Abstract
The receptor tyrosine kinase Flk-1 is essential for embryonic blood vessel development and for tumor angiogenesis. To identify upstream transcriptional regulators of Flk-1, the gene regulatory elements that mediate endothelium-specific expression in mouse embryos were characterized. By mutational analysis, binding sites for SCL/Tal-1, GATA, and Ets transcription factors located in theFlk-1 enhancer were identified as critical elements for the endothelium-specific Flk-1 gene expression in transgenic mice. c-Ets1, a transcription factor that is coexpressed withFlk-1 during embryonic development and tumor angiogenesis, activated the Flk-1 promoter via 2 binding sites. One of these sites was required for Flk-1 promoter function in the embryonic vasculature. These results provide the first evidence that SCL/Tal-1, GATA, and Ets transcription factors act upstream ofFlk-1 in a combinatorial fashion to determine embryonic blood vessel formation and are key regulators not only of the hematopoietic program, but also of vascular development.
Introduction
The formation of a functional vascular system is a prerequisite to fulfill the metabolic demands of the growing vertebrate embryo and of many solid tumors. During development, a primitive vascular system is formed by the differentiation of mesodermal precursor cells into vascular endothelial cells, a process termed vasculogenesis.1 During the subsequent process of angiogenesis, endothelial cells proliferate and sprout to form capillaries, which eventually differentiate into mature blood vessels.2 Although vasculogenesis and angiogenesis are orchestrated by a plenitude of genes, the receptor tyrosine kinase Flk-1 plays a pivotal role in both processes.1,2 This has been demonstrated by a targeted null mutation of Flk-1 that completely prevents the differentiation of endothelial cells from their precursors in vivo3 and by overexpression of a dominant-negative form of Flk-1 that inhibits tumor angiogenesis.4,5 Flk-1 is a high-affinity receptor for the endothelial cell mitogen vascular endothelial growth factor (VEGF) and for certain other VEGF family members.6 VEGF and its receptors Flt-1 (fms-like tyrosine kinase-1, VEGF receptor-1) and Flk-1 (fetal liver kinase-1, VEGF receptor-2) represent the first endothelial cell–specific signal transduction system known to be activated during embryonic development, as the inactivation of any of these genes leads to defective vasculogenesis.3,7-9 In addition to its function in endothelial cell differentiation,Flk-1 is required for primitive and definitive hematopoiesis.10 This reflects the common origin of endothelial and blood cells from a population of bipotential precursor cells, the hemangioblasts.1 11 The onset ofFlk-1 expression in mesodermal precursor cells is thought to mark the establishment of the hemangioblastic lineage during embryonic development.
Flk-1 is expressed almost exclusively on endothelial cells and their precursors during vasculogenesis and angiogenesis.12,13Recently, the transcriptional regulatory elements of the murineFlk-1 gene and its human homologue KDR have been cloned and characterized in vitro14,15 and in vivo.16 So far, the transcription factors Sp1 and hypoxia-inducible factor-2α (HIF-2α) (also known as HRF, EPAS-1, and HLF) have been identified by in vitro studies as regulators of the promoters for Flk-1 or its human homologue, kinase-insert domain containing receptor (KDR).16-18 Because the Flk-1 promoter alone is not capable of directing endothelium-specific reporter gene expression in transgenic mice,16 it is unlikely that the endothelium specificity of Flk-1 expression is directed by Sp1 and HIF-2α only. Thus, the transcriptional mechanisms that govern the distinct expression of Flk-1 in the embryonic vasculature remain unknown.
Several transcription factors such as c-Ets1, GATA-2, SCL/Tal-1, and HIF-2α are predominantly expressed in endothelial cells or their precursors and are therefore candidate regulators of Flk-1expression.19-25 SCL/Tal-1 is coexpressed withFlk-1 in hemangioblasts11,26 and regulatesFlk-1 expression in zebrafish.27 This functional link between SCL/Tal-1 and Flk-1 was not observed in SCL/Tal-1−/− mice.28 GATA-2 has been shown to regulate the promoter activity of several endothelium-specific genes such as platelet endothelial cell adhesion molecule-1 (PECAM-1) and endothelin-1 in vitro29-31; however, the in vivo significance of the GATA binding sites that were identified in these studies remained unclear. Putative Ets binding sites are involved in the endothelium-specific expression of the Tie-1and Tie-2 genes.32,33 Moreover, c-Ets1 has been shown to regulate the promoter of Flt-1 in vitro.34 Despite the expression of Ets, GATA, or SCL/Tal-1 transcription factors in endothelial cells or their progenitors, their role in early murine vascular development is poorly understood. Gene targeting experiments have revealed their function in hematopoiesis35-37 but did not provide evidence of their involvement in embryonic blood vessel differentiation, perhaps because of the functional redundancy of related family members. However, transgenic rescue of the hematopoietic defects in SCL/Tal-1–deficient mice revealed that this factor is also critical for developmental angiogenesis in the yolk sac.28 We therefore investigated whether these transcription factors are involved in early embryonic vascular development by regulating the expression of Flk-1, the earliest known marker for endothelial cells.
We have recently identified gene regulatory elements ofFlk-1 that reproduced most properties of the endogenousFlk-1 expression in transgenic mice.16 The endothelial cell type specificity of these sequences was mediated by an autonomous endothelium-specific enhancer located in the first intron ofFlk-1. In contrast, the Flk-1 promoter was not sufficient for endothelium-specific reporter gene expression, but rather contributed to a strong and position-independent reporter gene expression in transgenic mice. In the present study, we have characterized critical transcription factor binding sites located in the Flk-1 intron enhancer. A minimal sequence of 430 bp from the Flk-1 first intron was necessary and sufficient for endothelium-specific reporter gene expression in transgenic mouse embryos. Two SCL/Tal-1 motifs in this minimal enhancer were required for high-level and uniform endothelial expression in transgenic mice. The mutation of a GATA site rendered the enhancer completely inactive in vivo. Analysis of protein–DNA interactions on the Flk-1intron enhancer demonstrated a specific binding of SCL/Tal-1 and of a GATA factor to these sites. Moreover, cell type–specific interaction of nuclear proteins with the putative Ets binding site that was required for the cell-type specificity of the enhancer during embryonic development was observed. In addition, c-Ets1 activated theFlk-1 promoter via 2 previously undiscovered Ets sites, one of them being functional during embryonic development.
Materials and methods
Plasmid construction and DNA sequence analysis
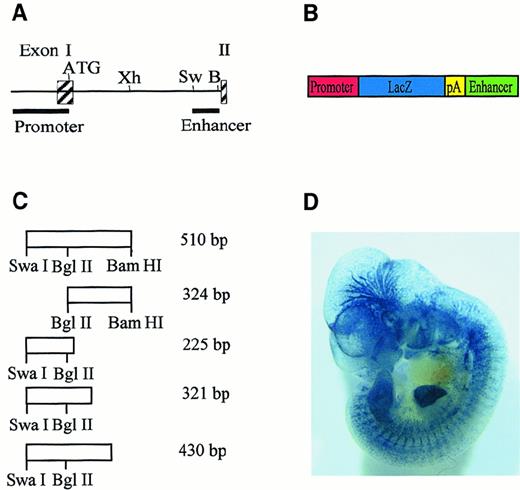
The pGL-2 (Promega, Madison, WI)-based LacZ reporter gene construct containing the Flk-1 promoter from bp −640 to bp +299 relative to the transcriptional start site was constructed as described previously.16 Deletion fragments of the 510-bpSwaI/BamHI fragment from the Flk-1first intron were amplified by polymerase chain reaction (PCR), as described,15 using the oligonucleotide enhfor as a forward primer. For amplification of the 225-bp, 321-bp, or 430-bp intron fragments (Figure 1A), the primers −225rev, −321rev, or −430rev were used as reverse primers, respectively. Primers used were as follows: enhfor, 5′-GGGGATCCTA AATGTGCTGT CTTTAGAAGCC-3′; −225rev, 5′-CCCGTCGACC CACTGACATT TCTTGTAAAGC-3′; −321rev, 5′-CCCGTCGACG TGGAGTTCCT GTTTTCCTGCG-3′; and −430rev, 5′-CCCGTCGACG GATTGACTTT GCCCCAGTCCC-3′. The PCR products were inserted into the SalI and BamHI sites of the pGL-2–based LacZ reporter gene construct containing theFlk-1 promoter, as described previously.16 The 324-bp intron fragment was a BglII/BamHI fragment derived from the 510-bp SwaI/BamHI fragment from the Flk-1 first intron and was inserted by blunt-end cloning into the blunted BamHI site of the LacZ reporter gene construct. pKSenh was constructed by insertion of the 430-bp intron fragment into the BamHI and SalI sites of pBSIIKS(+) (Stratagene, La Jolla, CA).
A 430-bp fragment from the
Flk-1 first intron is sufficient for endothelium-specific reporter gene expression in transgenic mice.(A) Partial structure and restriction-enzyme map of the murineFlk-1 locus. Exons are represented by shaded boxes. The positions of the 939-bp promoter fragment and the 510-bpSwaI/BamHI enhancer fragment are indicated. Abbreviations for restriction enzymes are Xh, XhoI; Sw,SwaI; B, BamHI. (B) Structure of reporter gene constructs. The LacZ reporter gene (blue) is flanked by aFlk-1 promoter fragment spanning bp −640 to bp +299 (red) and subfragments of the 510-bp SwaI/BamHI intron fragment (green). Transcription terminates at a simian virus (SV40) polyadenylation signal (yellow). (C) Structure of intron fragments and their position in the 510-bp SwaI/BamHI intron fragment. Fragment lengths are indicated. (D) Whole-mount LacZ-stained embryo. This embryo is transgenic for a reporter gene construct in which the LacZ gene is under control of the 939-bp Flk-1promoter and the 430-bp enhancer fragment.
A 430-bp fragment from the
Flk-1 first intron is sufficient for endothelium-specific reporter gene expression in transgenic mice.(A) Partial structure and restriction-enzyme map of the murineFlk-1 locus. Exons are represented by shaded boxes. The positions of the 939-bp promoter fragment and the 510-bpSwaI/BamHI enhancer fragment are indicated. Abbreviations for restriction enzymes are Xh, XhoI; Sw,SwaI; B, BamHI. (B) Structure of reporter gene constructs. The LacZ reporter gene (blue) is flanked by aFlk-1 promoter fragment spanning bp −640 to bp +299 (red) and subfragments of the 510-bp SwaI/BamHI intron fragment (green). Transcription terminates at a simian virus (SV40) polyadenylation signal (yellow). (C) Structure of intron fragments and their position in the 510-bp SwaI/BamHI intron fragment. Fragment lengths are indicated. (D) Whole-mount LacZ-stained embryo. This embryo is transgenic for a reporter gene construct in which the LacZ gene is under control of the 939-bp Flk-1promoter and the 430-bp enhancer fragment.
Flk-1 promoter deletion fragments were amplified by PCR as described15 using the following forward primers: wt, 5′-GGGGTACCTT CTGGACCGAC CCAGCCAGG-3′; delets1, 5′-GGGGTACCCA ACCGAAATGT CTTCTAGGG-3′; delets2, 5′-GGGGTACCCC GCCCGGCACA GTTCCGGGG-3′; delets3, 5′-GGGGTACCGC GTGGGAAACC GGGAAACCC-3′; delets4, 5′-GGGGTACCAA ACCTGGTATC CAGTGGGGG-3′; delets5, 5′-GGGGTACCGG GGGGCGTGGC CGGACGCAG-3′; and delets6, 5′-CCGGTACCAC GCAGGGAGTC CCCACCCC-3′. The primer +16rev (5′-GGGAAGCTTG ACTCAGGGCA GAAAGAGAGC-3′) was included as a reverse primer. PCR products were cloned into theAcc65I and HindIII sites of pGL-2 containing the luciferase reporter gene. The PCR product generated with primers wt and +16 rev was also cloned into the Acc65I andHindIII sites of pGL-2 containing the LacZ reporter gene, and the 430-bp intron fragment was cloned into the BamHI andSalI sites.
To generate reporter constructs containing the thymidine kinase (TK) promoter, the 145-bp XhoI/NcoI fragment from ptk-32 spanning the TK promoter was inserted by blunt-end cloning into the blunted XhoI site of pGL-2 containing either the luciferase or LacZ reporter gene cassettes to generate pTKLuc and pTKLacZ, respectively. pTKEnh was generated by cloning the 430-bp Flk-1 intron fragment into theSalI and BamHI sites of pTKLacZ.
The chicken c-Ets1 cDNA expression vector was pSG5c-Ets1p68.38 Mouse c-Ets2 cDNA was amplified by PCR as described for the HIF-2α cDNA16 using primers ets2 5′ (5′-GGGGATCCGG CGCGATGAAT GACTTTGG-3′) and ets2 3′ (5′-CCCTCGAGTC TTCTGTATCA GGCTGG-3′). The PCR product was cloned into the BamHI and XhoI sites of pcDNA3 (Invitrogen, Carlsbad, CA).
The nucleotide sequence of all constructs was determined on an Applied Biosystems 373 automated sequencer (PE Biosystems, Weiterstadt, Germany). The search for potential transcription factor binding sites was performed using the on-line software MatInspector (http://transfac.gbf-braunschweig.de). Site-directed mutagenesis of reporter gene constructs was performed with the QuikChange Site-Directed Mutagenesis kit (Stratagene).
Cell culture and transfection analysis
All cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. For cotransfection assays, A293 cells were split 1:2 and transfected 18 hours later with 2.5 μg of DNA, complexed with 10 μL of Superfect (Qiagen, Hilden, Germany). For the analysis of luciferase constructs containing Flk-1promoter sequences only, 1 μg of the luciferase reporter construct and 0.5 μg of pTKLacZ were cotransfected with 1 μg of c-Ets1 or c-Ets2 expression vectors, respectively, or pcDNA3 as control. For the analysis of LacZ constructs containing Flk-1 enhancer sequences, 1 μg of the LacZ reporter construct and 0.5 μg of pTKLuc were cotransfected with 1 μg of GATA-2, c-Ets1, or c-Ets2 expression vectors, respectively, or pcDNA3 as control. After 24 hours, reporter gene activities were determined as described.16 For each extract, the reporter gene activity corresponding to the construct containing Flk-1 regulatory sequences was normalized to the activity corresponding to pTKLuc or pTKLacZ, respectively. The normalized reporter gene activity of the control transfections was arbitrarily set at 1. Each cotransfection experiment was performed at least 6 times.
Nuclear extract preparation, electrophoretic mobility shift assay, and in vitro DNAseI footprint
Nuclear extracts of bovine aortic endothelial (BAE) cells,39 TK1,40 and endothelioma cells41 were prepared as described.42,43 For the electrophoretic mobility shift assay, 5 μL of e.END40 (a murine embryonic endothelioma cell line; kindly provided by Dr Urban Deutsch) nuclear extract (approximately 10 μg) was preincubated for 30 minutes on ice in 19 μL of buffer (20 mmol/L HEPES, pH 7.3, 0.5 mmol/L dithiothreitol, 10% glycerol, 1 μg poly-dIdC, 0.025% NP40) either without or with competitor (2 pmol) or with 1.5 μL of anti-SCL/Tal-1 antiserum.44 After this, 1 μL of32P-labeled probe (10 fmol) was added, and after incubation for 30 minutes at room temperature, the complexes were resolved by electrophoresis in a 5% polyacrylamide gel (29:1; 0.5 × Tris-bovate). Oligonucleotides used were as follows (only the upper strands are shown; see Table1 for the changes in the corresponding mutant oligonucleotides): G72-W, 5′-CACAGCATGA TAAAAGACAA; S232-W, 5′-AGATCATCAG ATGGAGGTTC; S358-W, 5′-TTGTGACCAT CTGCCCATTC; SCL-W (Geneka, Montreal, Canada), 5′-ACCTGAACAG ATGGTCGGCT; SCL-M, 5′-ACCTGAATTG ATGGTCGGCT; and GATA-W (Santa Cruz Biotech, Santa Cruz, CA), 5′-CACTTGATAA CAGAAAGTGA TAACTCT. In vitro DNAseI footprinting on supercoiled DNA was performed as described45 using pKSenh as plasmid DNA, 10 to 100 μg of nuclear extracts or bovine serum albumin (BSA; Serva, Heidelberg, Germany), and 10 ng of DNAseI (Amersham Pharmacia Biotech, Uppsala, Sweden). DNAseI cleavage products were detected by PCR as described45 using the 32P-radiolabeled primer enhfor. Parallel sequencing of pKSenh with the radiolabeled primer enhfor was performed with the Sequenase kit (United States Biochemical, Cleveland, OH). The products of the footprint and sequencing reaction were detected on a sequencing gel, as described.45
Summary of the in vivo activity of different mutated enhancer constructs
| Mutated site enhancer . | Wild-type sequence . | Mutant sequence . | TG . | EC . | ET . |
|---|---|---|---|---|---|
| wt | wt | wt | 15 | 6 | 4 |
| GATA bp 43 | AGATAC | AGgggC | 10 | 3 | 3 |
| GATA bp 72 | TGATAA | TGgggA | 9 | 0 | 4 |
| GATA bp 264 | GGATAC | GGgggC | 9 | 5 | 2 |
| SCL/Tal-1 bp 232 | TCAGAT | TCcccT | 12 | 7 | 6 |
| SCL/Tal-1 bp 358 | GCAGAT | GCcccT | 11 | 5 | 5 |
| Ets bp 308 | AGGAAC | AcGcgC | 10 | 0 | 3 |
| Ets bp 260 | GTCCCG | GcgcgG | 13 | 4 | 0 |
| Mutated site enhancer . | Wild-type sequence . | Mutant sequence . | TG . | EC . | ET . |
|---|---|---|---|---|---|
| wt | wt | wt | 15 | 6 | 4 |
| GATA bp 43 | AGATAC | AGgggC | 10 | 3 | 3 |
| GATA bp 72 | TGATAA | TGgggA | 9 | 0 | 4 |
| GATA bp 264 | GGATAC | GGgggC | 9 | 5 | 2 |
| SCL/Tal-1 bp 232 | TCAGAT | TCcccT | 12 | 7 | 6 |
| SCL/Tal-1 bp 358 | GCAGAT | GCcccT | 11 | 5 | 5 |
| Ets bp 308 | AGGAAC | AcGcgC | 10 | 0 | 3 |
| Ets bp 260 | GTCCCG | GcgcgG | 13 | 4 | 0 |
The number of embryos is indicated. To exclude stage-specific effects, transgenic embryos from at least 2 different stages between E10 and E12 were analyzed. Wt indicates wild type; TG, number of transgenic embryos analyzed; EC, number of transgenic embryos showing endothelium-specific reporter gene expression; ET, number of transgenic embryos showing ectopic reporter gene expression. Embryos were transgenic for LacZ reporter gene constructs containing the 939-bp Flk-1 promoter and the 430-bp minimal enhancer bearing mutations in putative transcription factor binding sites. The position of putative transcription factor binding sites in the 430-bp enhancer is depicted in Figure 2.
Generation and analysis of transgenic mice
Results
A 430-bp minimal enhancer contained in the first intron ofFlk-1 is sufficient for endothelial cell–specific reporter gene expression
We have recently demonstrated that 5′-flanking sequences ofFlk-1 alone are not sufficient for endothelium-specific reporter gene expression in vivo.16 However, a 510-bpSwaI/BamHI fragment from the first intron of theFlk-1 gene conferred endothelium-specific gene expression to the 939-bp Flk-1 promoter in transgenic mouse embryos. The intron sequences acted as an endothelial cell–type specific autonomous enhancer. To characterize the minimal sequences contained in the 510-bp enhancer that are required for endothelium-specific reporter gene expression in vivo, we created LacZ reporter constructs containing the 939-bp Flk-1 promoter and subfragments of the 510-bp enhancer (Figure 1A,B). Transgenic mouse embryos were generated with the reporter gene constructs and were analyzed for LacZ expression during the critical phase of embryonic blood vessel growth between embryonic day 10 and day 12 (E10-E12). Among the enhancer subfragments tested (Figure 1C), only a 430-bp fragment could reproducibly target LacZ expression to vascular endothelial cells (Figure 1D; Table1). Reporter gene activity was observed in blood vessels that originated by vasculogenesis, such as the liver vasculature, and vessels that originated by angiogenesis, such as brain vessels (Figure 1D). The deletion of 186 bp spanning the 5′ end of the 430-bp fragment rendered the reporter gene construct inactive in vivo, as did the deletion of 109 bp at the 3′ end of the 430-bp fragment (Table 2), indicating a requirement of these regions for endothelial cell–specific transcription mediated by the Flk-1regulatory elements. Therefore, the regulatory elements sufficient for endothelium-specific reporter gene expression in transgenic mouse embryos are contained in a 430-bp minimal enhancer from theFlk-1 first intron.
Summary of the in vivo activity of Flk-1enhancer fragments
| Enhancer fragment (bp) . | TG . | EC . |
|---|---|---|
| 324 | 8 | 0 |
| 225 | 9 | 1 |
| 321 | 8 | 0 |
| 430 | 15 | 6 |
| Enhancer fragment (bp) . | TG . | EC . |
|---|---|---|
| 324 | 8 | 0 |
| 225 | 9 | 1 |
| 321 | 8 | 0 |
| 430 | 15 | 6 |
The number of embryos is indicated. Embryos were transgenic for LacZ reporter gene constructs containing the 939-bp Flk-1promoter spanning bp −640 to bp +299, and intron fragments depicted in Figure 1C. TG indicates number of transgenic embryos analyzed; EC, number of transgenic embryos showing endothelium-specific reporter gene expression.
Importance of SCL/Tal-1 and GATA transcription factor binding sites for the in vivo function of the 430-bp Flk-1minimal enhancer
One hallmark of the Flk-1 minimal enhancer is the presence of multiple putative binding sites for GATA and SCL/Tal-1 transcription factors (Figure 2). These factors have been proposed to play a role in vasculogenesis and angiogenesis21 24-28 and are therefore candidate regulators of Flk-1 expression. To study the function of these GATA and SCL/Tal-1 sites in Flk-1 expression, we mutated them individually in a LacZ reporter gene construct (Figure 1B) containing the 939-bp Flk-1 promoter and the 430-bp minimal enhancer and tested the reporter gene constructs in transgenic mouse embryos (Table 1). Based on microscopic inspection of whole-mount LacZ-stained embryos, the mutation of the SCL/Tal-1 binding motifs located at bp 232 or bp 358 of the minimal enhancer (Figure 2) led in both cases to nonhomogeneous and reduced reporter gene activity in E11-E12 transgenic embryos (Figure 3A-D) as compared with the activity of the wild-type construct (Figure 1D). To confirm that SCL/Tal-1 interacts with these sites in endothelial cells, we performed electrophoretic mobility shift assays using nuclear extracts from a mouse endothelioma cell line. Complexes formed when nuclear extracts were incubated with oligonucleotides spanning SCL/Tal-1 sites at bp 232 or at bp 358 (Figure4A,B). Complex formation was mediated by the SCL/Tal-1 sites because it could be inhibited by an excess of the wild-type but not the mutant oligonucleotides. Competition was also observed with a wild-type but not a mutant consensus SCL-binding site. Finally, when the binding reactions were incubated with an antiserum specific for mouse SCL/Tal-1, the predominant complex (Figure 4A,B; arrowheads) was supershifted (Figure 4A,B; asterisk). This indicates that SCL/Tal-1, by interacting with the SCL/Tal-1 sites in theFlk-1 minimal enhancer, contributes to a strong and uniform vascular reporter gene expression at least in midgestation mouse embryos. However, the frequency of transgenic embryos expressing LacZ in the vasculature and the endothelium specificity of reporter gene expression were not altered by any of the SCL/Tal-1 mutations (Table 1).
Nucleotide sequence and putative transcription factor binding sites of the 430-bp
Flk-1 enhancer. Sequences matching known transcription factor binding sites are underlined. The sequence of the 430-bp Flk-1 minimal enhancer is deposited in GenBank (accession number AF 153058).
Nucleotide sequence and putative transcription factor binding sites of the 430-bp
Flk-1 enhancer. Sequences matching known transcription factor binding sites are underlined. The sequence of the 430-bp Flk-1 minimal enhancer is deposited in GenBank (accession number AF 153058).
Mutational analysis of SCL/Tal-1 and GATA motifs in the
Flk-1 minimal enhancer in transgenic mouse embryos. Mutations were introduced into the indicated transcription factor sites shown in Figure 2, as presented in Table 1. The LacZ reporter gene is expressed under the control of the 939-bpFlk-1 promoter and mutants of the 430-bp minimal enhancer. Different representative transgenic embryos are shown after LacZ staining. (A,B) Mutation of the SCL/Tal-1 site (bp 232). (C,D) Mutation of the SCL/Tal-1 site (bp 358). (E) Mutation of the GATA site (bp 43). (F,G) Mutation of the GATA site (bp 72). (H) Mutation of the GATA site (bp 264).
Mutational analysis of SCL/Tal-1 and GATA motifs in the
Flk-1 minimal enhancer in transgenic mouse embryos. Mutations were introduced into the indicated transcription factor sites shown in Figure 2, as presented in Table 1. The LacZ reporter gene is expressed under the control of the 939-bpFlk-1 promoter and mutants of the 430-bp minimal enhancer. Different representative transgenic embryos are shown after LacZ staining. (A,B) Mutation of the SCL/Tal-1 site (bp 232). (C,D) Mutation of the SCL/Tal-1 site (bp 358). (E) Mutation of the GATA site (bp 43). (F,G) Mutation of the GATA site (bp 72). (H) Mutation of the GATA site (bp 264).
Electrophoretic mobility shift assays demonstrating specific binding of SCL/Tal-1 and a GATA factor to the
Flk-1 minimal enhancer. SCL/Tal-1–containing complexes interact with SCL/Tal-1 motifs at bp 232 (A) and at bp 358 (B), and a GATA factor interacts with the GATA motif at bp 72 (C) of the minimal enhancer. Antis indicates antiserum; comp, competitor oligonucleotide; W, wild-type; M, mutant.
Electrophoretic mobility shift assays demonstrating specific binding of SCL/Tal-1 and a GATA factor to the
Flk-1 minimal enhancer. SCL/Tal-1–containing complexes interact with SCL/Tal-1 motifs at bp 232 (A) and at bp 358 (B), and a GATA factor interacts with the GATA motif at bp 72 (C) of the minimal enhancer. Antis indicates antiserum; comp, competitor oligonucleotide; W, wild-type; M, mutant.
Mutation of the GATA sites located at bp 43 and bp 264 (Figure 2; Table 1) did not alter the reporter gene expression level or frequency (Figure 3E,H; Table 1). Hence, these sites are not required for the in vivo activity of the minimal enhancer at the developmental stages examined (E10-E12). In contrast, mutation of the GATA site located at bp 72 (Figure 2) led to a complete loss of the endothelium specificity of the minimal enhancer in vivo, as only a weak and ectopic reporter gene expression was observed (Figure 3F,G; Table 1). When nuclear extracts prepared from endothelioma cells were incubated with an oligonucleotide spanning this GATA motif, a complex was observed whose formation was inhibited by oligonucleotides containing a GATA consensus motif but not by an oligonucleotide containing the mutated GATA sequence (Figure 4C). Moreover, a specific shift was observed with extracts prepared from A293 cells transfected with an expression vector encoding GATA-2 (data not shown), a member of the GATA family that is highly expressed in endothelial cells. Thus, the GATA site at bp 72 of the minimal enhancer can functionally interact with GATA-2 or other members of the GATA family.
An Ets transcription factor binding site is required for the endothelium specificity of the Flk-1 minimal enhancer
To identify additional functional transcription factor binding sites on the Flk-1 minimal enhancer that specifically interact with nuclear proteins from endothelial cells, we performed a DNAseI footprint analysis of the Flk-1 minimal enhancer sequence. Nuclear extracts from BAE cells and, as a negative control, from TK1 lymphoma cells were tested for their ability to form nucleoprotein complexes with the 430-bp Flk-1 minimal enhancer. The only difference in the binding pattern of both nuclear extracts was observed on a putative Ets site located at bp 308 of the minimal enhancer (Figure 5A). Although proteins from both extracts protected the core sequence of this site from DNAseI digestion, only proteins from the BAE extract induced a DNAseI-hypersensitive site adjacent to the Ets site (Figure 5A). This indicates a cell type–specific difference in protein binding to the Ets site. To test whether this difference reflects a functional requirement of the Ets site for the endothelium specificity of the minimal enhancer, we mutated the site in a LacZ reporter gene construct containing the 939-bp Flk-1 promoter and the 430-bp minimal enhancer (Figure 1B). The analysis of embryos transgenic for this construct confirmed that the Ets site is required for the endothelium-specific Flk-1 expression, as the embryos expressed the reporter gene only ectopically (Figure 5B,C; Table 1). In contrast, the mutation of a putative Ets site located at bp 260 of the minimal enhancer (Figure 2) had no effect on the endothelium specificity of reporter gene expression (Figure 5D; Table 1).
The Ets site (bp 308) is required for the endothelium specificity of the
Flk-1 minimal enhancer. (A) DNAseI footprint. The 430-bp Flk-1 minimal enhancer subcloned into pBluescript KS was incubated with increasing amounts (triangles) of BSA or nuclear extracts from BAE or TK1 cells, and digested with DNAseI. The cleavage products were analyzed as described in “Materials and methods” on a sequencing gel. Sequencing reactions terminated at the indicated bases were loaded in parallel. The position of the Ets site (bp 308) of the minimal enhancer is indicated. Arrows indicate changes in the DNAseI cleaving pattern. HS indicates DNAseI hypersensitive site. (B,C) Different LacZ-stained mouse embryos transgenic for a construct containing the 939-bp Flk-1 promoter and the 430-bp minimal enhancer with a mutation in the Ets site at bp 308. (D) LacZ expression in a mouse embryo transgenic for a construct containing the 939-bpFlk-1 promoter and the 430-bp minimal enhancer with a mutation in the Ets site at bp 260.
The Ets site (bp 308) is required for the endothelium specificity of the
Flk-1 minimal enhancer. (A) DNAseI footprint. The 430-bp Flk-1 minimal enhancer subcloned into pBluescript KS was incubated with increasing amounts (triangles) of BSA or nuclear extracts from BAE or TK1 cells, and digested with DNAseI. The cleavage products were analyzed as described in “Materials and methods” on a sequencing gel. Sequencing reactions terminated at the indicated bases were loaded in parallel. The position of the Ets site (bp 308) of the minimal enhancer is indicated. Arrows indicate changes in the DNAseI cleaving pattern. HS indicates DNAseI hypersensitive site. (B,C) Different LacZ-stained mouse embryos transgenic for a construct containing the 939-bp Flk-1 promoter and the 430-bp minimal enhancer with a mutation in the Ets site at bp 308. (D) LacZ expression in a mouse embryo transgenic for a construct containing the 939-bpFlk-1 promoter and the 430-bp minimal enhancer with a mutation in the Ets site at bp 260.
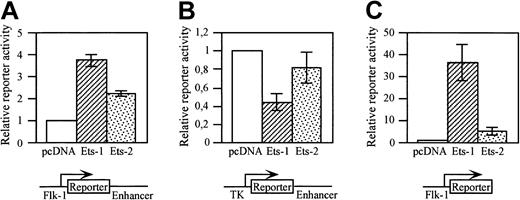
c-Ets1 is expressed in endothelial cells and activates the promoters of several endothelium-specific genes.19,20,34,47 In particular, c-Ets1, but also c-Ets2, activates the promoter of the VEGF receptor tyrosine kinase Flt-1in vitro.34 We therefore studied whether the putative Ets site located at bp 308 of the minimal enhancer can functionally interact with c-Ets1 or c-Ets2. A293 cells were cotransfected with a LacZ reporter gene construct containing the 939-bp Flk-1promoter and the minimal enhancer, and with expression vectors for c-Ets1 or c-Ets2, respectively, or pcDNA3 as control. Both Ets factors increased the reporter gene activity (Figure6A). However, when the Flk-1promoter sequences in the reporter gene construct were exchanged by the herpes simplex virus TK minimal promoter, the reporter gene activity was not increased by c-Ets1 or c-Ets2 in cotransfected A293 cells, but was decreased (Figure 6B). This indicates that c-Ets1 or c-Ets2 cannot activate the minimal enhancer in vitro.
The
Flk-1 promoter is activated by c-Ets1 and c-Ets2.A293 cells were cotransfected with LacZ (A,B) or luciferase (C) reporter gene constructs containing Flk-1 gene regulatory elements and with expression vectors for c-Ets1, c-Ets2, or pcDNA3 as control. The structures of the reporter gene constructs are shown in the bottom panels. (A) Cotransfection with a LacZ reporter gene construct containing the 939-bp Flk-1 promoter and the 430-bp minimal enhancer. (B) Cotransfection with a LacZ reporter gene construct containing the TK promoter and the 430-bp minimal enhancer. (C) Cotransfection with a luciferase reporter gene construct containing the 939-bp Flk-1 promoter. Relative promoter activities were determined as described in “Materials and methods.” The promoter activities of the control transfections were arbitrarily set at 1. The relative induction levels shown in A and C are not comparable because of the different stabilities of the different reporter gene products (β-galactosidase versus luciferase).
The
Flk-1 promoter is activated by c-Ets1 and c-Ets2.A293 cells were cotransfected with LacZ (A,B) or luciferase (C) reporter gene constructs containing Flk-1 gene regulatory elements and with expression vectors for c-Ets1, c-Ets2, or pcDNA3 as control. The structures of the reporter gene constructs are shown in the bottom panels. (A) Cotransfection with a LacZ reporter gene construct containing the 939-bp Flk-1 promoter and the 430-bp minimal enhancer. (B) Cotransfection with a LacZ reporter gene construct containing the TK promoter and the 430-bp minimal enhancer. (C) Cotransfection with a luciferase reporter gene construct containing the 939-bp Flk-1 promoter. Relative promoter activities were determined as described in “Materials and methods.” The promoter activities of the control transfections were arbitrarily set at 1. The relative induction levels shown in A and C are not comparable because of the different stabilities of the different reporter gene products (β-galactosidase versus luciferase).
Ets factors regulate Flk-1 promoter activity
We next investigated whether the Flk-1 promoter can be activated by these Ets factors. A293 cells were cotransfected with a luciferase reporter gene construct containing the 939-bpFlk-1 promoter including the 5′UTR, and with c-Ets1 or c-Ets2 expression plasmids or pcDNA3 as control. Both Ets factors increased the reporter gene activity of this construct (Figure 6C), indicating that the 939-bp Flk-1 promoter fragment contains functional Ets binding sites.
The analysis of this DNA sequence revealed 6 putative Ets binding sites in the 5′ flanking region of Flk-1 (Figure7A). To determine which of these putative binding sites are functional, we generated a series of 5′-end deletions of a Flk-1 promoter fragment extending from bp −640 to bp +16 and fused the resulting fragments to a luciferase reporter gene in the pGL-2 vector (Figure 7B). A293 cells were cotransfected with the promoter deletion constructs and with a c-Ets1 expression vector or pcDNA3, respectively. The deletion of the Ets sites E#3 and E#6 led to a significant drop of the c-Ets1–mediated stimulation of reporter gene activity (Figure 7B) from 13.15-fold to 6.64-fold and 5.31-fold to 3.5-fold, respectively. In contrast, deletion of the motifs E#1, E#2, E#4, and E#5 had no significant influence on the increase of reporter gene activity mediated by c-Ets1. To confirm that c-Ets1 can functionally interact with the c-Ets1 motifs E#3 and E#6, we individually mutated both sites in the Flk-1 promoter spanning bp −640 to bp +299. The wild-type or mutant promoter fragments were fused to the luciferase reporter gene in the pGL-2 vector (Figure 7C). A293 cells were cotransfected with these reporter gene constructs and with a c-Ets1 expression vector or pcDNA3. The 31.4-fold increase in promoter activity of the wild-type construct by c-Ets1 dropped to 17.47-fold in the case of the E#3 mutant and to 12.54-fold in the case of the E#6 mutant (Figure 7C). This indicates that both the E#3 and the E#6 Ets sites can functionally interact with c-Ets1.
The
Flk-1 promoter contains 2 functional Ets sites.(A) Nucleotide sequence and putative Ets binding sites of theFlk-1 promoter spanning bp −640 to +16. Sequences matching putative Ets binding sites are underlined. The nucleotide sequence of the Flk-1 promoter spanning bp −640 to bp +299 is deposited in GenBank (accession number AF 153057). (B) 5′-Deletion analysis of the Flk-1 promoter. Luciferase reporter gene constructs were cotransfected with either a c-Ets1 expression vector (black bars) or pcDNA3 (gray bars) as control. Values that are significantly (P < .05, Student t test) below the corresponding values of the previous construct are marked with an asterisk. (C) Mutational analysis of the Ets sites E#3 and E#6. Luciferase reporter gene constructs containing the 939-bpFlk-1 promoter with the wild-type sequence or mutations in the Ets sites E#3 or E#6 (black cross) were cotransfected with either a c-Ets1 expression vector (black bars) or pcDNA3 (gray bars). The sequence 5′-CGGA-3′ of the Ets sites E#3 or E#6 (see Figure 6) was mutated to 5′-CccA-3′. The activation of both mutant promoters by c-Ets1 was significantly lower (P < .05; Studentt test) than the activation of the wild-type promoter by c-Ets1. The promoter activities of the pcDNA3 transfections in A and B were arbitrarily set at 1, and promoter activities were determined as described in “Materials and methods.” The relative induction levels shown in B and C are not directly comparable because the 5′-UTR is lacking in the constructs used for the analysis in B.
The
Flk-1 promoter contains 2 functional Ets sites.(A) Nucleotide sequence and putative Ets binding sites of theFlk-1 promoter spanning bp −640 to +16. Sequences matching putative Ets binding sites are underlined. The nucleotide sequence of the Flk-1 promoter spanning bp −640 to bp +299 is deposited in GenBank (accession number AF 153057). (B) 5′-Deletion analysis of the Flk-1 promoter. Luciferase reporter gene constructs were cotransfected with either a c-Ets1 expression vector (black bars) or pcDNA3 (gray bars) as control. Values that are significantly (P < .05, Student t test) below the corresponding values of the previous construct are marked with an asterisk. (C) Mutational analysis of the Ets sites E#3 and E#6. Luciferase reporter gene constructs containing the 939-bpFlk-1 promoter with the wild-type sequence or mutations in the Ets sites E#3 or E#6 (black cross) were cotransfected with either a c-Ets1 expression vector (black bars) or pcDNA3 (gray bars). The sequence 5′-CGGA-3′ of the Ets sites E#3 or E#6 (see Figure 6) was mutated to 5′-CccA-3′. The activation of both mutant promoters by c-Ets1 was significantly lower (P < .05; Studentt test) than the activation of the wild-type promoter by c-Ets1. The promoter activities of the pcDNA3 transfections in A and B were arbitrarily set at 1, and promoter activities were determined as described in “Materials and methods.” The relative induction levels shown in B and C are not directly comparable because the 5′-UTR is lacking in the constructs used for the analysis in B.
To examine whether the Ets sites E#3 and E#6 contribute to the function of the Flk-1 promoter during embryonic development, we individually mutated both sites in a LacZ reporter gene construct containing the Flk-1 promoter spanning bp −640 to bp +299 and the 430-bp minimal enhancer. Transgenic embryos were generated with the mutant or wild-type constructs, and the LacZ expression in transgenic embryos was analyzed at E10.5 or E11.5 (Table3). When compared with the construct containing the wild-type promoter (Figure8A), the mutation of the Ets site E#3 resulted in a reduced reporter gene activity in the vasculature of transgenic embryos (Figure 8B). In contrast, mutation of the Ets site E#6 did not alter the LacZ expression level at the developmental stages examined (Figure 8C). These results indicate that the Ets site E#3 in the Flk-1 promoter is required for high-level endothelial gene expression during embryonic development.
In vivo effect of Ets-site mutations in theFlk-1 promoter
| Mutated site . | Wild-type sequence . | Mutant sequence . | TG . | EC . |
|---|---|---|---|---|
| wt | wt | wt | 15 | 6 |
| E#3 | TTCCGG | TTggGG | 14 | 4 |
| E#6 | CCGGAC | CCccAC | 13 | 7 |
| Mutated site . | Wild-type sequence . | Mutant sequence . | TG . | EC . |
|---|---|---|---|---|
| wt | wt | wt | 15 | 6 |
| E#3 | TTCCGG | TTggGG | 14 | 4 |
| E#6 | CCGGAC | CCccAC | 13 | 7 |
The number of embryos is indicated. Embryos were transgenic for LacZ reporter gene constructs containing the 939-bp Flk-1promoter with mutations in the indicated Ets sites and the 430-bp minimal enhancer. Abbreviations are shown in Table 2. The position of the Ets sites in the Flk-1 promoter is depicted in Figure 6.
The Ets site E#3 is required for the activity of the
Flk-1 promoter in transgenic mouse embryos. (A) An embryo transgenic for a construct containing the 939-bpFlk-1 promoter and the 430-bp minimal enhancer, showing a uniform and strong vascular LacZ expression. (B,C) LacZ expression in mouse embryos transgenic for constructs containing the 939-bpFlk-1 promoter with a mutation in either the Ets site E#3 (B) or in the Ets site E#6 (C), and the 430-bp minimal enhancer.
The Ets site E#3 is required for the activity of the
Flk-1 promoter in transgenic mouse embryos. (A) An embryo transgenic for a construct containing the 939-bpFlk-1 promoter and the 430-bp minimal enhancer, showing a uniform and strong vascular LacZ expression. (B,C) LacZ expression in mouse embryos transgenic for constructs containing the 939-bpFlk-1 promoter with a mutation in either the Ets site E#3 (B) or in the Ets site E#6 (C), and the 430-bp minimal enhancer.
Discussion
Flk-1 is the first endothelial receptor tyrosine kinase known to be expressed on endothelial cell precursors and plays a central role in regulating early embryonic vascular development and tumor angiogenesis. Upstream regulators of Flk-1 can therefore be considered as master regulators of vasculogenesis and angiogenesis. To elucidate the transcriptional control mechanisms that regulate the cell type specificity of Flk-1 expression, we have characterized the gene regulatory elements of Flk-1 in vivo that were isolated in a previous study.16 A 430-bp fragment of theFlk-1 intron was sufficient to confer endothelium-specific reporter gene expression to the 939-bp Flk-1 promoter. Deletions on both ends of this fragment abolished the cell type–specific enhancer activity, indicating that a complex composition of transcription factor binding sites is present within this fragment. In combination with the 939-bp Flk-1 promoter, the 430-bp minimal enhancer could target reporter gene expression to blood vessels that originated either by vasculogenesis or by angiogenesis. This suggests that the expression of Flk-1 is mediated by the same regulatory elements during both processes of blood vessel formation.
Despite the strong activity of the Flk-1 enhancer in the endothelium of transgenic mice, in which the enhancer is integrated into chromatin, this element exhibits only little activity in transiently transfected endothelial cells.16 It therefore appears that the Flk-1 enhancer belongs to a class of enhancers that need to be integrated into chromatin to exhibit their activity. Transcription factors binding to this class of enhancers, which includes the CD34 and MyoD enhancers,48,49 act by influencing the chromatin structure rather than by interacting with the basal transcription machinery, the model described for classic enhancers.50 Transcription factors that bind to theFlk-1 enhancer are therefore expected to cause only little, if any, increase in the Flk-1 enhancer activity in transiently transfected cells, making it difficult to study the transcription factor requirement of the Flk-1 enhancer by transient cotransfections. In contrast, the in vivo analysis of mutations in transcription factor binding sites of the Flk-1enhancer provides a powerful approach to study the function of transcription factors in Flk-1 gene expression.
Two binding sites for the transcription factor SCL/Tal-1 were identified as important elements of the 430-bp minimal enhancer because their individual mutations resulted in a decreased reporter gene activity and nonhomogeneous LacZ expression pattern in the vasculature of transgenic mouse embryos. These results suggest that the function of the SCL/Tal-1 sites and their interacting factor is to increase the transcriptional activity of Flk-1 and that the minimal enhancer is probably active in a given endothelial cell. This hypothesis is supported by the observations that the overexpression of SCL/Tal-1 increases the number of Flk-1–expressing endothelial precursor cells in transgenic zebrafish embryos and that SCL/Tal-1 specifies vascular progenitors in zebrafish.26,27 SCL/Tal-1 has also been suggested to function during hemangioblast differentiation in mice because SCL/Tal-1 is expressed in both endothelial and blood cell precursors in blood islands, and in hemangioblasts.11,25 However, although yolk sac vascularization is defective in SCL/Tal-1−/−mice, endothelial cell differentiation proceeds normally.28 It has therefore been suggested that a functional redundancy of SCL/Tal-1–related factors might explain the differentiation of endothelial cells in SCL/Tal-1−/−mice.35 37 Our results provide the first direct evidence that SCL/Tal-1 acts upstream of Flk-1 in higher vertebrate embryos via the Flk-1 minimal enhancer.
The mutation of a GATA site rendered the Flk-1 minimal enhancer completely inactive, as the mutant enhancer could not confer endothelium-specific reporter gene expression in transgenic mouse embryos. We provide evidence that a GATA factor can functionally interact with this GATA site in vitro. This suggests that GATA factors determine the cell type specificity of Flk-1 expression. GATA factors were initially identified as regulators of the hematopoietic program.35 However, the GATA factors GATA-2, GATA-3, GATA-4, and GATA-5 are also expressed in endothelial cells.21,51-53 In addition, GATA-2 has been demonstrated to activate the promoters of endothelium-specific genes for endothelin-1 and PECAM-1 in vitro.29-31 Therefore, GATA-2 has been suggested to play a role in endothelial cell differentiation. However, no endothelial target genes of GATA factors have been identified in vivo. To our knowledge, our data provide the first evidence that Flk-1 is a target gene for GATA factors during murine embryonic vascular development.
In addition to the functional GATA site, the endothelium specificity of the Flk-1 minimal enhancer is also determined by an Ets site. As is the case for Flk-1, the in vivo activities of the endothelium-specific regulatory elements ofTie-1 and Tie-2 are also dependent on Ets sites.32,33 This suggests a common requirement for Ets sites in endothelial gene expression. Several Ets factors such as c-Ets1, NERF, and Fli-1 are highly expressed in endothelial cells19,20,54 55 and are therefore candidate regulators of endothelial genes such as Flk-1. However, the nature of the factor that activates the minimal enhancer via the Ets site remains unknown.
In contrast to the minimal enhancer, the Flk-1 promoter was activated by c-Ets1 via 2 Ets sites in vitro. However, only one of these Ets sites was required for high-level reporter gene expression during embryonic development. Our findings do not preclude the possibility that the other functional Ets site may be functional during other developmental stages or under pathologic conditions such as tumor angiogenesis, in which c-Ets1 is highly expressed in endothelial cells.56 c-Ets1 is coexpressed with Flk-1 in blood islands and blood vessels during embryonic development.19,20 Our experiments suggest that the high activity of the Flk-1 promoter in transfected endothelial cells15 and in vivo16 is in part determined by the 2 functional Ets sites in the Flk-1 promoter. This hypothesis is supported by our finding that the individual mutation of both Ets sites led to a reduction of Flk-1 promoter activity in transfected endothelial cells (A. Kappel, unpublished observations).
c-Ets1 has also been shown to activate the promoter ofFlt-134 and the promoters of proteases that are expressed by endothelial cells during angiogenesis, such as urokinase-type plasminogen activator and matrix metalloproteinase-1.57 Because VEGF receptors and proteases are required for angiogenesis, these data suggest that c-Ets1 or related factors regulate the expression of a group of genes that convert endothelial cells from a resting to an angiogenic phenotype.
Loss-of-function experiments failed to address a function of c-Ets1 and GATA transcription factors in the embryonic vasculature, probably because of the redundant function of other members of these transcription factor families expressed in endothelial cells.35-37 This redundant expression of transcription factors with similar functions makes it difficult to address the role of a single family member in the regulation of a target gene by loss-of-function experiments. In contrast, the mutation of specific binding sites for these transcription factor families in endothelial target genes such as Flk-1 prevents the action of all family members. The in vivo analysis of mutated transcription factor binding sites in the regulatory elements of key regulator genes of vasculogenesis and angiogenesis, such as Flk-1, therefore provides a valuable tool to study the role of transcription factor families during endothelial cell differentiation and vascular development.
Our results provide the first direct evidence that GATA, Ets, and SCL/Tal-1 transcription factors are not only key regulators of hematopoiesis,35-37 but also specify endothelial cell differentiation and vascular development by regulating Flk-1expression. Hence, similar transcriptional programs might regulate the establishment of the endothelial and blood cell lineages during embryonic development, reflecting a common origin of endothelial and blood cells.
Acknowledgments
We thank Dr Catherine Porcher for providing SCL/Tal-1 antiserum; Dr Dietmar von der Ahe, Haemostasis Unit, Kerckhoff Klinik, Bad Nauheim, Germany, for kindly providing pSG5c-Ets1p68; and Dr Felix Müller-Holtkamp and Michael Walker for generating transgenic mice.
Supported in part by the Bundesministerium für Bildung und Forschung, Deutsche Krebshilfe, Sonderforschungsbereich 397, and the Howard Hughes Medical Institute.
In memoriam Werner Risau (1953-1998).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Georg Breier, Max-Planck-Institute for Physiological and Clinical Research, Parkstrasse 1, 61231 Bad Nauheim, Germany; e-mail: g.breier@kerckhoff.mpg.de.







This feature is available to Subscribers Only
Sign In or Create an Account Close Modal