Abstract
Chronic granulomatous disease (CGD) is a primary immunodeficiency caused by defects in any one of 4 genes encoding phagocyte NADPH oxidase subunits. Unlike other CGD subtypes, in which there is great heterogeneity among mutations, 97% of affected alleles in patients previously reported with A470 CGD carry a single mutation, a GT deletion (ΔGT) in exon 2 of the p47-phox gene, NCF-1. This unusually high incidence results from recombination events between NCF-1and its highly homologous pseudogenes, in which ΔGT originates. In 50 consecutive patients with A470 CGD, 4 were identified who were heterozygous for ΔGT in NCF-1, and for the first time, 2 were identified whose DNA appeared normal at this position. To avoid co-amplification of pseudogene sequence and to enable the identification of mutations in these patients, allele-specific polymerase chain reaction was used to amplify alleles not containing ΔGT. In each of the 4 patients who were heterozygous for ΔGT, an additional novel mutation was identified. These were 2 missense mutations, G125 → A in exon 2 (predicting Arg42 → Gln) and G784 → A in exon 8 (Gly262 → Ser), and 2 splice junction mutations at the 5′ end of intron 1, gt → at and gtg → gtt. The first of 2 patients who appeared normal at the GT position was a compound heterozygote with the G125 → A transition on one allele and a deletion of G811 on the other. In the second of these patients, only a single defect was detected, G574 → A, which predicts Gly192 → Ser but is likely to result in defective splicing because it represents the final nucleotide of exon 6.
Introduction
Chronic granulomatous disease (CGD) is an uncommon inherited disorder of the innate immune system arising from defects in any of 4 genes encoding protein components of the phagocyte NADPH oxidase. In stimulated normal phagocytes, this enzyme system catalyzes the one-electron reduction of oxygen to form superoxide. Superoxide itself has little microbicidal activity, but its toxic derivatives (eg, hydrogen peroxide, hypohalous acids, and hydroxyl radical) are potent microbicides and are essential for killing many invading microorganisms. CGD is characterized by an absence or, more rarely, very low levels of superoxide production by the patient's phagocytes. Affected patients consequently have recurrent, sometimes fatal, bacterial and fungal infections. The incidence of CGD is approximately 1 in 250 000 persons and is normally diagnosed in infancy or early childhood.1
NADPH oxidase is composed of at least 5 unique protein components. The electron transporting center of the oxidase, flavocytochromeb558, consists of 2 integral membrane proteins, p22-phox and gp91-phox, localized to the plasma membrane and the membranes of specific granules. Three soluble proteins, p40-phox, p47-phox, and p67-phox, are found in the cytosol of resting phagocytes as a multiprotein complex, probably associated with the submembranous cytoskeleton (reviewed in DeLeo and Quinn2 and Robinson and Badwey3). Activation of electron flow from NADPH through the FAD and heme groups of the flavocytochrome also requires the guanosine triphosphate-binding protein Rac (Rac1 or Rac2) and involves the association of the soluble components with the flavocytochrome subunits through multiple protein–protein interactions (reviewed in Clark4).
Approximately 65% of CGD is inherited in an X-linked manner and is caused by mutations in CYBB, the gene encoding the gp91-phox subunit of flavocytochromeb558. The remaining patients inherit the disease in an autosomal recessive mode, through mutations in the genes for p22-phox (CYBA), p47-phox (NCF-1), or p67-phox (NCF-2).1,5 6 The 4 forms of the disease are referred to as X91, A22, A47, and A67 CGD, with addition of the superscripts +, −, or 0 to indicate a normal level, a reduced level, or an absence of the affected oxidase component, respectively.
Mutations in NCF-1, the gene for p47-phox, account for approximately 23% of all CGD cases.1 In contrast to X91, A22, and A67 CGD, in which there is a high degree of heterogeneity among mutations, many of them family-specific,5-9 a single common mutation has been reported in 60 patients worldwide with A470 CGD and was identified in 97% of the affected alleles.5,10-13Fifty-six (93%) of these persons were homozygous for a dinucleotide deletion (ΔGT) at a GTGT repeat at the beginning of exon 2, which predicts a frameshift and premature stop codon at amino acid 51.10 In one patient who was heterozygous for ΔGT, a single nucleotide deletion (G502) was also observed and remains the only non-ΔGT mutation reported in A470 CGD before this study.11
The reason for the predominance of the ΔGT mutation in A470 CGD became clearer when it was discovered that at least 2 p47-phox pseudogenes exist, each of which contains the GT deletion. The pseudogenes (referred to here as ψNCF-1) co-localize with the functional p47-phox gene to chromosome 7q11.23 and, in addition to the GT deletion, contain 9 exonic single base pair differences to the functional gene and at least 140 intronic differences.14The presence of pseudogenes closely linked to the functionalNCF-1 gene, together with the presence within each gene of multiple recombination hot spots (eg, Alu repeats, Chi sequence, and human mini-satellite repeats) suggests that the predominance of the ΔGT mutation in A470 CGD is caused by recombination events between the p47-phox functional gene and its pseudogenes.13 14
While performing genetic analysis of a series of 50 consecutive A470 CGD patients (28 of the patients were included in a recent study of the pseudogene13), we identified 6 who showed a normal sequence and pseudogene sequence at the beginning of exon 2 and, therefore, clearly were not homozygous for the GT deletion. Because identification of the specific mutations in these persons provides the only basis for detecting carriers among siblings and other family members and for performing prenatal diagnosis, we undertook this study to identify mutations in the uncharacterized alleles. Furthermore, knowledge of the mutations in NCF-1 can provide insights into the structure and function of p47-phox and the complex relation between the gene and its pseudogenes.13
Patients, materials, and methods
CGD patients and families
Blood samples were obtained from CGD patients and their family members with appropriate institutional consent and were sent by the referring physicians to the investigators at The Scripps Research Institute. Protocols and consent forms for the collection of control blood samples were approved by the Human Subjects Committee of the Scripps Office for the Protection of Research Subjects.
Patient 1 is a 25-year-old woman. Her early clinical history and diagnosis have been documented in a previous study, in which her phagocyte NADPH oxidase was reported to have a diminished affinity for its substrate.15 Subsequent biochemical studies localized the defect to a cytosolic component of the oxidase (patient 5 in Curnutte et al16), and we have demonstrated the absence of p47-phox by immunoblotting.
Patient 2 is the 22-year-old daughter of unrelated parents of European origin. She was diagnosed with CGD at age 9 years by a negative NBT test and the absence of superoxide generation. Her CGD has followed a relatively mild clinical course, with a history of recurrent oral ulcers, recurrent skin infections, and abscesses. Her neutrophils contain normal levels of flavocytochrome b558 by spectroscopy and p67-phox by immunoblot but showed a deficiency of p47-phox in a cell-free complementation assay. This was confirmed in this study by immunoblotting.
Patient 3 was diagnosed with CGD by a negative NBT test and extremely low superoxide production (approximately 3% of normal) in a whole cell, cytochrome c assay.17 He died at the age of 2 years from pneumonia caused by Aspergillus. Immunoblots of neutrophils, purified with very low yield from blood collected immediately after death, were uninformative because of proteolysis (only p22-phox could be detected with any degree of certainty). Analysis of his genomic DNA failed to reveal any abnormalities in the entire coding regions of CYBA,CYBB, and NCF-2, leading us to believe that he had A470 CGD. This was confirmed by sequencing the p47-phox gene (see below).
Patient 4 is the 6-year-old son of unrelated Hispanic parents, with no known family history of the disease. He was diagnosed with CGD at age 5 years after he was brought for treatment of a disseminatedNocardia infection. The patient's parents and 2 healthy male siblings had normal NADPH oxidase activity as measured by flow cytometry. The patient's phagocytes had normal levels of gp91-phox, p22-phox, and p67-phox, but an absence of p47-phox.
Patient 5 is the 20-year-old son of unrelated Hispanic parents. He was diagnosed with CGD at 18 months after having recurrent fevers and right upper lobe pneumonia. Subsequently, he has had otitis media, Aspergillus pneumonia, stomatitis, and viral bronchitis. Findings on the NBT slide test were negative, and his intact neutrophils failed to generate superoxide (patient 7 in Curnutte et al16). This study also showed a normal level of flavocytochrome b558 in his neutrophils and localized the defect to the cytosol. The absence of p47-phoxwas demonstrated by immunoblotting.
Patient 6 is a male of Pakistani origin. He was first studied at the age of 12 years, after diagnosis of CGD by the absence of reactivity in an NBT assay. His mother and 2 male siblings had normal findings by NBT test. A flow cytometric assay using DCF revealed a very low level of hydrogen peroxide production by the patient's PMA-stimulated neutrophils compared to normal control cells. This is consistent with a deficiency of p47-phox, which was subsequently confirmed by immunoblotting of neutrophil extracts. The patient had normal levels of p67-phox by immunoblot and flavocytochromeb558 by spectrophotometry.
Preparation of neutrophils, functional assays, and immunoblotting
Neutrophils were purified from peripheral blood by dextran sedimentation, hypotonic lysis of erythrocytes, and centrifugation through Ficoll-Hypaque.18 The NBT slide test, measurement of superoxide generation, and determination of flavocytochromeb558 were performed as previously described.19 Protein immunoblotting and preparation of antibodies specific for each of the NADPH oxidase components have also been described elsewhere.8,19 20
Preparation of DNA and oligonucleotide primers
Genomic DNA was isolated from whole blood stored in EDTA using the Puregene DNA Isolation Kit (Gentra Systems, Minneapolis, MN) or an Applied Biosystems DNA extractor. Custom synthesized oligonucleotide primers were purchased from Sigma-Genosys and are listed in Table1.
Primers used for PCR amplification of NCF-1
| Primers for gDNA analysis . | Primers for cDNA analysis . |
|---|---|
| 5′L gcctggataacaaagcaaga | cDNA1F ATGGGGGACACCTTCATCCGTCACA |
| 5′L2 gaactgtgatccaccactgcact | cDNA1R AGGGCGATGTGACGGATGAAGGT |
| 5′L3 cggaagaggtgacatttga | cDNA3F GAGGCAGGGGCGATCAATC |
| 5′R1 ggaaaagggtttacacttgt | cDNA4R GTCATCAGGGCGCACCTTGAA |
| il-3′F ggtccacgtttgtgccct | cDNA6F CCCATCATCCTGCAGACGTA |
| 1L agcgacttcctctttcca | cDNA7F GTGGTTCTGTCAGATGAAAGC |
| 1R ctgtccctcccgtattcc | cDNA7R CTCAGGGTCTTCCGTCTCGT |
| 2LA caatcctctgggcttcct | cDNA9F GCCCAACGCCAGATCAAGC |
| 2RA ccctttaactgtggtctgg | cDNA9R CCCCGCTTGATCTGGCGTTG |
| 2LB2 gtgcacacagcaaagcctct | cDNA11R CAGACGCCAGCTTCCGCTTG |
| 2RB2 ctaaggtccttcccaaagggt | |
| 3L ccaatctcgtgcttttccaa | |
| 3R gccaatgaccccctgaca | |
| 4L ccctctcgggcttgacct | |
| 4R3 gcaaaacacagaaagtccca | |
| 5L ccagtgcctcttcttcctc | |
| 5R ggtctcctcagggtgtcc | |
| 6L ggcagcagactcaagatg | |
| 6R cttgtggctgtgggttcc | Allele-specific primers |
| 7L gcccctcagtcacattcc | |
| 7R ctcccacacccctacagc | GTGT ctttcccccagGTGTACAT gDNA |
| 8L gctgggccgacctcacact | GTGT-R TCACCAGGAACATGTACAC gDNA/cDNA |
| 8R ctggctggggctgactca | cDNAGTGT CCCAGCCAGCACTATGTGT cDNA |
| 9LA cagaggcaggtgtccggttg | |
| 9RA tgcctctcctgccaggctga | |
| 9L gccctgaaaccctctcctc | |
| 9R cagagcgaacccgtggtg | |
| 10L ggggtgcccatctgagtcc | |
| 10RA tccctctgattccgccctc | |
| 10R ggtcccgcccgctactct | |
| 11LA agagcgcggagcaggagtt | |
| 11LB2 agcgctgtgggcggggccagtgtg | |
| 11R gctgccctcggcgtccag |
| Primers for gDNA analysis . | Primers for cDNA analysis . |
|---|---|
| 5′L gcctggataacaaagcaaga | cDNA1F ATGGGGGACACCTTCATCCGTCACA |
| 5′L2 gaactgtgatccaccactgcact | cDNA1R AGGGCGATGTGACGGATGAAGGT |
| 5′L3 cggaagaggtgacatttga | cDNA3F GAGGCAGGGGCGATCAATC |
| 5′R1 ggaaaagggtttacacttgt | cDNA4R GTCATCAGGGCGCACCTTGAA |
| il-3′F ggtccacgtttgtgccct | cDNA6F CCCATCATCCTGCAGACGTA |
| 1L agcgacttcctctttcca | cDNA7F GTGGTTCTGTCAGATGAAAGC |
| 1R ctgtccctcccgtattcc | cDNA7R CTCAGGGTCTTCCGTCTCGT |
| 2LA caatcctctgggcttcct | cDNA9F GCCCAACGCCAGATCAAGC |
| 2RA ccctttaactgtggtctgg | cDNA9R CCCCGCTTGATCTGGCGTTG |
| 2LB2 gtgcacacagcaaagcctct | cDNA11R CAGACGCCAGCTTCCGCTTG |
| 2RB2 ctaaggtccttcccaaagggt | |
| 3L ccaatctcgtgcttttccaa | |
| 3R gccaatgaccccctgaca | |
| 4L ccctctcgggcttgacct | |
| 4R3 gcaaaacacagaaagtccca | |
| 5L ccagtgcctcttcttcctc | |
| 5R ggtctcctcagggtgtcc | |
| 6L ggcagcagactcaagatg | |
| 6R cttgtggctgtgggttcc | Allele-specific primers |
| 7L gcccctcagtcacattcc | |
| 7R ctcccacacccctacagc | GTGT ctttcccccagGTGTACAT gDNA |
| 8L gctgggccgacctcacact | GTGT-R TCACCAGGAACATGTACAC gDNA/cDNA |
| 8R ctggctggggctgactca | cDNAGTGT CCCAGCCAGCACTATGTGT cDNA |
| 9LA cagaggcaggtgtccggttg | |
| 9RA tgcctctcctgccaggctga | |
| 9L gccctgaaaccctctcctc | |
| 9R cagagcgaacccgtggtg | |
| 10L ggggtgcccatctgagtcc | |
| 10RA tccctctgattccgccctc | |
| 10R ggtcccgcccgctactct | |
| 11LA agagcgcggagcaggagtt | |
| 11LB2 agcgctgtgggcggggccagtgtg | |
| 11R gctgccctcggcgtccag |
Initial genotyping of A470 CGD patients
For the initial molecular analysis of all A470 CGD patients, we amplified and sequenced exon 2 from genomic DNA, using primers 2LB2 and 2RB2, to determine whether they had the prevalent ΔGT/ΔGT genotype. For this polymerase chain reaction (PCR), an initial denaturation for 3 minutes at 94°C was followed by 30 cycles of 94°C for 30 seconds, 62°C for 30 seconds, and 72°C for 30 seconds, with a 7-minute extension at 72°C.
Allele-specific PCR and sequencing
To overcome problems resulting from the presence of co-amplified p47-phox pseudogenes, which lead to a high ratio of pseudogene-to-functional gene sequence when analyzing fragments amplified from genomic DNA, we used an allele-specific PCR strategy to amplify only alleles containing GTGT at the start of exon 2. Reactions were performed using the Expand Long Template system (Roche Molecular Biochemicals, Indianapolis, IN) in a GeneAmp PCR system 9600 (Perkin Elmer/ABI, Norwalk, CT). The p47-phoxfunctional gene alleles (non-ΔGT containing) were amplified using 3 allele-specific reactions (Figure 1). DNA from a homozygous ΔGT patient was used as a negative control in each set of allele-specific PCRs to verify that no ΔGT product was obtained.
Location of oligonucleotide primers used for allele-specific amplification of
NCF-1. Solid arrows below the gene indicate the direction and approximate position of primers used for allele-specific amplification of the p47-phox gene. Broken arrows indicate primers used for sequencing to ensure that only GTGT-containing alleles had been amplified. The 2 mutations shown above the gene, C → T in intron 1 and GTGT → GT in exon 2, are specific for ψNCF-1 (see text).
Location of oligonucleotide primers used for allele-specific amplification of
NCF-1. Solid arrows below the gene indicate the direction and approximate position of primers used for allele-specific amplification of the p47-phox gene. Broken arrows indicate primers used for sequencing to ensure that only GTGT-containing alleles had been amplified. The 2 mutations shown above the gene, C → T in intron 1 and GTGT → GT in exon 2, are specific for ψNCF-1 (see text).
The first PCR encompassed exon 1 through the last nucleotide of intron 1 and was accomplished using a forward primer 5′ of exon 1 (1 L; Table 1) and a reverse allele-specific primer from exon 2 (GTGT-R). Denaturation for 3 minutes at 95°C was followed by 10 cycles of amplification at 94°C for 10 seconds, 58°C for 30 seconds, 68°C for 2 minutes 15 seconds, and 20 cycles at 94°C for 10 seconds, 58°C for 30 seconds, and 68°C for 2 minutes 15 seconds with cycle elongation of 20 seconds per cycle. A final extension was performed for 7 minutes at 68°C. The success of the allele specificity of the amplification was checked by sequencing the reaction product with a forward primer 5′ of exon 2 (i1-3′F) into the end of the allele-specific primer sequence to confirm that only the GTGT-containing sequence was present and that there was a C rather than a T in intron 1, 122 bp upstream from the 5′ end of exon 2. C and T at this position are specific for NCF-1 and ψNCF-1, respectively.13
The second PCR used an allele-specific forward primer (GTGT) and a reverse intron 4 primer (4R3), covering the sequence from 4 bp downstream of the GTGT at the start of exon 2 and extending into intron 4. An initial denaturation for 3 minutes at 94°C, was followed by 10 cycles at 94°C for 30 seconds, 60°C for 30 seconds, 68°C for 1 minute 30 seconds, then 20 cycles of 94°C for 30 seconds, 60°C for 30 seconds, and 68°C for 1 minute 30 seconds plus cycle elongation of 20 seconds per cycle. A final extension was performed for 7 minutes at 68°C. The allele specificity of this amplification was confirmed by sequencing with a reverse primer from intron 2 (2RB2) through the allele-specific primer sequence to ensure that no ΔGT-containing sequence was present. Exons 2 through 4 were sequenced directly in this DNA fragment. To check for mutations in the 4 bp immediately following GTGT, the sequence of the PCR product used for the initial genotyping (see above) was examined.
The third PCR also used the primer GTGT, but with a reverse exonic primer from exon 11 (cDNA11R), covering the sequence downstream from the start of exon 2 and extending partially into exon 11. An initial denaturation for 3 minutes at 94°C was followed by 10 cycles at 94°C for 15 seconds, 65°C for 30 seconds, 68°C for 6 minutes 45 seconds, then 20 cycles of 94°C for 15 seconds, 65°C for 30 seconds, 68°C for 6 minutes 45 seconds plus cycle elongation of 20 seconds per cycle. A final extension was performed for 7 minutes at 68°C. The allele specificity of the reaction was confirmed with the primer 2RB2, as above. The large size of this PCR product made direct sequencing of the remaining exons difficult; therefore one-fiftieth completed reaction mix was used as a template for amplification of individual exons 4 through 10. (Exon 4 was amplified to check that this nested PCR gave the same result as direct sequencing of the GTGT/4R3 product.) These PCR reactions were performed using AmpliTaq DNA polymerase with 10× buffer II (Perkin Elmer/ABI) in 2.5 mM MgCl2, 0.125 mM each dNTP, 90 ng each primer, and 2.5 U AmpliTaq. Amplification reactions were performed under the following conditions: an initial denaturation for 3 minutes at 94°C was followed by 30 cycles at 95°C for 30 seconds, 60°C for 15 seconds, and 72°C for 15 seconds, followed by a 7-minute extension at 72°C.
Allele-specific RT-PCR
In some instances, total RNA was isolated from whole blood using the RNeasy Blood Mini Kit (Qiagen, Valencia, CA) and reverse transcription (RT)-PCR was performed using the SuperScript Preamplification System for first-strand cDNA synthesis (Life Technologies, Grand Island, NY). Two allele-specific PCRs were performed. For the first, from exon 1 to exon 2, we used an exon 1 primer (cDNA1F) and the same reverse primer as for genomic DNA (GTGT-R). An initial denaturation for 3 minutes at 94°C was followed by 30 cycles at 94°C for 15 seconds, 58°C for 15 seconds, and 72°C for 15 seconds, followed by a 7-minute extension at 72°C. For the second RT-PCR, from exon 2 to exon 11, we used a different forward allele-specific primer than for the genomic PCR (cDNAGTGT) but the same exon 11 reverse primer. An initial denaturation for 3 minutes at 95°C was followed by 10 cycles of 94°C for 15 seconds, 60°C for 30 seconds, 68°C for 50 seconds, then 20 cycles of 94°C for 15 seconds, 60°C for 30 seconds, and 68°C for 50 seconds plus cycle elongation of 20 seconds per cycle. This was followed by a 7-minute extension at 68°C. RT-PCR products were checked for allele specificity by sequencing into the allele-specific primer sequences to ensure that only the GTGT sequence was present. For this purpose, primers cDNA1F and cDNA4R were used for exon 1 to 2 and exon 2 to 11 fragments, respectively.
Non-allele–specific PCR and sequencing
Using the above protocols, we were able to check the entire coding sequence plus intron/exon borders of NCF-1, with the exception of the 5′ promoter region and the region 3′ of the exon 11 primer. To cover these regions, non-allele–specific PCR from genomic DNA was performed to amplify the promoter and exon 11. We observed no differences between the alleles of NCF-1 and ψNCF-1 in these regions, either in the current or the previous study,14 and they generated an unambiguous sequence. For the 5′ promoter region, reactions were performed using the Expand Long Template System (Roche Molecular Biochemicals), under the following conditions: an initial denaturation at 94°C for 3 minutes was followed by 10 cycles of 94°C for 10 seconds, 65°C for 30 seconds, and 68°C for 1 minute, then 20 cycles of 94°C for 10 seconds, 65°C for 30 seconds, and 68°C for 1 minute with a cycle elongation of 20 seconds. This was followed by a 7-minute extension at 68°C. For exon 11 the reaction was performed in the following buffer: 33.5 mM Tris-HCl (pH 8.8), 8.3 mM (NH4)2SO4, 3.35 mM MgCl2, 85 μg/mL BSA, 5% DMSO, 0.125 mM each dNTP, 90 ng each primer (11LA, 11R), 2.5 U AmpliTaq polymerase, 500 ng gDNA. An initial denaturation at 94°C for 3 minutes was followed by 30 cycles of 94°C for 5 seconds and 70°C for 1 minute, followed by a 7-minute extension at 72°C. The primer used for sequencing was 11LB2.
In all instances, PCR-amplified fragments were purified using a QIAquick PCR purification kit (Qiagen) and analyzed by direct sequencing in both directions using the ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction Kit (PE Applied Biosystems, Foster City, CA) and an ABI Prism 310 Genetic Analyzer. The fidelity of the amplified DNA was confirmed by comparing it to the published normal sequence and to normal control DNA sequenced during this study. Except for the reported mutations, one silent polymorphism (see “Results and discussion” on patient 3), and one novel difference to the published sequence seen consistently in all normal alleles of NCF-1(see below), no nucleotide changes were observed. In addition, mutations found in patients were also detected in their carrier parents (in whom parental DNA samples were available), confirming that each nucleotide change was inherited and not the result of a PCR-induced error. Sequence numbering in this report is based on the convention that +1 corresponds to A of the ATG initiator codon. This is 12 nucleotides less than the numbering of the cDNA sequence for p47-phox deposited in GenBank (accession numbers M25665 andM26193).
Results and discussion
Molecular genetic analysis of 50 patients with confirmed (or, in one case, suspected) A470 CGD revealed most patients (44) to be homozygous for the GT deletion at the beginning of exon 2 and to exhibit only the pseudogene sequence (Figure2E and Roesler et al13 and Görlach et al14). The remaining 6 patients showed normal sequence and pseudogene sequence at this position, indicating that they differed from the prevalent genotype. Furthermore, these 6 patients could be split into 2 groups, based on the ratio of electropherogram peak heights for the pseudogene and functional gene sequence in exon 2. In nonaffected persons, the mean ratio is approximately 2:1, consistent with the presence of 2 copies of ψNCF-1.14,21 22 Patients 5 and 6 appeared to be normal in this respect (peak height ratio of approximately 2:1) (compare panels A and B, Figure 2), suggesting that their absence of p47-phox (Table 2) was not due to ΔGT on either allele of NCF-1. In contrast, patients 1 to 4 appeared to be heterozygous for the ΔGT mutation because their peak height ratios were approximately 5:1, similar to obligate heterozygous carriers (GTGT/ΔGT) of the predominant form (ΔGT/ΔGT) of A470 CGD (compare panels C and D, Figure 2).
Identification of A470 CGD patients who were not homozygous for the common ΔGT mutation.
Genomic DNA from a nonaffected donor, a classic (ΔGT/ΔGT) A470 CGD patient and carrier parent, and patients 2 and 5 was amplified using primers 2LB2 and 2RB2, which do not differentiate between NCF-1 and ψNCF-1. Electropherograms show the first 10 nucleotides of exon 2. In the nonaffected person (A), co-amplification of functional gene and pseudogene DNA was evident because the sequences diverged after the initial GT. In contrast, the ΔGT/ΔGT A470 CGD patient showed only pseudogene sequence (E). The obligate heterozygous carrier of this common form of the disease exhibited both NCF-1 and ψNCF-1sequence (C) but with a much higher pseudogene–functional gene ratio than seen in normal DNA. Patients 1 to 4 (only patient 2 is shown; D) had electropherograms similar to those of the carrier, indicating they were heterozygous for the ΔGT mutation. Patients 5 and 6 (only patient 5 is shown; B) were similar to the normal control, suggesting their p47-phox deficiency was not associated with a GT deletion in NCF-1.
Identification of A470 CGD patients who were not homozygous for the common ΔGT mutation.
Genomic DNA from a nonaffected donor, a classic (ΔGT/ΔGT) A470 CGD patient and carrier parent, and patients 2 and 5 was amplified using primers 2LB2 and 2RB2, which do not differentiate between NCF-1 and ψNCF-1. Electropherograms show the first 10 nucleotides of exon 2. In the nonaffected person (A), co-amplification of functional gene and pseudogene DNA was evident because the sequences diverged after the initial GT. In contrast, the ΔGT/ΔGT A470 CGD patient showed only pseudogene sequence (E). The obligate heterozygous carrier of this common form of the disease exhibited both NCF-1 and ψNCF-1sequence (C) but with a much higher pseudogene–functional gene ratio than seen in normal DNA. Patients 1 to 4 (only patient 2 is shown; D) had electropherograms similar to those of the carrier, indicating they were heterozygous for the ΔGT mutation. Patients 5 and 6 (only patient 5 is shown; B) were similar to the normal control, suggesting their p47-phox deficiency was not associated with a GT deletion in NCF-1.
Summary of biochemical and sequence data in 6 patients with A470 CGD
| Patient (sex) . | Mutation(s) . | CGD phenotype . | Nucleotide change . | Amino acid change . | |
|---|---|---|---|---|---|
| p47-phox by immunoblot . | production . | ||||
| 1 (F) | Deletion | Absent | 0 | GT73-74 deleted | Frameshift, stop at codon 51 |
| Splice site | 5′ intron 1 gt → at | Splicing defect | |||
| 2 (F) | Deletion | Absent | 0 | GT73-74 deleted | Frameshift, stop at codon 51 |
| Splice site | 5′ intron 1 gtg → gtt | Splicing defect | |||
| 3 (M) | Deletion | −* | <3% | GT73-74 deleted | Frameshift, stop at codon 51 |
| Missense | G784 → A | Gly262 → Ser | |||
| 4 (M) | Deletion | Absent | 0 | GT73-74 deleted | Frameshift, stop at codon 51 |
| Missense | G125 → A | Arg42 → Gln | |||
| 5 (M) | Missense | Absent | 0 | G125 → A | Arg42 → Gln |
| Deletion | G811 deleted | Frameshift | |||
| 6 (M) | Missense or splice site (?homozyg) | Absent | 0 | G574 → A | Gly192 → Ser or splicing defect (see text) |
| Patient (sex) . | Mutation(s) . | CGD phenotype . | Nucleotide change . | Amino acid change . | |
|---|---|---|---|---|---|
| p47-phox by immunoblot . | production . | ||||
| 1 (F) | Deletion | Absent | 0 | GT73-74 deleted | Frameshift, stop at codon 51 |
| Splice site | 5′ intron 1 gt → at | Splicing defect | |||
| 2 (F) | Deletion | Absent | 0 | GT73-74 deleted | Frameshift, stop at codon 51 |
| Splice site | 5′ intron 1 gtg → gtt | Splicing defect | |||
| 3 (M) | Deletion | −* | <3% | GT73-74 deleted | Frameshift, stop at codon 51 |
| Missense | G784 → A | Gly262 → Ser | |||
| 4 (M) | Deletion | Absent | 0 | GT73-74 deleted | Frameshift, stop at codon 51 |
| Missense | G125 → A | Arg42 → Gln | |||
| 5 (M) | Missense | Absent | 0 | G125 → A | Arg42 → Gln |
| Deletion | G811 deleted | Frameshift | |||
| 6 (M) | Missense or splice site (?homozyg) | Absent | 0 | G574 → A | Gly192 → Ser or splicing defect (see text) |
Blots were uninformative (see text).
Our initial attempts to identify specific defects in NCF-1in 5 of these 6 patients were hindered by the presence of the highly homologous ψNCF-1 genes.13,14 When the gene was amplified exon by exon using intronic primers and the DNA fragments were sequenced, the high ratio of pseudogene to functional gene sequence made it difficult to identify nucleotide changes. This is exemplified in Figure 3 (bottom right panel), in which the small peak representing an altered nucleotide (T) on one allele of NCF-1 in patient 2 is barely detectable. The dominant peak at this position represents the normal G on the ΔGT allele of NCF-1 and in co-amplified DNA strands from the 2 copies of the pseudogene. We therefore developed an allele-specific PCR protocol that amplified only GTGT-containing alleles from genomic DNA. Using this strategy (plus non-allele–specific amplification of the 5′ regulatory region and exon 11), we obtained unambiguous sequence for the entire coding region of the gene, including at least 6 bp of intronic sequence at the 3′ and 15 bp at the 5′ ends of each exon. This allowed us to identify 6 novel mutations in NCF-1 in 6 unrelated A470 CGD patients. Biochemical and mutational data for these patients are summarized in Table 2. In DNA from nonaffected persons, we also identified one nucleotide change from the published sequence of NCF-1, in addition to those previously noted.14 The single new change, A621 → G, was detected in all normal alleles examined (14 or more) and is silent at the amino acid level.
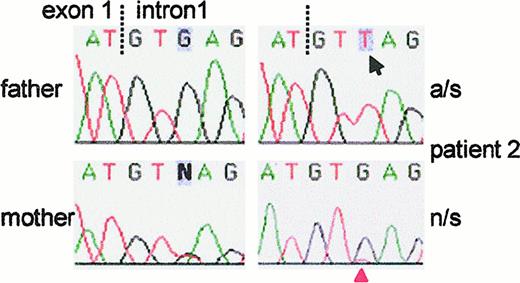
Sequence analysis of the exon 1/intron 1 junction in the GTGT-containing allele of
NCF-1 in patient 2. The patient's G → T mutation at the +3 position of the 5′ (donor) splice site of intron 1 is easily observed after allele-specific (a/s) PCR to amplify only the GTGT-containing allele (arrow, upper right panel). The patient's father, who carried the ΔGT mutation on the other allele (not shown), had the normal G at this position. The patient's mother was heterozygous for the G → T mutation. In the patient's non-allele–specific (n/s) PCR product, the mutant T was barely detectable above background (red arrowhead, bottom right panel) and was dominated by the normal G arising in the ΔGT allele ofNCF-1 and in ψNCF-1 DNA.
Sequence analysis of the exon 1/intron 1 junction in the GTGT-containing allele of
NCF-1 in patient 2. The patient's G → T mutation at the +3 position of the 5′ (donor) splice site of intron 1 is easily observed after allele-specific (a/s) PCR to amplify only the GTGT-containing allele (arrow, upper right panel). The patient's father, who carried the ΔGT mutation on the other allele (not shown), had the normal G at this position. The patient's mother was heterozygous for the G → T mutation. In the patient's non-allele–specific (n/s) PCR product, the mutant T was barely detectable above background (red arrowhead, bottom right panel) and was dominated by the normal G arising in the ΔGT allele ofNCF-1 and in ψNCF-1 DNA.
In patients 1 and 2, sequencing of GTGT-containing alleles identified different splice site mutations at the 5′ end of intron 1. In patient 1, the invariant G at position +1 of the intron was mutated to A (data not shown). In patient 2, the mutation changed the nucleotide at position +3 of the consensus donor splice site from G → T. This is clearly seen in Figure 3 (upper right panel), which shows the sequence obtained from the allele-specific PCR product. The father, who carried ΔGT on his other allele (not shown), had the normal G at this position, whereas the mother had both G and T (and was not a carrier for ΔGT), demonstrating that the patient had inherited the defective GTGT allele from her mother (Figure 3). A (57%) and G (39%) are most common at the +3 position of mammalian donor splice sites, with C and T occurring rarely (2%).23 Replacement of G with T at this position decreases the consensus value (CV) of the exon 1/intron 1 splice junction of NCF-1 from 80.3 to 73.8, calculated according to Shapiro and Senapathy.23 This change appears to be sufficient to cause aberrant splicing because the patient's phagocytes were devoid of p47-phox protein. RNA from the patient was unavailable for study, but RT-PCR analysis of maternal RNA revealed only normal message, presumably transcribed from her unaffected allele. It is possible that any abnormal transcript from the affected allele was unstable or was not amplified in our allele-specific PCR. Genetic disease caused by a mutation at the +3 position of the 5′ splice site is relatively uncommon but not novel; a G → T mutation at +3 of a donor splice site in the β-spectrin gene (which decreases the CV from 88.9 to 82.1) causes hereditary elliptocytosis,24 and a similar defect, an A → T mutation at position +3 of intron 5 of CYBB, causes X910 CGD.25
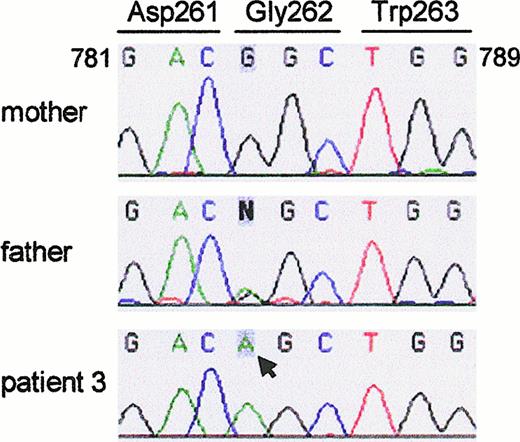
Sequence analysis of the GTGT-containing allele in the genomic DNA of patient 3 revealed a single nucleotide change, G784 → A in exon 8, that predicts the amino acid change Gly262 → Ser (Figure4). The mutation was carried on the paternal allele; the father's sequence revealed both A and G at this position. The mother had the normal G at nucleotide 784 of her GTGT-containing allele (Figure 4), and carried the ΔGT mutation on her other allele (not shown). The mother's functional (GTGT) allele contained a single nucleotide change from the published sequence, C345 → T, which is silent at the amino acid level (Leu115 → Leu) (not shown). The patient (and his father) showed only C at this position, confirming that the patient had only one GTGT-containing allele and that it was paternal in origin. This change previously was considered specific for a pseudogene subset,14 but we have identified it in an additional 3 of 20 GTGT-containing alleles analyzed, indicating that it is a silent polymorphism in NCF-1. Gly262 lies in the C-terminal SH3 domain of p47-phox, immediately adjacent to a highly conserved Trp.26 It is likely that the amino acid change destabilizes the protein, but immunoblotting experiments were inconclusive (see “Patients, materials, and methods”).
Sequence analysis of the GTGT-containing allele of
NCF-1 (exon 8) in genomic DNA of patient 3.Amplification and sequencing of the GTGT-containing allele revealed a G784 → A mutation in patient 3 (arrow). The mutation changes codon 262 from GGC to AGC (Gly → Ser). The patient's mother only showed the normal nucleotide G (and carried the ΔGT mutation on her other allele; not shown), whereas the father had both G and A at this position.
Sequence analysis of the GTGT-containing allele of
NCF-1 (exon 8) in genomic DNA of patient 3.Amplification and sequencing of the GTGT-containing allele revealed a G784 → A mutation in patient 3 (arrow). The mutation changes codon 262 from GGC to AGC (Gly → Ser). The patient's mother only showed the normal nucleotide G (and carried the ΔGT mutation on her other allele; not shown), whereas the father had both G and A at this position.
The mutation in the GTGT-containing allele of NCF-1 in patient 4 was a G125 → A transition that changes codon 42 from CGG to CAG and predicts the replacement of Arg with Gln (not shown, but see discussion of patient 5 in whom this mutation was also found). The patient's mother and one of his brothers showed both G and A at this position, indicating that they had 2 GTGT-containing alleles and carried the mutation on one of them. His father and second brother were normal at this position but appeared to be carriers for ΔGT.
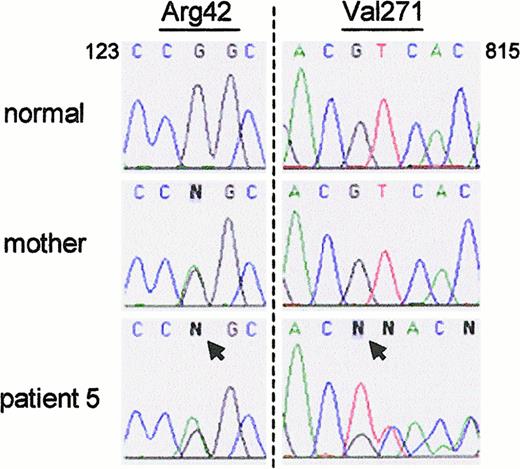
The data in Figure 2 suggested that in patients 5 and 6, ΔGT was confined to the pseudogenes, as in nonaffected persons, and therefore that their CGD did not arise from this deletion on either allele ofNCF-1. This was confirmed in patient 5, who was found to be a compound heterozygote for 2 non-ΔGT mutations after allele-specific amplification of NCF-1. One allele contained the same G125 → A transition that was found in patient 4 (Figure5). This allele was presumably inherited from his mother; she was also heterozygous for the defect (Figure 5). (DNA from the patient's father was unavailable for study.) Because the patient's neutrophils (and those from patient 4) were devoid of p47-phox, the missense mutation must lead to either unstable mRNA or unstable protein. The patient's second mutation was a deletion of a single base, G811, that caused a frameshift starting at residue 271 and led to a premature stop codon at 375 (Figure 5). DNA from the patient's mother showed the normal sequence at this position.
Patient 5 was a compound heterozygote for 2 non-ΔGT mutations.
(left panel) The patient and his mother were both heterozygous for a G125 → A transition in exon 2 (arrow) that predicts the amino acid change Arg42 → Gln. (right panel) The patient was also heterozygous for the deletion of G811 (arrow); sequence from normal and mutant alleles can be seen after nucleotide C810. The patient's mother was normal at this position.
Patient 5 was a compound heterozygote for 2 non-ΔGT mutations.
(left panel) The patient and his mother were both heterozygous for a G125 → A transition in exon 2 (arrow) that predicts the amino acid change Arg42 → Gln. (right panel) The patient was also heterozygous for the deletion of G811 (arrow); sequence from normal and mutant alleles can be seen after nucleotide C810. The patient's mother was normal at this position.
Insufficient DNA was available from patient 6 for allele-specific PCR, but non-allele–specific amplification and sequencing of the patient's genomic DNA revealed a single nucleotide change from the published sequence. The G574 → A transition identified in exon 6 (Figure6) would result in the change of codon 192 from GGT → AGT and predict the conservative replacement of Gly with Ser. Alternatively, because G574 is the final nucleotide of exon 6, the transition could result in a splicing defect. The relative peak heights at nucleotide 574 suggest that the patient is homozygous for this change in NCF-1 (with the G at this position originating only in ψNCF-1). Although no other change was identified in the patient's sequence that could not be attributed to known differences between NCF-1 and ψNCF-1, we cannot exclude the possibility that an additional undetected mutation exists on the second allele of NCF-1. Unfortunately, neither an additional sample from the patient nor DNA from the patient's parents was available for study, and there was no information regarding consanguinity in the family. Interestingly, Gly192 lies next to the conserved Trp in the N-terminal SH3 domain of p47-phox, corresponding exactly to Gly262 in the C-terminal SH3 domain, which is also changed to Ser by the mutation identified in patient 3. However, we cannot be certain that the absence of p47-phox in patient 6 results from the Gly192 → Ser missense mutation. The final nucleotide of an exon (position −1 of the consensus donor splice site) is most commonly G (approximately 78%) and rarely A (10%).23 24 As noted above, G574 is the final nucleotide of exon 6, and its mutation to A decreases the CV for this splice junction (position −3 to +6) from 74.0 to 62.1; the mean CV for splice junctions in NCF-1 is 82.9 ± 8.6 (SD). Therefore, it is perhaps more likely that aberrant splicing during processing of the RNA transcript is the primary cause of p47-phox deficiency in this patient rather than protein instability resulting from the change of Gly192 → Ser.
Sequence analysis of the NCF-1 exon 6/intron 6 junction in genomic DNA of patient 6.
Non-allele–specific amplification and sequencing of genomic DNA from patient 6 revealed a single nucleotide change from the normal sequence, a G574 → A transition (arrow). The mutation is likely to disrupt splicing because it alters the final nucleotide of exon 6, but it also predicts a change of Gly192 → Ser.
Sequence analysis of the NCF-1 exon 6/intron 6 junction in genomic DNA of patient 6.
Non-allele–specific amplification and sequencing of genomic DNA from patient 6 revealed a single nucleotide change from the normal sequence, a G574 → A transition (arrow). The mutation is likely to disrupt splicing because it alters the final nucleotide of exon 6, but it also predicts a change of Gly192 → Ser.
The high degree of homology between the p47-phox gene and its pseudogenes, which results in the co-amplification of DNA strands from NCF-1 and ψNCF-1 with most oligonucleotide primers, greatly complicates the molecular analysis of families affected by A470 CGD. In most CGD patients who have a p47-phox deficiency, amplification and sequencing of exon 2 will reveal the absence of GTGT-containing sequence and therefore identify them as having the common ΔGT/ΔGT genotype (eg, Figure2E). However, diagnosis of the carrier state (GTGT/ΔGT) among their family members is challenging because genomes of all nonaffected persons include DNA sequences with the GT deletion within ψNCF-1. The ratio of electropherogram peak heights can be a guide (compare panels A and C, Figure 2), but this cannot be considered definitive because the number of pseudogene copies may vary in some persons (Heyworth et al, unpublished observation). The p47-phox pseudogenes also complicated the identification of novel mutations in the rare subset of A470 CGD patients described in this study who were either heterozygous or homozygous for non-ΔGT mutations. As we show here, this can be overcome by using allele-specific primers for DNA amplification, which allows nucleotide changes in NCF-1 to be detected against a background of unambiguous sequence. With this technique we were able to confirm that patients 1 to 4 were compound heterozygotes for ΔGT and one additional mutation and to identify patients 5 and 6 as the first reported patients with A470 CGD not resulting from ΔGT on at least one allele of NCF-1. In all these patients, identifying the specific mutations provides the only means of detecting carriers in other family members and providing prenatal diagnosis in mothers at risk.
Acknowledgments
We thank Dr Pablo J. Patiño for preliminary work on DNA from patient 5. We also thank the referring physicians and the CGD patients and their families for providing blood samples.
Supported by National Institutes of Health grants CA68276 (P.G.H.), AI24838 (A.R.C.), DK54369 (P.E.N.), and RR00833 (to the General Clinical Research Center at The Scripps Research Institute). This is manuscript 13273-MEM of The Scripps Research Institute.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Paul G. Heyworth, Dept of Molecular and Experimental Medicine, MEM-241, The Scripps Research Institute, 10550 North Torrey Pines Rd, La Jolla, CA 92037; e-mail:heyworth@scripps.edu.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal