cycle EBV proteins. Surprisingly, the size of EBV-specific T-cell populations measured by limiting dilution did not differ detectably between patients receiving unmanipulated bone marrow grafts and those receiving T-cell–depleted grafts.
Haque et al36 evaluated the frequency of EBV-specific CTLs in solid organ recipients before and after EBV-specific CTL infusions using limiting dilution analysis. In the few patients studied, they detected very low levels of EBV CTL pre-infusion in one patient but none in the other patients studied. After treatment with autologous EBV-specific CTL lines, the precursor frequency increased after the third infusion and correlated inversely with the EBV viral load. Peripheral blood stem cell allografts deliver more CD3+ T lymphocytes by 1-2 logs than traditional bone marrow allografts,37,38 and this may contribute to the rapid EBV-specific immune recovery seen in these patients. It has been argued that limiting dilution analysis does not detect all antigen-specific T cells, only those that are capable of undergoing further division.39 Thus, it is possible that many of the T cells we detected by tetramer staining are incapable of in vitro growth. However, our finding that EBV-specific T-cell populations can rapidly increase in size in response to a greater viral burden suggests that the T-cell populations we are measuring have the potential to divide in vivo.
In patients receiving T-cell depleted BMT, the incidence of PTLD, which can approach 10%, is related to the degree of HLA disparity, rigorous T-cell depletion approaches that do not remove donor B cells, or the use of OKT3 or ATG.8 The adoptive transfer of donor lymphocytes has resulted in the remarkable regression of PTLD.12 T cell lines generated in vitro by restimulation of donor T lymphocytes with autologous B-LCL have also been adoptively transferred and have resulted in the regression of PTLD.13,40,41 A recent report from Gustafsson et al42 correlated EBV genome titer with prophylactic adoptive transfer of EBV-specific T cell lines. They showed that in T-cell–depleted stem cell allograft recipients, transfer of EBV-specific T cells resulted in a decrease in the viral titer and protection from PTLD. Our study confirms the finding by Callan and colleagues20 that the frequency of EBV-specific T cells in healthy EBV+ individuals is remarkably high, exceeding 5% of CD8 T cells in some individuals. The high frequency of these cells suggests that HLA tetramers may be useful for the direct purification of EBV-specific T cells. This may be used for the prophylactic supplementation of T-cell–depleted stem cell allografts or therapeutic treatment of patients with PTLD without the cost and time involved to generate EBV-specific T cell lines or the GVHD risk involved with giving unmanipulated donor lymphocyte infusions.
Acknowledgments
We would like to thank Jan Kotora, RN, and the nurses in the bone marrow unit for collection of samples and excellent care of these patients in relation to this project.
Supported by a Center of Excellence Grant from the John A. Hartford Foundation; a Swebilius Cancer Research Award; E.G.P. is the recipient of a Donaghue Investigator Award from the Donaghue Foundation of Hartford, CT.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Eric G. Pamer, Infectious Diseases Service, Memorial Sloan Kettering Cancer Center, 1275 York Ave, New York, NY 10021; e-mail: pamere@mskcc.org.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal