Abstract
Cutaneous T-cell lymphomas (CTCL) comprise a heterogeneous group of lymphoproliferative disorders that are characterized by an accumulation of T-lymphocytes in the skin and occasionally in blood known as Sézary syndrome (SS). In most cases the dominant clone displays T-helper 2 cytokines. Because IFN-γ is a natural inhibitor of T-helper 2 cells and IFN-α is frequently used in CTCL, the impact of IFNs on SS-derived purified clonal T-helper 2 cells was studied using anti-Vβ antibodies. Moreover, IFNs are known to mediate virus resistance in normal cells. The isolated clonal CD4+ cells, but not the nonclonal CD4+ cells, appeared resistant to IFN-γ and IFN-α stimulation in terms of human leukocyte antigen up-regulation and MxA induction caused in part by alterations in Stat-1 molecule mRNA and IFNγR1 mRNA transcription. The IFN resistance of the patient-derived clonal cells was then targeted by vesicular stomatitis virus infection after IFN-α priming, resulting in selective viral replication in clonal cells. In contrast, nonclonal cells of the same patient showed IFN-dependent MxA expression, which is a major mediator protein of viral protection. The IFN resistance of the dominant T-helper 2 cells might be important for lymphomagenesis. Interferon signaling deficiencies can be targeted for purging patients' cells in vitro. Furthermore, this approach may allow specific molecular interventions, resulting in the efficient treatment of CTCL and other IFN-resistant neoplasms such as lung cancer.
Introduction
Primary cutaneous lymphomas are the second most common group of extranodal non-Hodgkin lymphomas after primary gastrointestinal lymphomas.1 They have highly characteristic clinical features and are considered prototype malignancies because they can be recognized clinically very early. In addition, because of their locations, they are well accessible for the study of immune and molecular biology. Most cutaneous lymphomas are low-grade T-cell lymphomas such as mycosis fungoides and Sézary syndrome (SS).2 These neoplasms are not curable, even using aggressive chemotherapy and radiotherapy.3 The malignant lymphocytes mostly express the T-cell markers CD2, CD3, CD4, and CD5. Often they lack CD7, and they display T-helper 2 (Th2) cytokines such as IL-5 and IL-10 and express the IFN-γ receptor chain 2 (IFNγR2).4 IFN-α, alone or in combination with retinoids or psoralen plus ultraviolet A irradiation, is helpful, especially in early disease.5 Interferons are known for their pleiotropic effects. Besides the up-regulation of major histocompatibility complex molecules and the inhibition of cell proliferation and tumor growth, IFN can induce resistance to viral replication in all cells.6 Interferons exert their effects by inducing overlapping sets of effector proteins through specific cell surface receptors (IFNαR and IFNγR). In this context, a cross-talk between IFN-γ and IFN-α/β signaling components has been reported recently.7 It has been shown that the IFNαR1 chain favors the association of the IFNγR1 and the IFNγR2 chains.
Because of the beneficial clinical impact of IFN-α and the inhibitory potential of IFN-γ on benign human and murine Th2 lymphocytes,8 we studied the effects of IFNs on CTCL-derived tumor cells purified by FACS-sorting from the peripheral blood of 4 patients with SS.4 This subtype of cutaneous T-cell lymphoma is characterized by an accumulation of malignant T-lymphocytes in skin and peripheral blood.2 Furthermore, we evaluated the susceptibility of these SS cells to viral infections with vesicular stomatitis virus (VSV).
Patients and methods
Patients
SS was diagnosed using standard clinical criteria (erythroderma, lymphadenopathy), histologic criteria (subepidermal lymphoid infiltrate showing strong epidermotropism and expressing a T-helper cell phenotype), hematologic criteria (more than 1000 SS cells/mL), and an elevated CD4–CD8 ratio.9-12 All patients were seronegative for human T-cell lymphocytic virus-I antibodies by enzyme-linked immunosorbent assay. Of the 4 patients whose T-cell clones were identified and investigated, 2 were in stage III (T4 N1 M0) and 2 were in stage IVa (T4 N3 M0) disease.13 They did not receive any systemic therapy in the last 4 weeks before blood was taken. However, topical steroids had to be applied to control itching.
Molecular biology studies for the T-cell receptor γ chain,14 Southern blot analysis, or both documented the presence of a dominant T-cell clone in the peripheral blood of all patients. In all patients, a dominant CD4+Vβ+ (double-positive) clone was detected by screening the peripheral blood cells with a panel of anti-Vβ antibodies.4,11,15,16 The double-positive populations of all patients demonstrated low expression levels for CD3.17
The percentage of CD4+ Vβ+ lymphocytes of the total number of T-helper lymphocytes was 69.6% for patient R, 81.3% for patient X, 73.8% for patient P, and 85% for patient C when blood was taken for functional analysis. The absolute number of double-positive T-helper cells was followed over years. Patients P and C had progressive and finally lethal disease associated with increasing percentage and absolute number of CD4+ Vβ+cells. Patient X had stable disease reflected by a constant number of double-positive T-helper lymphocytes, whereas patient R experienced partial remission with a decrease of the double-positive CD4+ Vβ+ population during treatment with PUVA and IFN-α.18
Purification of clonal T cells
In all patients, approximately 1 × 106 clonal CD4+ T cells and approximately 1 × 106nonclonal T-helper cells were prepared by FACS sorting (FACSVantage; Becton Dickinson, San Jose, CA) using fluorescence-labeled antibodies against the clonal Vβ-chains (anti-TCR Vβ3.1, anti-TCR Vβ5a, anti-TCR Vβ5.3, anti-TCR Vβ6.7; Immunotech, Marseille, France). The purity and viability after sorting was greater than 97% (assessed by immunohistochemistry on cytospins stained with anti-CD4 and anti-Vβ monoclonal antibodies [mAbs]).4
Electrophoretic mobility shift assay
Electrophoretic mobility shift assays were performed as described elsewhere.19 Double-stranded p32-ATP–Labeled oligonucleotides (30 000 cpm) containing the SIE (sis-inducible element) were incubated with 3 μg nuclear extracts of the investigated cells in binding buffer consisting of 10 mM HEPES, pH 7.9, 60 mM KCl, 4% Ficoll, 1 mM dithiothreitol, and 1 mM EDTA, pH 8.0. Two micrograms poly-deoxy-inosinic-deoxy cytidylic acid was used as competitor for unspecific DNA-binding activities. Total volume of the reaction was 30μL.
Flow cytometry
Approximately 106 isolated clonal T cells were analyzed by FACS using anti-HLA–ABC mAb W6.32 (DAKO Diagnostics, Zug, Switzerland), anti-IFNγR2 mAb Hub159 (Genzyme Diagnostics, Kings Hill, United Kingdom), and mouse IgG2a (Ancell, Bayport, MN) and hamster IgG (Becton Dickinson) as isotype controls. As secondary reagents, fluorescence-conjugated rabbit antimouse polyclonal antibodies (DAKO), biotin-labeled goat antihamster polyclonal mAbs (CALTAG, Burlingame, CA), and fluorescence-conjugated streptavidin (DAKO) were used, respectively. Cells were incubated with antibodies for 2 hours on ice in 2% fetal calf serum–phosphate-buffered saline, then washed with phosphate-buffered saline and fixed in 0.5% formaldehyde.
Reverse transcription–polymerase chain reaction
Cell pellets from purified clonal T cells were resuspended in buffer A (10 mM HEPES, 10 mM KCl, 1 mM EDTA, 1 mM EGTA) and lysed by vortexing with the addition of 1/16 vol 10% NP40. The supernatant was added to an equal volume of buffer B (7 M urea, 1% sodium dodecyl sulfate, 0.35 M NaCl). After one phenol–chloroform extraction and one chloroform extraction, the RNA was precipitated with ethanol–glycogen and dissolved in RNAase-free water. If more than 1 million cells could be extracted, the RNA was quantified by reading at A260. Amounts between 2 and 4μg RNA were used to synthesize cDNA (M-MuLV Reverse Transcriptase; New England Biolabs, Beverly, MA). If fewer cells were available, all RNA was reverse-transcribed to 20 to 40μL cDNA.
Polymerase chain reaction (PCR) was performed with the incubation buffer supplied with the Taq DNA polymerase (Boehringer Mannheim, Mannheim, Germany), with PCR–DIG-labeling nucleotide mix (Boehringer Mannheim), and with 2.0 μM oligonucleotide primers. The cDNA probes were first amplified with primers for β-actin to test the quality and quantity of the cDNA. Only satisfactory cDNA was then subjected to PCR with primers for IFNαR1 and 2, IFNγR1 and 2, JAK1, and Stat-1, respectively. Primers were selected using the Oligo Primer Analysis Software, version 4.0 (National Biosciences, Plymouth, MN). Primer sequences were sent to a gene bank to exclude cross-binding to other published sequences.20 PCR was performed with Perkin Elmer 9600 GenAmp cyclers. DNA fragments were amplified with 30 cycles and an annealing temperature of 55°C. An aliquot of PCR product was electrophoresed on a 1.6% agarose gel and visualized by ethidium bromide staining. In all PCR reactions, positive controls from various cell lines and water as negative control were included.
Virus infection
Patients' peripheral blood mononuclear cells (PBMCs) (1 × 106 cells/well) were either treated with 1000 U IFN-α for 15 hours or left untreated. Subsequently, the PBMCs were infected with 5 infectious particles of VSV per cell for 10 hours. The cells were harvested, fixed with acetone, and immunostained with antibodies specific for MxA, TCR Vβ (Immunotech), and VSV.
Results
IFN-α and IFN-γ did not up-regulate typically IFN-induced surface molecules
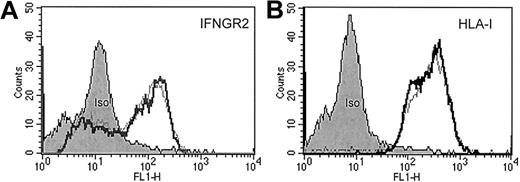
Stimulation of patient-derived clonal T cells with IFN-α or IFN-γ neither induced the up-regulation of IFNγR2 and human leukocyte antigen (HLA) class I (Figure1) nor the up-regulation of intercellular adhesion molecule 1 and HLA class II (data not shown). IFN-γ has been shown to suppress IFNγR2 expression representing a mechanism of cellular desensitization in which the ligand down-regulates expression of a receptor subunit.21 In CTCL-derived tumor cells, however, it had no impact on IFNγR2 surface levels (Figure1).
FACS analysis of sorted CD4+/Vβ5.3+ clonal T cells from patient C.
Approximately 106 isolated malignant T cells were incubated either with (fat lines) or without (thin lines) IFN-γ (500 U/mL) for 40 hours at 37°C in a 5% CO2 atmosphere. Then cells were washed and stained with anti-IFNγR2 antibodies (A), anti-HLA class I antibodies (B), and appropriate isotype-control antibodies for protein surface expression.
FACS analysis of sorted CD4+/Vβ5.3+ clonal T cells from patient C.
Approximately 106 isolated malignant T cells were incubated either with (fat lines) or without (thin lines) IFN-γ (500 U/mL) for 40 hours at 37°C in a 5% CO2 atmosphere. Then cells were washed and stained with anti-IFNγR2 antibodies (A), anti-HLA class I antibodies (B), and appropriate isotype-control antibodies for protein surface expression.
IFN-α does not stimulate the DNA binding of signal transducer and activator of transcription factors
Interferon-α activates the transcription factors Stat-1 and Stat-2 to bind to specific regulating DNA sequences within the promoters and enhancers of their target genes. To see whether IFN-α also stimulates the DNA binding of Stat-1 or Stat-2 in SS cells, we performed electrophoresis mobility shift assays using nuclear extracts from sorted nonmalignant CD4+ cells, malignant CD4+ cells from patients with SS, and the SIE (sis-inducible element) oligonucleotide. Figure2 shows that IFN-α induces a DNA–protein complex in sorted nonmalignant CD4+ cells (compare lanes 1 and 2) but not in the sorted malignant CD4+ cells from patient R (lanes 3 and 4). Similar results were obtained with extracts from 2 other patients, with the exception that some of them contained a constitutive DNA–protein complex that could not be stimulated by IFN-α and was not recognized by antibodies specific for Stat-1 to Stat-6 (data not shown). Accordingly, the IFN inducible MxA-protein was only detected in nonclonal “normal” T lymphocytes (Figure 3).
Impact of IFN-α on the binding of Stat factors to the SIE.
Nonmalignant CD4+ T cells (lanes 1 and 2) from patient R respond with the induction of DNA binding molecules to IFN stimulation (lane 2). The dominant Vβ CD4+ T-cell population from patient R (lanes 3 and 4) fails to induce DNA binding molecules on stimulation (lane 4). No IFN treatment (lanes 1 and 3); treatment with 500 U IFN-α for 30 minutes (lanes 2 and 4).
Impact of IFN-α on the binding of Stat factors to the SIE.
Nonmalignant CD4+ T cells (lanes 1 and 2) from patient R respond with the induction of DNA binding molecules to IFN stimulation (lane 2). The dominant Vβ CD4+ T-cell population from patient R (lanes 3 and 4) fails to induce DNA binding molecules on stimulation (lane 4). No IFN treatment (lanes 1 and 3); treatment with 500 U IFN-α for 30 minutes (lanes 2 and 4).
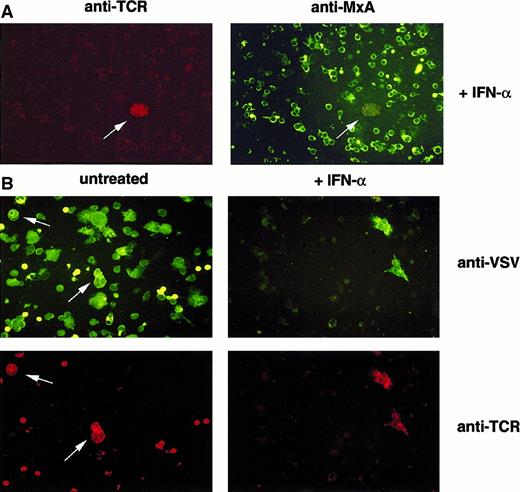
Clonal T cells fail to establish MxA-dependent antiviral status and allow selective viral replication.
Immunofluorescence analysis of VSV-infected PBMCs from patients with CTCL. (A) PBMCs were either treated with IFN-α (+IFN-α) or left untreated (untreated). (B) PBMCs were subsequently infected with VSV. Cells were harvested, fixed with acetone, and immunostained using antibodies specific for MxA and TCR Vβ6.7 (A) or VSV and TCR Vβ6.7 (B).
Clonal T cells fail to establish MxA-dependent antiviral status and allow selective viral replication.
Immunofluorescence analysis of VSV-infected PBMCs from patients with CTCL. (A) PBMCs were either treated with IFN-α (+IFN-α) or left untreated (untreated). (B) PBMCs were subsequently infected with VSV. Cells were harvested, fixed with acetone, and immunostained using antibodies specific for MxA and TCR Vβ6.7 (A) or VSV and TCR Vβ6.7 (B).
Transcriptional down-regulation of IFNγR1 and Stat-1 in clonal T cells
Interferon signal transduction involves specific receptor molecules,22,23 Janus kinases (JAK1/2, Tyk2),24,25 and several Stat factors.26 FACS analysis of IFN receptors with antibodies recognizing the extracellular domains, reverse transcription (RT)–PCR analysis, or both for biologically relevant mRNA segments of molecules involved in signal transduction (IFNαR1/2, IFNγR1/2, JAK1, Stat-1) was applied to illuminate the molecular alterations in the IFN signaling. We focus on JAK1 and Stat-1 because these molecules are involved in IFN-α/β and IFN-γ signal transduction cascades.23 By RT-PCR, we observed mRNA down-regulation of IFNγR1 and Stat-1 in 2 patients (Table 1).
Transcription of IFN signal-associated genes in clonal and nonclonal cells of patients with Sézary syndrome
| Patients . | IFNαR1 . | IFNαR2 . | IFNγR1 . | IFNγR2 . | Stat-1 . | JAK1 . |
|---|---|---|---|---|---|---|
| R (nonclonal) | + | + | + | + | ± | + |
| R (clonal Vβ6.7+) | + | + | − | + | − | ± |
| X (nonclonal) | + | + | + | + | + | + |
| X (clonal Vβ5a+) | + | + | + | + | ± | + |
| P (clonal Vβ3.1) | + | + | + | + | + | + |
| C (PBMC, 85% Vβ5.3+) | ± | + | − | − | − | ± |
| Patients . | IFNαR1 . | IFNαR2 . | IFNγR1 . | IFNγR2 . | Stat-1 . | JAK1 . |
|---|---|---|---|---|---|---|
| R (nonclonal) | + | + | + | + | ± | + |
| R (clonal Vβ6.7+) | + | + | − | + | − | ± |
| X (nonclonal) | + | + | + | + | + | + |
| X (clonal Vβ5a+) | + | + | + | + | ± | + |
| P (clonal Vβ3.1) | + | + | + | + | + | + |
| C (PBMC, 85% Vβ5.3+) | ± | + | − | − | − | ± |
RT-PCR analysis: +, strong band; ±, faint band; −, no band.
Susceptibility of SS-derived clonal T cells to VSV infection
Because IFN-α–induced MxA proteins are major mediators of resistance against infections with RNA viruses,27,28 we investigated whether the observed IFN resistance could be targeted by an IFN-sensitive virus. Mixed-cell populations containing the CTCL-derived cell line SeAx29 and PBMCs from healthy donors were stimulated with IFN-α and infected with VSV. VSV, which is known to be sensitive to the action of IFNs,30 was able to infect a high percentage of mononuclear cells in the absence of IFN-α. On IFN-α treatment, viral replication was restricted to the CTCL-derived SeAx cells (data not shown). The same experiment was then performed with PBMCs derived from patients with SS that contained a various proportion of malignant T-lymphocytes identified by their clonal Vβ-receptor usage. After incubation of the patients' PBMCs with IFN-α and infection of the cell culture with VSV, we detected VSV antigens selectively in the malignant T-cell population (Figure 3, Table 2).
Susceptibility of CTCL-derived tumor cells to VSV is independent of IFN-α treatment
| Patients . | −IFN-α . | +IFN-α . | ||||||
|---|---|---|---|---|---|---|---|---|
| Total cells . | Infected . | % Infected . | MxA . | Total cells . | Infected . | % Infected . | MxA . | |
| R (nonclonal) | 273 | 97 | 36 | ± | 287 | 11 | 4 | +++ |
| R (clonal Vβ6.7+) | 102 | 45 | 44 | − | 115 | 49 | 43 | − |
| X(nonclonal) | 255 | 86 | 34 | ± | 216 | 25 | 12 | ++ |
| X (clonal Vβ5a+) | 95 | 52 | 44 | − | 103 | 44 | 43 | − |
| P (nonclonal) | 236 | 91 | 39 | ± | 267 | 5 | 2 | +++ |
| P (clonal Vβ3.1+) | 100 | 39 | 39 | − | 85 | 31 | 36 | − |
| Patients . | −IFN-α . | +IFN-α . | ||||||
|---|---|---|---|---|---|---|---|---|
| Total cells . | Infected . | % Infected . | MxA . | Total cells . | Infected . | % Infected . | MxA . | |
| R (nonclonal) | 273 | 97 | 36 | ± | 287 | 11 | 4 | +++ |
| R (clonal Vβ6.7+) | 102 | 45 | 44 | − | 115 | 49 | 43 | − |
| X(nonclonal) | 255 | 86 | 34 | ± | 216 | 25 | 12 | ++ |
| X (clonal Vβ5a+) | 95 | 52 | 44 | − | 103 | 44 | 43 | − |
| P (nonclonal) | 236 | 91 | 39 | ± | 267 | 5 | 2 | +++ |
| P (clonal Vβ3.1+) | 100 | 39 | 39 | − | 85 | 31 | 36 | − |
Immunofluorescence analysis of MxA: +++, strong expression in >95% of cells; ++, clearly detectable expression in >95% of cells; ±, expression in 1% to 3% of cells; −, no expression detectable.
Discussion
Our data clearly indicate that CTCL-derived tumor cells displaying Th2 cytokines4 present elementary defects in IFN signaling. Because none of these patients had been treated with IFN before the investigations, we are convinced that these defects, with their consequent IFN unresponsiveness, are early events in the lymphomagenesis of these Th2 neoplasms. However, there is also one report of a CTCL-derived cell line cultured in the presence of IFN that acquired IFN resistance by reduced Stat-1 expression.31Because normal Th2 cells can be easily inhibited by IFN-γ,8 IFN-γ resistance might be an elegant pathway for Th2 tumor cells to avoid inhibition by reactive Th1 lymphocytes.
Endogenous IFN-γ and functional IFN-γ signal transduction are necessary for an appropriate immune response against infectious agents32-34 and are essential for tumor surveillance against other tumors as shown in IFNγR and Stat-1–deficient mice.35-37 Indeed, one third of human melanoma and nonadenocarcinoma lung tumor cell lines showed a quantitative reduction in IFN-γ sensitivity, and 24% of lung adenocarcinoma lines were totally unresponsive to IFN-γ transduction.37 We are convinced that IFN resistance is present in a substantial proportion of human malignancies, implying molecular treatment modalities targeting this deficiency.
Viral oncolysis was recently investigated after exploiting the differences in the physiology of tumor cells and surrounding normal tissue. A mutant adenovirus (Onyx-015) lacking the E1b region was reported to selectively replicate and destroy cells deriving from various p53-deficient tumors. The E1b protein binds to and inactivates the p53 protein.38 Moreover, the Onyx-015 virus showed significant antitumoral efficacy in nude mice.39-41 In addition, the fact that human reovirus preferentially infects and replicates in cells with an activated Ras-signaling cascade renders it a good candidate for selective oncolysis.42 Indeed, the intratumoral injection of human reovirus into Ras-transformed cells resulted in tumor regression in several mouse tumor models.43 Recently published data44 presented a nude-mouse model in which SK-MEL3, a melanoma cell line, was targeted by VSV after IFN treatment, resulting in the killing of tumor cells. In contrast, we studied fresh patient-derived PBMCs containing clonal CD4+ populations and showed a specific viral protection of nonclonal cells on IFN-α treatment, whereas CTCL cells bearing the clonal T-cell receptor became highly infected.
None of the currently used treatments, including radiotherapy or aggressive chemotherapy, demonstrated an impact on overall survival in patients with CTCL.3 Therefore, new biologically based treatment approaches have to be developed for these disfiguring and ultimately lethal diseases. Our data prove that the IFN resistance of CTCL-derived tumor cells can be targeted to allow the selective replication of lysing viral infectious particles in tumor cells in vitro after incubation with IFN-α.
Our simple and effective approach to target the pathologic IFN unresponsiveness of lymphoma T cells can be used for purging bone marrow or peripheral blood-derived stem cells of patients with CTCL before ablative chemotherapy and radiotherapy in vitro. It is probable that a similar approach can be even followed in vivo by injecting IFN-sensitive viral particles into patients after IFN priming. This approach could be, but may not be, accompanied by the use of genetically engineered viral vectors. A possible candidate for the use in humans is the VSV used in our experiments because neutralizing antibodies against VSV are not present in most humans. This approach is expected to be easy and safe and might realize a tumor-specific molecular intervention. If it is successful in patients with CTCL, it may be well applicable for other IFN-resistant tumors, such as lung cancer.
Acknowledgments
We thank E. Ludwig (Institute for Clinical Immunology) and E. Niederer (Institute for Biomedical Engineering at ETH Zurich) for excellent support with cell sorting and FACS analysis and E. Laine (Department of Dermatology, University Hospital Zurich) for cDNA preparation and technical support.
Supported by grant SKL 983-02-2000 from the Swiss Cancer League and the Stiftung für wissenschaftliche Forschung, University Zürich.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Reinhard Dummer, Department of Dermatology, University Hospital Zurich, Gloriastrasse 31, CH-8091 Zürich, Switzerland; e-mail: dummer@derm.unizh.ch.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal