Abstract
Reporter mouse strains are important tools for monitoring Cre recombinase-mediated excision in vivo. In practice, excision may be incomplete in a given population due to threshold level or variegated expression of Cre. Hence, it is desirable in many experimental contexts to isolate cells that have undergone excision to assess the consequences of gene ablation. To generate alternative reporter mice, an enhanced green fluorescent protein (EGFP) gene was targeted to the retroviral-trapped ROSA26 locus. Upon Cre-mediated excision of “Stop” sequences, EGFP was expressed ubiquitously during embryogenesis and in adult tissues (including T cells, B cells, and myeloid cells). Using this new reporter strain, separation of excised from nonexcised cells in vitro was achieved in thymocytes in a noninvasive manner based on activated EGFP expression. This new EGFP reporter strain should facilitate a variety of conditional gene-targeting experiments, including the functional studies of hematopoietic cells in lineage-specific knockout mice.
Introduction
Conditional gene targeting using Cre-loxP elements is dependent on the generation of mice that express Cre recombinase in a cell type-specific or stage-specific pattern.1,2 To evaluate Cre-expressing mice during embryogenesis and in adult tissues, reporter mice have been made such that the expression of a reporter gene is activated only after Cre-mediated excision of a “Stop” fragment.3 Previously 2 similar reporter strains for detecting Cre-mediated excision were obtained by targeting wild-type or proviral ROSA26 loci.4-6Such improved reporter lines offer several important features for monitoring Cre-mediated excision events in vivo. In these mice, expression of the germline-excised βgeo or lacZgene is ubiquitous during embryogenesis and in adult tissues. In addition, mosaic (stochastic) expression or silencing has not been observed for the germline-excised βgeo reporter gene in any cell lineage.
Because of the transgenic nature of many Cre-expressing mice, the excision of gene fragment flanked by loxP sites (referred to as floxed gene) by transgenic Cre recombinase is often incomplete in a given lineage. As a result, selection for or against unexcised cells could occur in vivo. In instances where hematopoietic cells with targeted mutations are analyzed, isolation of cells that have undergone Cre-mediated excision is required and followed by functional studies in vitro. EGFP provides a noninvasive marker for cell isolation because its detection does not require fixation or hypotonic permeabilization of cells, which is required for the detection of lacZactivity. We reasoned that an EGFP reporter allele whose expression is blocked by a loxP-flanked Stop fragment might function as an indicator for Cre-mediated excision when used together with other floxed alleles in the same cell. In the current study, cDNA encoding the enhanced green fluorescent protein (EGFP) variant was targeted to the proviral ROSA26 locus to generate a new reporter line. After interbreeding of the founder line with 3 different Cre-expressing lines, ubiquitous, inducible, and lineage-specific activation of EGFP was obtained. In addition, we demonstrated that specific isolation of cells undergoing complete excision at a loxP-flanked test allele could be achieved based on the activation of ROSA26-EGFPf/+ allele in T lymphocytes.
Study design
Construction of targeting vector
An 0.8-kb XbaI–NotI fragment of pEGFP (Clontech, Palo Alto, CA) containing translational starting/terminating signals of EGFP was placed 5′ to the βgeo sequence of pSAβgeo vector between SpeI andHindIII sites.7 A Stop fragment from cACT-XstopXnZ was placed 3′ to a selection cassette containing phosphoglycerate kinase (PGK)-Puror and PGK-cytosine deaminase (CD) genes flanked by loxPsites.3,6 Gene targeting in embryonic stem (ES) cells was performed as described previously.6
Fluorescence-activated cell sorter analysis
Cells were labeled with phycoerythrin-conjugated antibodies (Pharmingen) and analyzed on a FACScalibur (Becton Dickinson) machine. Cell suspensions from the thymus of ROSA26-EGFPf/+;Testf/+;lck-cre mouse were subjected to high-speed cell sorting using Cytomation's MOFLO machine. EGFP+ and EGFP− fractions were individually collected.
Induced activation of floxed ROSA26-EGFP allele by Mx-cre
Mice were injected intraperitoneally with 100 to 300 μg polyI-polyC (Sigma) every other day from days 14 to 30 after birth.8 EGFP expression in different hematopoietic lineages was analyzed by FACS analysis.
Results and discussion
Targeting of EGFP cDNA to proviral ROSA26 locus
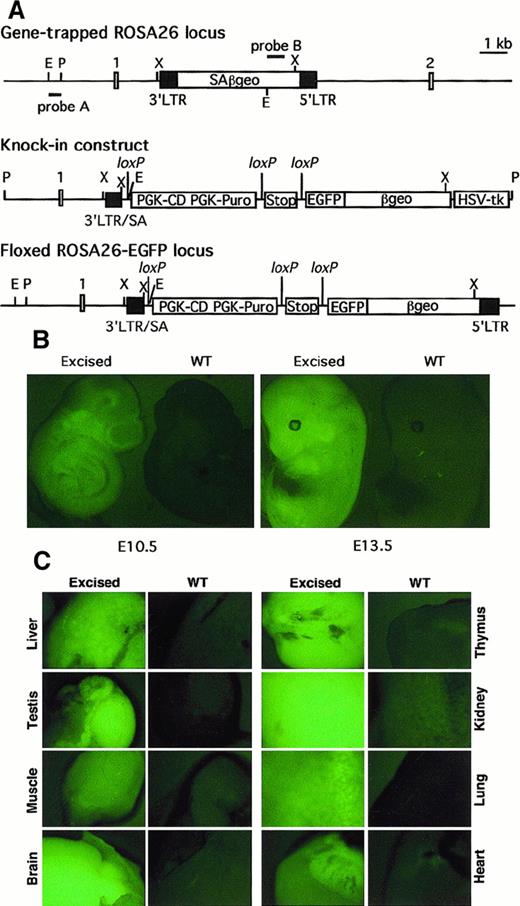
To generate a loxP-flanked (floxed) ROSA26-EGFP founder line, EGFP cDNA was placed between Stop and βgeosequences in the knockin targeting construct shown in Figure1A. Of 136 Puror/ganciclovirr clones obtained from electroporated ROSA26 ES cells, 3 were correctly targeted as judged by Southern blot analysis. Heterozygous ROSA26-EGFPf/+ mice generated with targeted ES cells appeared normal and were fertile.
Targeting of EGFP cDNA to the proviral ROSA26 locus and its expression after Cre-mediated excision of Stop and PGK-CD/PGK-Puro sequences.
(A) Structures of gene-trapped ROSA26 locus, targeting construct, and floxed ROSA26-EGFP locus. The positions of selection cassettes, the Stop fragment, and 3 loxP sites with the same orientation are indicated. E, EcoRI; P, PacI, X,XbaI, SA, splicing acceptor. Positions of probes used to identify and confirm structures are also indicated. (B) EGFP expression in E10.5 and E13.5 wild-type (WT) and excised ROSA26-EGFP embryos. (C) EGFP expression in organs in 1-week-old wild-type or excised ROSA26-EGFP mouse.
Targeting of EGFP cDNA to the proviral ROSA26 locus and its expression after Cre-mediated excision of Stop and PGK-CD/PGK-Puro sequences.
(A) Structures of gene-trapped ROSA26 locus, targeting construct, and floxed ROSA26-EGFP locus. The positions of selection cassettes, the Stop fragment, and 3 loxP sites with the same orientation are indicated. E, EcoRI; P, PacI, X,XbaI, SA, splicing acceptor. Positions of probes used to identify and confirm structures are also indicated. (B) EGFP expression in E10.5 and E13.5 wild-type (WT) and excised ROSA26-EGFP embryos. (C) EGFP expression in organs in 1-week-old wild-type or excised ROSA26-EGFP mouse.
Ubiquitous activation of loxP-flanked ROSA26-EGFP during embryogenesis and in postnatal organs
To examine excision-activated EGFP expression in vivo, the ROSA26-EGFPf/+ mice were interbred withGATA1-cre transgenic mice in which Cre is expressed during early embryogenesis.6 Embryonic day (E)7.0 to E15.5 embryos with a germline-excised ROSA26-EGFP allele showed widespread fluorescence in both the embryo proper and the yolk sac, whereas embryos with a ROSA26-EGFPf/+ allele displayed only background fluorescence as wild-type embryos. Representative E10.5 and E13.5 embryos are shown in Figure 1B.
Expression of EGFP was also investigated in hematopoietic lineages of 2-week-old mice. In a heterozygous mouse with a germline-excised ROSA26-EGFP allele, almost all T cells, B cells, and myeloid cells from thymus, spleen, lymph node, and bone marrow expressed detectable EGFP compared with cells from a ROSA26-EGFPf/+ mouse (Figure2A and data not shown). Erythroid cells from fetal blood of E12.5 embryos and adult spleen and bone marrow failed to exhibit detectable EGFP by FACS analysis (data not shown). Expression of EGFP was further examined in different organs from 1-week-old newborn mice. As shown in Figure 1C, all organs from a mouse with an excised allele showed obvious fluorescence compared with those from controls. The ubiquitous expression pattern of the excised ROSA26-EGFP allele mirrored that of the proviral ROSA26 locus itself.
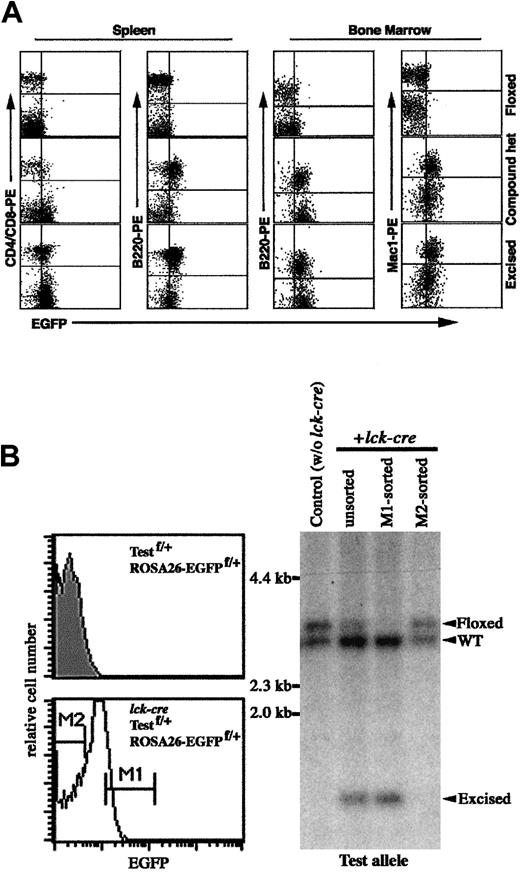
Inducible and cell-type–specific activation of
loxP-flanked ROSA26-EGFP allele byMx-cre or lck-cretransgenes. (A) Flow cytometric analysis of EGFP expression in cells from ROSA26-EGFPf/+ and ROSA26-EGFPf/+;Mx-cre mice injected with pI-pC. A mouse with excised ROSA26-EGFP allele was used as a positive control. Fluorescence intensity was represented on log scale. (B) The enrichment of thymocytes with excised test allele based on the activated expression of EGFP in ROSA26-EGFPf/+;Testf/+;lck-cremouse. (Left) Histogram of EGFP expression in thymocytes of heterozygous mouse with lck-cre (nonshaded) or withoutlck-cre (shaded). M1 and M2 indicate the EGFP+and EGFP− fractions of thymocytes from triple-compound mouse that are sorted. (Right) Southern blot analysis for the excision of the test floxed allele. Genomic DNAs were digested withSpeI and BglII. The positions of DNA bands representing WT, floxed, and excised test alleles are indicated.
Inducible and cell-type–specific activation of
loxP-flanked ROSA26-EGFP allele byMx-cre or lck-cretransgenes. (A) Flow cytometric analysis of EGFP expression in cells from ROSA26-EGFPf/+ and ROSA26-EGFPf/+;Mx-cre mice injected with pI-pC. A mouse with excised ROSA26-EGFP allele was used as a positive control. Fluorescence intensity was represented on log scale. (B) The enrichment of thymocytes with excised test allele based on the activated expression of EGFP in ROSA26-EGFPf/+;Testf/+;lck-cremouse. (Left) Histogram of EGFP expression in thymocytes of heterozygous mouse with lck-cre (nonshaded) or withoutlck-cre (shaded). M1 and M2 indicate the EGFP+and EGFP− fractions of thymocytes from triple-compound mouse that are sorted. (Right) Southern blot analysis for the excision of the test floxed allele. Genomic DNAs were digested withSpeI and BglII. The positions of DNA bands representing WT, floxed, and excised test alleles are indicated.
Inducible activation of loxP-flanked ROSA26-EGFP in hematopoietic cells
To test inducible activation, ROSA26-EGFPf/+ mice were bred with inducible Mx-cre transgenic mice.8Compound heterozygous mice (ROSA26-EGFPf/+;Mx-cre) were injected repeatedly with pI-pC, which induces the expression of Mx1-driven Cre activity.8 Induced EGFP expression was assessed by FACS analysis of hematopoietic cells from injected mice. As shown in Figure2A, in the spleen of an injected compound mouse, almost all B220+ B cells were EGFP+, whereas only a fraction of single positive (CD4+ or CD8+) T cells were EGFP+. In the bone marrow of injected mice, B220+ B cells and Mac1+ myeloid cells were EGFP+. These results were supported by Southern blot analysis of DNA samples from thymocytes, splenocytes, bone marrow cells, and different organs (data not shown).
Comparison of relative accessibility of loxP-flanked test locus and the ROSA26-EGFP allele to Cre-mediated excision in thymocytes
To determine whether the floxed ROSA26-EGFP allele exhibits similar accessibility to Cre-mediated excision as other floxed loci, triple-compound heterozygous mice (ROSA26-EGFPf/+;Testf/+;lck-cre) were obtained by mating.9 EGFP expression in compound mice was detected in thymocytes and peripheral T cells but not in B cells and myeloid cells (data not shown). Because excision of the floxed ROSA26-EGFP locus by lck-cre is incomplete in thymocytes of the triple-compound mouse, thymocytes were then subjected to cell sorting based on EGFP expression level. EGFP+ and EGFP− thymocytes were collected, and genomic DNAs were analyzed for excision at the floxed testing locus by Southern blot analysis (Figure 2B). Nearly complete excision at the test locus was observed in sorted EGFP+ cells, whereas no significant excision was evident in sorted EGFP− cells. Therefore, in the one lineage examined (thymocytes), the accessibility of the floxed ROSA26-EGFP allele to Cre-mediated excision approximates that of a floxed test locus. Although it is important to compare the accessibility of other floxed alleles at a variety of chromosomal locations, enrichment for those cells in which excision has occurred should be greatly facilitated by isolation of the EGFP+ population.
The findings reported here also provide proof-of-principle for use of the targeted proviral ROSA26 locus for conditional gene activation in various settings in the intact mouse.10 For genes with functions in diverse cell types, it is desirable to develop a cost-effective and efficient approach by which a transgene may be activated in a temporally and spatially regulated fashion in virtually any cell lineage or tissue. To this end, a dormant transgene preceded by floxed Stop DNA sequences can be targeted first to the proviral ROSA26 locus and then maintained in the founder line. Its activation is achieved by breeding with different Cre-expressing mice with unique tissue specificity and inducibility. The pattern of transgene expression will be dependent on the timing and location of Cre expression in compound heterozygous mice. The increasingly available Cre-expressing mouse strains will be used to great advantage in conditional gene activation, as well as gene inactivation experiments in the future. Such approaches should facilitate the creation of animal models of human diseases for pathogenesis and therapy.
Acknowledgments
We thank Philippe Soriano, Elizabeth Robertson, Jamey D. Marth, David J. Anderson, Klaus Rajewsky, and Gary Silverman for reagents. The reporter strain will be deposited for distribution in the collection of the Jackson Laboratories.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Stuart H. Orkin, Department of Pediatrics, Division of Hematology/Oncology, Children's Hospital, Harvard Medical School, Boston, MA 02115; e-mail: orkin@rascal.med.harvard.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal