Abstract
To determine the incidence of leukemia-specific rearrangements, 60 cases of childhood acute myeloblastic leukemia and transient myeloproliferative disorder were screened with a novel multiplex reverse transcriptase–polymerase chain reaction (RT-PCR) assay, and the results were correlated with the cytogenetic findings. The RT-PCR assay detects 28 different fusion genes and more than 80 different fusion transcript variants. RNA was isolated from methanol/acetic acid–fixed cells that had been routinely prepared for cytogenetic analysis. Nine different fusion transcripts were found in 40% of the cases, whereas 78.3% of the cases had abnormal karyotypes. Two cases with a t(6;11) and an MLL/AF6 gene fusion were missed cytogenetically. Conversely, cytogenetic analysis revealed 10 other well-defined chromosome rearrangements. Although cytogenetic analysis reveals a much broader range of abnormalities, multiplex RT-PCR serves as quality control and provides the essential information for minimal residual disease studies. Moreover, discrepant findings lead to the detection of new rearrangements on the molecular genetic level.
Introduction
Approximately 50% of adult and 80% of childhood acute myeloblastic leukemias (AMLs) harbor nonrandom karyotype abnormalities that define subentities with unique biological and clinical features.1-4 (Interactive database,http://www.infobiogen.fr/services/chromcancer/.) Cytogenetic analysis provides a comprehensive overview of overall quantitative and qualitative karyotype abnormalities and reveals clonal changes and secondary abnormalities. The cloning of translocation-associated breakpoints has led to the identification of a variety of genes that normally regulate and control cell division, growth, differentiation, and apoptosis.5,6 The fusion of such genes either leads to their abnormal activation or generates novel chimeric genes with neoplastic properties.5,6 More than 50 leukemia-specific fusion genes have been defined already.4-6 The resulting hybrid transcripts provide the essential basis for the development of reverse transcriptase–polymerase chain reaction (RT-PCR) techniques for the molecular genetic detection of such rearrangements.7-15 So far, the majority of RT-PCR screening programs have searched for each of the most common fusion transcripts individually.7-15 This is particularly true for those transcripts found in AML, whereas the clinically most important acute lymphocytic leukemia (ALL)–specific abnormalities have been combined in several types of multiplex assays.16 17 However, the steadily increasing number of detectable abnormalities makes the conventional screening approaches for single specific fusion transcripts more and more impractical and obsolete.
With that in mind, Pallisgaard et al18 have recently presented a multiplex RT-PCR assay that facilitates the detection of 29 fusion genes and more than 80 breakpoint and splice variants. We have used a similar modified assay (Hemavision; DNA Technology, Aarhus, Denmark, for Bio-Rad Laboratories, Hercules, CA) to screen all childhood AML cases that were collected at our institution during a 5-year period. Our particular aim was to compare the results obtained by this assay with those from conventional cytogenetic analysis and to assess the diagnostic specificity and value of both techniques.
Study design
Patients
Between 1993 and 1998, 67 children with AML or transient myeloproliferative disorder (TMD) were diagnosed in Austria and registered at our institution. Cytogenetic analysis was performed on bone marrow aspirates or peripheral blood samples that were obtained from 64 of these patients. Sufficient material for the multiplex RT-PCR assay was available from 60 patients. This study was approved by the ethics committee of the Children's Cancer Research Institute, and informed consent was obtained from the patients or their parents.
Multiplex RT-PCR
Total RNA was isolated from 48 methanol/acetic acid–fixed cell samples that had been stored at −20°C and from 17 cryopreserved samples according to methods described previously.19 20 We centrifuged 200-500 μL fixed-cell suspension, and pellets were washed with ethanol. Then RNA was prepared using the RNeasy Mini Kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer's recommendations. Samples were analyzed using a multiplex RT-PCR assay (HemaVision) according to the manufacturer's instructions. The assay discriminates between 28 different fusion transcripts. Reverse transcription was performed with a mixture of translocation-specific complementary DNA (cDNA) primers and PCR amplification in 8 parallel nested multiplex master reactions. Each of the master solutions contains several primers that are specific for particular fusion transcripts and a pair of control primers that amplifies a ubiquitously expressed gene. Thus, each of the master reactions identifies various chromosomal aberrations that, due to the heterogeneity of the breakpoints on the genomic level and/or alternative splicing, generate different messenger RNA (mRNA) variants. To verify the presence of a specific fusion transcript in a positive multiplex reaction, a nested split-out analysis with individual translocation-specific primer sets was performed. The specific translocation and splice variant was classified by comparing the respective pattern with the interpretation table provided by the manufacturer.
Results and discussion
Cytogenetic analysis of 64 childhood AML and TMD samples found an abnormal clone in 51 (79.7%) samples of the cases and a normal chromosome complement in 13 (20.3%) samples of the cases, whereas 24 (39%) of 60 samples analyzed by the multiplex RT-PCR were positive for one of 9 different fusion transcripts (Table1). Examples of the multiplex RT-PCR results are shown in Figure1.
Characteristics, multiplex reverse transcriptase–polymerase chain reaction results, and cytogenetic data of 67 cases of childhood acute myeloblastic leukemia
| Case . | Age (y) . | Sex . | FAB . | Multiplex RT-PCR . | Fusion transcript . | Karyotype . |
|---|---|---|---|---|---|---|
| 1 | 0.2 | M | AUL | neg | 46,XY,t(X;8)(p11;q23)[30] | |
| 2 | 14.0 | M | AUL | ND | 46,XX,del(5)(q13q33)[20]* | |
| 3 | 10.4 | M | M0 | MLL/AF4 | MLLex7/AF4(1414) | 48,XX,t(4;11)(q21;q23),+4,+mar[1]/48,XX,idem,−4,+der(4)t(4;11)(q21;q23)[19] |
| 4 | 0.5 | F | M0 | neg | 46,XX[3]/46,XX,t(6;X)(q22;p22)[17] | |
| 5 | 1.0 | F | M0 | neg | 46,XX,t(X;17)(p21;q11),r(6p)[15] | |
| 6 | 1.4 | F | M0 | neg | 48,XX,+8,+21c[20] | |
| 7 | 2.9 | M | M0 | neg | 45,XY,−7[40] | |
| 8 | 1.3 | M | M0 | ND | ND | |
| 9 | 0.01 | F | M1 | neg | 46,XX[5]/47,XX,+21[15] | |
| 10 | 6.4 | M | M1 | neg | 46,XY[13]/47,XY,+?del(19)(p13),del(2)(q13-14),inc[cp11] | |
| 11 | 0.3 | M | M1 | neg | 47,XY,+der(19)[40]† | |
| 12 | 9.0 | M | M2 | AML1/MGT8 | 46,XY[4]/46,XY,t(8;21)(q22;q22)[16] | |
| 13 | 7.8 | M | M2 | AML1/MGT8 | 45,X,−Y,t(8;21)(q22;q22)[5] | |
| 14 | 11.7 | M | M2 | AML1/MGT8 | 46,XY[1]/45,X,−Y,t(8;21)(q22;q22)[15] | |
| 15 | 10.1 | M | M2 | AML1/MGT8 | 45,X,−Y,t(8;21)(q22;q22)[20] | |
| 16 | 3.8 | M | M2 | AML1/MGT8 | 45,X,−Y,t(8;21)(q22;q22)[20] | |
| 17 | 4.3 | F | M2 | AML1/MGT8 | 46,XX,t(8;21)(q22;q22)[20] | |
| 18 | 7.9 | F | M2 | AML1/MGT8 | 46,XX,t(8;21)(q22;q22)[17] | |
| 19 | 8.4 | F | M2 | MLL/ELL | MLLex7/ELL | 46,XX,t(11;19)(q23;p13)[10] |
| 20 | 12.1 | M | M2 | neg | 46,XY,del(5)(q14q34)[15]/45,idem,−Y[15] | |
| 21 | 15.2 | F | M2 | neg | 46,XX[15] | |
| 22 | 12.0 | F | M2 | neg | 46,XX[40] | |
| 23 | 15.8 | F | M2 | neg | 46,XX[20] | |
| 24 | 14.4 | F | M2 | neg | 46,XX[20] | |
| 25 | 3.0 | F | M2 | neg | 46,XX[19] | |
| 26 | 4.0 | M | M2 | neg | 46,XY[20] | |
| 27 | 9.6 | F | M2 | ND | ND | |
| 28 | 3.2 | F | M3 | PML/RARA | PMLex6/RARAex2 | 46,XX,t(15;17)(q22;q21)[20] |
| 29 | 13.8 | F | M3 | PML/RARA | PMLex6/RARAex2 | 46,XX[3]/46,XX,t(15;17)(q22;q21)[17] |
| 30 | 12.2 | M | M3 | PML/RARA | PMLex6/RARAex2 | 46,XY,t(4;9)(q31;q34)t(15;17)(q22;q21)[16] |
| 31 | 1.8 | M | M3V | PML/RARA | PMLex6/RARAex2 | 46,XY,t(15;17)(q22;q12),del(17)(p11),t(17;17)(q11;p13)[30] |
| 32 | 10.3 | F | M3V | PML/RARA | PMLex6/RARAex2 | 46,XX,t(15;17)(q22;q21)[20] |
| 33 | 14.0 | F | M4 | MLL/AF6 | MLLex7/AF6 | 46,XX[20] |
| 34 | 12.7 | F | M4 | MLL/AF6 | MLLex6/AF6 | 46,XX[6]/46,del(11)(q23)[20]/46,XX,idem,del(9)(q22),[2] |
| 35 | 1.8 | F | M4 | MLL/AF1q | MLLex6/AF1q | 46,XX[2]/46,XX,t(1;11)(q21;q23),t(12;12)(q13;p12)[18] |
| 36 | 11.9 | M | M4 | CBFB/MYH11 | type A | 46,XY,inv(16)(p13q22),t(16;17)(p10;q10)[20] |
| 37 | 0.8 | M | M4 | neg | MLL/? | 46,XY,der(11)add(11)(q23)[20]‡ |
| 38 | 8.8 | M | M4 | neg | MLL/? | 54,XY,+Y,+6,+7,+8,+8,t(11;17)(q23;q25),+19,+20,+21[20] |
| 39 | 2.1 | F | M4 | neg | MLL/? | 46,XX,t(11;17)(q23;q21)[20] |
| 40 | 12.2 | F | M4 | neg | 46,XX[23]/46,XX,t(3;5)(q26;q13-14)[2] | |
| 41 | 13.4 | M | M4 | neg | 46,XY[8]/45,X−Y[1]90,idemx2[10] | |
| 42 | 16.2 | M | M4 | neg | 46,XY[2]/46,XY,t(7;14)(p15;q32)[19] | |
| 43 | 1.2 | M | M4 | neg | MLL/new gene | 46,XY[13]/46,XY,inv(11)(q12q23)[7]1-153 |
| 44 | 2.8 | M | M4 | ND | 46,XY[17]/46,XY,inv(8)(p11q13)[3] | |
| 45 | 11.5 | M | M4 | ND | 46,XY[9]/46,XY,del(5)(q13q31),t(7;11)(p15;p12)[9] | |
| 46 | 2.0 | F | M4Eo | CBFB/MYH11 | type A | 46,XX[12]/47,XX,+8[2]/46,XX,inv(16)(p13q22)[6] |
| 47 | 11.9 | M | M4Eo | ND | 46,XY,inv(16)(p13q22),t(15;16)(q22;p13)inv(16)(p13q22)[20] | |
| 48 | 14.0 | M | M5a | MLL/AF9 | MLLex6/AF9A | 46,XY,t(9;11)(p21;q23)[4]/46,XY,idem,del(13)(q14)[16] |
| 49 | 16.4 | F | M5a | MLL/AF9 | MLLex6/AF9B | 46,XX,t(9;11)(p21;q23)[20] |
| 50 | 14.6 | M | M5a | MLL/AF9 | MLLex7/AF9A | 46,XY,t(9;11)(p21;q23)[20] |
| 51 | 4.5 | M | M5a | MLL/AF9 | MLLex7/AF9A | 47,XY,t(9;11)(p21;q23),+8[20] |
| 52 | 7.3 | M | M5a | MLL/AF10 | MLLex6/AF10(883) | 46,XY[2]/46,XY,t(10;11)(p12;q23),t(10;12)(q24;p13)[2]/47,XY,idem,+del(1)(p13)[16] |
| 53 | 10.8 | F | M5b | neg | 46,XX[25] | |
| 54 | 6.0 | M | M6 | neg | 46,XY[3]/46-48,XY,inv(1)(p21p31),+del(1)(q21),der(1)t(1;8)(p13;q10),−6,−7,del(7)(q33), del(8)(q12),−9,−10,−21,+r,+1−3mar[cp13] | |
| 55 | 0.8 | M | M7 | neg | 46,XY[20] | |
| 56 | 1.9 | M | M7 | neg | 48,XY,del(6)(q21q24),+21,+21c[20] | |
| 57 | 0.7 | M | M7 | neg | 47,XY,t(1;22)(p13;q13.1),t(3;19)(p23;p13),t(3;15)(q22;q22),+19[8]/46,XY,idem,−20[2] | |
| 58 | 1.7 | F | M7 | neg | 47,XX,inv(9)(p11q13)c,+21c[1]/47,XX,idem,der(4)t(1;4)(q23;p15),[17]/47,XX,idem,iso(7)(q10)[2] | |
| 59 | 1.4 | F | M7 | neg | 47,XX,+21c[7]/47,XX,der(7)t(1;7)(q22;p22),+21c[11]/47,XX,dup(1)(q22q44),+21c[2] | |
| 60 | 1.2 | M | M7 | neg | 47,XY,t(2;4)(q37;q28),+21c[20] | |
| 61 | 1.9 | M | M7 | neg | 47,XY,+21c[18]/48,XY,+8,+21c[1]/49,XY,+8,+12,+21c[1] | |
| 62 | 1.9 | M | M7 | neg | 47,XY,+21c[16]/48,XY,+8,+21c[4] | |
| 63 | 1.3 | F | M7 | neg | 47,XX,+21c[20] | |
| 64 | 0.01 | M | M7/TMD | neg | 47,XY,+21c[20] | |
| 65 | 0.02 | M | M7/TMD | neg | 47,XY,+21c[27] | |
| 66 | 0.01 | M | M7/TMD | neg | 47,XY,+21c[20] | |
| 67 | 0.01 | F | M7/TMD | ND | ND |
| Case . | Age (y) . | Sex . | FAB . | Multiplex RT-PCR . | Fusion transcript . | Karyotype . |
|---|---|---|---|---|---|---|
| 1 | 0.2 | M | AUL | neg | 46,XY,t(X;8)(p11;q23)[30] | |
| 2 | 14.0 | M | AUL | ND | 46,XX,del(5)(q13q33)[20]* | |
| 3 | 10.4 | M | M0 | MLL/AF4 | MLLex7/AF4(1414) | 48,XX,t(4;11)(q21;q23),+4,+mar[1]/48,XX,idem,−4,+der(4)t(4;11)(q21;q23)[19] |
| 4 | 0.5 | F | M0 | neg | 46,XX[3]/46,XX,t(6;X)(q22;p22)[17] | |
| 5 | 1.0 | F | M0 | neg | 46,XX,t(X;17)(p21;q11),r(6p)[15] | |
| 6 | 1.4 | F | M0 | neg | 48,XX,+8,+21c[20] | |
| 7 | 2.9 | M | M0 | neg | 45,XY,−7[40] | |
| 8 | 1.3 | M | M0 | ND | ND | |
| 9 | 0.01 | F | M1 | neg | 46,XX[5]/47,XX,+21[15] | |
| 10 | 6.4 | M | M1 | neg | 46,XY[13]/47,XY,+?del(19)(p13),del(2)(q13-14),inc[cp11] | |
| 11 | 0.3 | M | M1 | neg | 47,XY,+der(19)[40]† | |
| 12 | 9.0 | M | M2 | AML1/MGT8 | 46,XY[4]/46,XY,t(8;21)(q22;q22)[16] | |
| 13 | 7.8 | M | M2 | AML1/MGT8 | 45,X,−Y,t(8;21)(q22;q22)[5] | |
| 14 | 11.7 | M | M2 | AML1/MGT8 | 46,XY[1]/45,X,−Y,t(8;21)(q22;q22)[15] | |
| 15 | 10.1 | M | M2 | AML1/MGT8 | 45,X,−Y,t(8;21)(q22;q22)[20] | |
| 16 | 3.8 | M | M2 | AML1/MGT8 | 45,X,−Y,t(8;21)(q22;q22)[20] | |
| 17 | 4.3 | F | M2 | AML1/MGT8 | 46,XX,t(8;21)(q22;q22)[20] | |
| 18 | 7.9 | F | M2 | AML1/MGT8 | 46,XX,t(8;21)(q22;q22)[17] | |
| 19 | 8.4 | F | M2 | MLL/ELL | MLLex7/ELL | 46,XX,t(11;19)(q23;p13)[10] |
| 20 | 12.1 | M | M2 | neg | 46,XY,del(5)(q14q34)[15]/45,idem,−Y[15] | |
| 21 | 15.2 | F | M2 | neg | 46,XX[15] | |
| 22 | 12.0 | F | M2 | neg | 46,XX[40] | |
| 23 | 15.8 | F | M2 | neg | 46,XX[20] | |
| 24 | 14.4 | F | M2 | neg | 46,XX[20] | |
| 25 | 3.0 | F | M2 | neg | 46,XX[19] | |
| 26 | 4.0 | M | M2 | neg | 46,XY[20] | |
| 27 | 9.6 | F | M2 | ND | ND | |
| 28 | 3.2 | F | M3 | PML/RARA | PMLex6/RARAex2 | 46,XX,t(15;17)(q22;q21)[20] |
| 29 | 13.8 | F | M3 | PML/RARA | PMLex6/RARAex2 | 46,XX[3]/46,XX,t(15;17)(q22;q21)[17] |
| 30 | 12.2 | M | M3 | PML/RARA | PMLex6/RARAex2 | 46,XY,t(4;9)(q31;q34)t(15;17)(q22;q21)[16] |
| 31 | 1.8 | M | M3V | PML/RARA | PMLex6/RARAex2 | 46,XY,t(15;17)(q22;q12),del(17)(p11),t(17;17)(q11;p13)[30] |
| 32 | 10.3 | F | M3V | PML/RARA | PMLex6/RARAex2 | 46,XX,t(15;17)(q22;q21)[20] |
| 33 | 14.0 | F | M4 | MLL/AF6 | MLLex7/AF6 | 46,XX[20] |
| 34 | 12.7 | F | M4 | MLL/AF6 | MLLex6/AF6 | 46,XX[6]/46,del(11)(q23)[20]/46,XX,idem,del(9)(q22),[2] |
| 35 | 1.8 | F | M4 | MLL/AF1q | MLLex6/AF1q | 46,XX[2]/46,XX,t(1;11)(q21;q23),t(12;12)(q13;p12)[18] |
| 36 | 11.9 | M | M4 | CBFB/MYH11 | type A | 46,XY,inv(16)(p13q22),t(16;17)(p10;q10)[20] |
| 37 | 0.8 | M | M4 | neg | MLL/? | 46,XY,der(11)add(11)(q23)[20]‡ |
| 38 | 8.8 | M | M4 | neg | MLL/? | 54,XY,+Y,+6,+7,+8,+8,t(11;17)(q23;q25),+19,+20,+21[20] |
| 39 | 2.1 | F | M4 | neg | MLL/? | 46,XX,t(11;17)(q23;q21)[20] |
| 40 | 12.2 | F | M4 | neg | 46,XX[23]/46,XX,t(3;5)(q26;q13-14)[2] | |
| 41 | 13.4 | M | M4 | neg | 46,XY[8]/45,X−Y[1]90,idemx2[10] | |
| 42 | 16.2 | M | M4 | neg | 46,XY[2]/46,XY,t(7;14)(p15;q32)[19] | |
| 43 | 1.2 | M | M4 | neg | MLL/new gene | 46,XY[13]/46,XY,inv(11)(q12q23)[7]1-153 |
| 44 | 2.8 | M | M4 | ND | 46,XY[17]/46,XY,inv(8)(p11q13)[3] | |
| 45 | 11.5 | M | M4 | ND | 46,XY[9]/46,XY,del(5)(q13q31),t(7;11)(p15;p12)[9] | |
| 46 | 2.0 | F | M4Eo | CBFB/MYH11 | type A | 46,XX[12]/47,XX,+8[2]/46,XX,inv(16)(p13q22)[6] |
| 47 | 11.9 | M | M4Eo | ND | 46,XY,inv(16)(p13q22),t(15;16)(q22;p13)inv(16)(p13q22)[20] | |
| 48 | 14.0 | M | M5a | MLL/AF9 | MLLex6/AF9A | 46,XY,t(9;11)(p21;q23)[4]/46,XY,idem,del(13)(q14)[16] |
| 49 | 16.4 | F | M5a | MLL/AF9 | MLLex6/AF9B | 46,XX,t(9;11)(p21;q23)[20] |
| 50 | 14.6 | M | M5a | MLL/AF9 | MLLex7/AF9A | 46,XY,t(9;11)(p21;q23)[20] |
| 51 | 4.5 | M | M5a | MLL/AF9 | MLLex7/AF9A | 47,XY,t(9;11)(p21;q23),+8[20] |
| 52 | 7.3 | M | M5a | MLL/AF10 | MLLex6/AF10(883) | 46,XY[2]/46,XY,t(10;11)(p12;q23),t(10;12)(q24;p13)[2]/47,XY,idem,+del(1)(p13)[16] |
| 53 | 10.8 | F | M5b | neg | 46,XX[25] | |
| 54 | 6.0 | M | M6 | neg | 46,XY[3]/46-48,XY,inv(1)(p21p31),+del(1)(q21),der(1)t(1;8)(p13;q10),−6,−7,del(7)(q33), del(8)(q12),−9,−10,−21,+r,+1−3mar[cp13] | |
| 55 | 0.8 | M | M7 | neg | 46,XY[20] | |
| 56 | 1.9 | M | M7 | neg | 48,XY,del(6)(q21q24),+21,+21c[20] | |
| 57 | 0.7 | M | M7 | neg | 47,XY,t(1;22)(p13;q13.1),t(3;19)(p23;p13),t(3;15)(q22;q22),+19[8]/46,XY,idem,−20[2] | |
| 58 | 1.7 | F | M7 | neg | 47,XX,inv(9)(p11q13)c,+21c[1]/47,XX,idem,der(4)t(1;4)(q23;p15),[17]/47,XX,idem,iso(7)(q10)[2] | |
| 59 | 1.4 | F | M7 | neg | 47,XX,+21c[7]/47,XX,der(7)t(1;7)(q22;p22),+21c[11]/47,XX,dup(1)(q22q44),+21c[2] | |
| 60 | 1.2 | M | M7 | neg | 47,XY,t(2;4)(q37;q28),+21c[20] | |
| 61 | 1.9 | M | M7 | neg | 47,XY,+21c[18]/48,XY,+8,+21c[1]/49,XY,+8,+12,+21c[1] | |
| 62 | 1.9 | M | M7 | neg | 47,XY,+21c[16]/48,XY,+8,+21c[4] | |
| 63 | 1.3 | F | M7 | neg | 47,XX,+21c[20] | |
| 64 | 0.01 | M | M7/TMD | neg | 47,XY,+21c[20] | |
| 65 | 0.02 | M | M7/TMD | neg | 47,XY,+21c[27] | |
| 66 | 0.01 | M | M7/TMD | neg | 47,XY,+21c[20] | |
| 67 | 0.01 | F | M7/TMD | ND | ND |
For the fusion transcript data, the interpretation table in the HemaVision kit uses the old exon nomenclature of the MLL gene; exons 6-7 correspond to exons 9-10 of the new numbering according to Nilson et al.21
FAB indicates French-American-British; RT-PCR, reverse transcriptase–polymerase chain reaction; M, male; AUL, acute undifferentiated leukemia; neg, negative; ND, not determined; F, female.
FISH analysis revealed an additional t(5;11)(q35;p15).
FISH analysis revealed a cryptic t(7;12)(q36;p12).
Whole chromosome painting showed a t(11;17).
The inv(11)(q12q23) was detected by FISH analysis, and a new MLL fusion partner was cloned (Litzka et al, unpublished data, 2000).
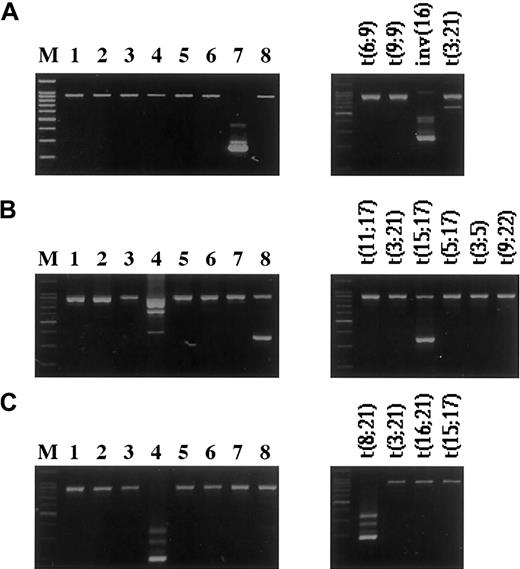
Examples of fusion genes detected by the multiplex RT-PCR.
Amplification products of 8 parallel multiplex RT-PCR reactions (left panels) and the corresponding split-out reactions (right panels). The product with the higher molecular weight represents the internal positive control, and the lower bands represent the specific fusion genes. (A) inv(16)(p13q22)—CBFB/MYH11 fusion gene. (B) t(15;17)(q21;q22)—PML/RARA. (Note: Due to the primer combinations for the detection of different breakpoints, a few of the translocations detected by the HemaVision kit can appear in more than one master reaction, as seen in lanes 4 and 8. In the right panel of B, split-out reactions correspond to master reaction no.8.) (C) t(8;21)(q22;q22)—AML1/MGT8 (M, 100-bp DNA Ladder (Promega).
Examples of fusion genes detected by the multiplex RT-PCR.
Amplification products of 8 parallel multiplex RT-PCR reactions (left panels) and the corresponding split-out reactions (right panels). The product with the higher molecular weight represents the internal positive control, and the lower bands represent the specific fusion genes. (A) inv(16)(p13q22)—CBFB/MYH11 fusion gene. (B) t(15;17)(q21;q22)—PML/RARA. (Note: Due to the primer combinations for the detection of different breakpoints, a few of the translocations detected by the HemaVision kit can appear in more than one master reaction, as seen in lanes 4 and 8. In the right panel of B, split-out reactions correspond to master reaction no.8.) (C) t(8;21)(q22;q22)—AML1/MGT8 (M, 100-bp DNA Ladder (Promega).
Of the 24 RT-PCR–positive cases, 22 (91.7%) cases had correlating cytogenetic findings. In 2 cases (nos. 33 and 34) with anMLL/AF6 fusion transcript, the presence of the t(6;11)(q27;q23) had been missed cytogenetically. In samples from cases Nos. 33 and 34, only normal metaphases and a clone with a del(11)(q23) were found, respectively. Due to the location of the breakpoints in the telomeric regions of the chromosomes and submicroscopic deletions that occur in approximately 20% to 30% of such cases, this cryptic translocation is difficult to identify by cytogenetic and fluorescence in situ hybridization (FISH) analysis.22
Of particular interest are cases with 11q23 abnormalities and rearrangements of the MLL gene, which account for 5% to 10% of acquired karyotype changes in childhood and adult acute leukemias and myelodysplastic syndromes. At least 40 different 11q23 translocations have been described cytogenetically, and 23 MLLfusion partner genes have already been identified.4Our group of patients included 14 (23%) cases with 11q23 abnormalities. Thirteen (20.3%) cases with 8 different structural abnormalities were identified cytogenetically. They consisted of 6 different translocations that involved chromosomes 1, 4, 9, 10, 17, and 19 as well as one inversion and one del(11)(q23). Using the multiplex RT-PCR, 10 positive cases with 6 different fusion transcripts were found.
Despite the fact that the multiplex RT-PCR kit allows the detection of 10 of the currently 23 cloned MLL fusion partners, we nevertheless encountered 4 cases with an involvement of theMLL gene that remained undetected in the RT-PCR analysis. In case nos. 38 and 39 at t(11;17)(q23;q21∼q25) and in case no. 37 additional chromosome material at 11q23 was found by cytogenetic analysis. In all 3 cases, whole chromosome painting and FISH using MLL-specific PAC clones confirmed the presence of a t(11;17) and involvement of the MLL gene (data not shown).23Although the multiplex RT-PCR detects 2 t(11;17)-associated fusion transcripts, MLL/AF17 andPLZF/RARA, neither of them was present in the samples. Moreover, 2 of these cases were also negative for the only other currently known MLL fusion partner on 17q25 (MLL/MSF) (data not shown). These data suggest the presence of a cluster of MLL fusion partners on 17q similar to those already known at 10p11.2-12 (ABII andAF10) and 19p13.1-13.3 (ENL,ELL/MEN, and EEN).4 In the fourth case (no. 43), with a paracentric inv(11)(q12q23) andMLL involvement, we were able to clone a new MLLfusion partner (Litzka et al, unpublished data, 2000).
Two other cytogenetically detected specific chromosome rearrangements, t(7;11)(p15;p13) (case no. 45) and inv(8)(p11p13) (case no. 44), have molecular genetic equivalents in the form of NUP98/HOXA9 andMOZ/TIF2 fusion genes, respectively.4 However, these fusion genes are not covered by the RT-PCR kit. In addition, 2 other translocations that are specific for particular subsets of myeloid neoplasms, t(1;22)(p13;q13) (case no. 57) and t(5;11)(q35;p15) (case no. 2), were encountered.24,25 FISH analysis also revealed a case with a cryptic translocation t(7;12)(q36;p12) (case no. 11) in an infant in whom cytogenetic analysis had only discovered a marker chromosome 19.26 All these translocations are currently not analyzable on the molecular genetic level because the involved genes have not been cloned yet. Finally, we observed a t(3;5)(q26;q13-14) (case no. 40) with breakpoints that differed from those generating the RT-PCR–detectable fusion geneNPM/MLF1.4
The results of the comparative karyotype and RT-PCR analyses prove that they are complimentary techniques and are both indispensable for the evaluation of the disease-specific genetic features of myeloid malignancies. Although an increasing number of reciprocal rearrangements are detectable by molecular genetic means, karyotyping still provides the most comprehensive overview that is not obtainable by any other method. The demonstration of particular fusion transcripts by RT-PCR, on the other hand, is essential for subsequent molecular genetic follow-up and minimal residual disease studies.
Acknowledgments
The authors gratefully acknowledge U. Horcika, E. Lang, B. Nistler, T. Pass, H. Pirc-Danoewinata, and B. Ulm for the excellent cytogenetic analysis.
Supported by the Österreichische Kinderkrebshilfe and private donations.
Dedicated to Prof Helmut Gadner on the occasion of his 60th birthday.
Submitted July 12, 2000; accepted September 28, 2000.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Oskar A. Haas, CCRI, St Anna Children's Hospital, Kinderspitalgasse 6, A-1090, Vienna, Austria; e-mail:o.a.haas@magnet.at.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal