Point mutations in the granulocyte colony-stimulating factor receptor (G-CSFR) gene have been linked to the development of secondary leukemia in patients with congenital neutropenia (CN). This report presents data on a now 18-year-old patient with CN who has received G-CSF treatment since 1989 and who developed acute myeloid leukemia (AML) in 1998. To evaluate whether there is an association between the occurrence of point mutations of the G-CSFR gene and development of secondary AML, DNA/messenger RNA of neutrophils and mononuclear cells from this patient were analyzed at different time points by polymerase chain reaction and subsequent cloning by DNA sequencing of representative numbers of individual clones. Findings suggest an increasing instability of the G-CSFR gene in time as judged by increasing numbers of mutations proposed to be one important step in the development of AML in this patient.
Introduction
We describe the time course of a stepwise acquisition of different granulocyte colony-stimulating factor receptor(G-CSFR) gene mutations in a patient with severe congenital neutropenia (CN, Kostmann syndrome). In this patient, the occurrence of these mutations significantly correlated with the transformation of CN into acute myeloid leukemia (AML).
Congenital neutropenia is characterized by a maturation arrest of myeloid progenitor cells at the promyelocyte-myelocyte stage, resulting in absence or low levels of mature neutrophils. Patients with CN usually develop recurrent localized or systemic bacterial and fungal infections, beginning in early infancy. CN treatment with granulocyte colony-stimulating factor (G-CSF) results in an increase of absolute neutrophil count (ANC), significantly fewer infections, and improvement of quality of life.1
Study design
We report data on an 18-year-old girl who was diagnosed with CN in 1982, at the age of 7 months. In 1989, the patient presented with a history of severe bacterial infections and an ANC persistently below 0.2 × 109/L. G-CSF treatment was initiated with 3 μg/kg per day subcutaneously and her ANC increased to above 1.0 × 109/L. During the following 9 years of G-CSF treatment, the patient developed normally and did clinically well without severe infections.
At a routine hospital visit in March 1998, her white blood count was 5.1 × 109/L and contained 11% myeloid blast cells. Acute myeloid leukemia (AML-M5b) was diagnosed by morphologic criteria. One third of the leukemic cells demonstrated monosomy 7 and two thirds showed a tetraploid karyotype. Treatment was initiated according to the AML-BFM-98 protocol,2 followed by bone marrow transplantation (BMT). Bone marrow was donated by her haplo-identical (2 loci mismatched) half-brother in July 1998.
Results and discussion
Patients with CN have an increased risk of approximately 10% to develop myelodysplastic syndrome (MDS) or AML or both.1,3,4 Acquired nonsense mutations in a critical region between nucleotide (nt) 2360 and nt 2430 of theG-CSFR gene encoding an intracellular part of the G-CSFR protein, which contributes to myeloid proliferation and maturation signaling, are thought to be involved in the leukemogenesis in these patients.3,5,6 These G-CSFR gene mutations introduce a stop codon, potentially leading to a loss of parts of the carboxyterminal domain of the receptor protein (Figure1). Interestingly, receptor proteins truncated in this way were shown to increase cell proliferation with arrest of differentiation.5
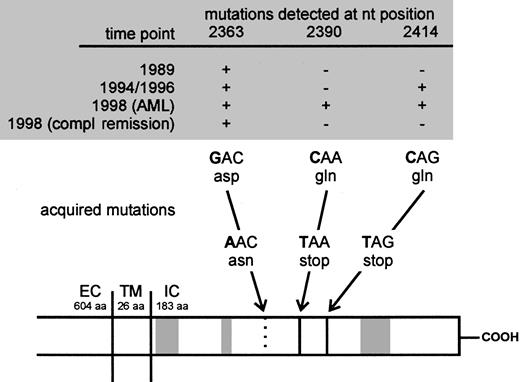
Sequence of acquisition of mutations in the CN patient demonstrated on a schematic presentation of the G-CSFR.
Arrows indicate positions of acquired point mutations of the G-CSFR mRNA at nt 2363, 2390, and 2414. The mutations either lead to an amino acid exchange from aspartate (asp) to asparagine (asn; nt 2363) or from glutamine (gln) to stop codons (TAA and TAG; nt 2390 and 2414). Gray boxes indicate the homologous domains (box 1-3) conserved in the cytokine receptor family. EC indicates extracellular; TM, transmembrane; IC, intracellular domain; compl, complete; aa, amino acids; nt, nucleotide.
Sequence of acquisition of mutations in the CN patient demonstrated on a schematic presentation of the G-CSFR.
Arrows indicate positions of acquired point mutations of the G-CSFR mRNA at nt 2363, 2390, and 2414. The mutations either lead to an amino acid exchange from aspartate (asp) to asparagine (asn; nt 2363) or from glutamine (gln) to stop codons (TAA and TAG; nt 2390 and 2414). Gray boxes indicate the homologous domains (box 1-3) conserved in the cytokine receptor family. EC indicates extracellular; TM, transmembrane; IC, intracellular domain; compl, complete; aa, amino acids; nt, nucleotide.
To evaluate a potential association between point mutations in the critical region of the G-CSFR gene and secondary AML, we examined DNA of bone marrow or messenger RNA (mRNA) of polymorphonuclear cells (PMCs) and mononuclear cells (MNCs) of our patient. Samples from different time points were analyzed by polymerase chain reaction (PCR) amplification of a region between nt 2306 and 2906 and subsequent cloning followed by DNA sequencing of representative numbers of individual Escherichia coli clones (Table1; nucleotide numbering refers to Fukunaga and colleagues7). Analyses of genomic DNA (gDNA) from 1989, a time point before initiation of G-CSF therapy, revealed a point mutation at nt 2363 leading to an exchange from aspartate to asparagine. Up to now this mutation could only be shown in myeloid cells of 2 other CN patients (M.G., unpublished results, January 2001). None of the tested normal donors or patients with other leukemic or preleukemic disorders demonstrated this point mutation. We detected an additional G-CSFR gene mutation at nt 2414 leading to a stop codon and to a truncated G-CSFR protein in samples from May 1994 (Figure 1). At this time, clonal heterogeneity was confirmed by detection of clones harboring either the mutation at nt 2363 or 2414 alone, or a combination of both mutations. Samples of MNC RNA from April 1996 revealed similar results (Table 1). The sequence of acquisition of mutations, in particular at that stage, when the second mutation (C2414T) is present in clones with as well as without the nt 2363 mutation (May 1994, April 1996) suggests that the nt 2414 mutation occurred in different hematopoietic cell clones (with and without the nt 2363 mutation). Another possibility could be that the nt 2414 mutation occurred in the nt 2363 clone, but more or less simultaneously on the wild-type allele and the mutated allele.
Sequence analyses of different clinical stages in a patient with congenital neutropenia who developed acute myeloid leukemia
| . | CN . | CN/AML . | During chemotherapy June 1998 . | Complete remission July 1998 . | After BMT August 1999 . | |||
|---|---|---|---|---|---|---|---|---|
| Time point . | Nov 1989* . | May 1994 . | April 1996 . | March 1998 . | ||||
| Sample material | BM gDNA | PB-PMC RNA | PB-MNC RNA | PB-PMC RNA | PB-MNC RNA | PB-PMC RNA | PB-PMC RNA | PB-PMC RNA |
| Total number of clones tested (100%) | 25 | 27 | 26 | 32 | 28 | 26 | 25 | 28 |
| No. of mutated clones | ||||||||
| nt 2390 C → T† | — | — | — | 2 (6%) | — | — | — | — |
| nt 2414 C → T | — | 3 (11%) | 2 (8%) | 1 (3%) | 1 (4%) | — | — | — |
| nt 2363 G → A plus nt 2414 C → T | — | 16 (59%) | 7 (27%) | 10 (31%) | 12 (43%) | — | — | — |
| nt 2363 G → A | 15 (60%) | 3 (11%) | 6 (23%) | 5 (16%) | — | 10 (38%) | 11 (44%) | — |
| Wild-type sequence | 10 (40%) | 5 (19%) | 11 (42%) | 14 (44%) | 15 (53%) | 16 (62%) | 14 (56%) | 28 (100%) |
| . | CN . | CN/AML . | During chemotherapy June 1998 . | Complete remission July 1998 . | After BMT August 1999 . | |||
|---|---|---|---|---|---|---|---|---|
| Time point . | Nov 1989* . | May 1994 . | April 1996 . | March 1998 . | ||||
| Sample material | BM gDNA | PB-PMC RNA | PB-MNC RNA | PB-PMC RNA | PB-MNC RNA | PB-PMC RNA | PB-PMC RNA | PB-PMC RNA |
| Total number of clones tested (100%) | 25 | 27 | 26 | 32 | 28 | 26 | 25 | 28 |
| No. of mutated clones | ||||||||
| nt 2390 C → T† | — | — | — | 2 (6%) | — | — | — | — |
| nt 2414 C → T | — | 3 (11%) | 2 (8%) | 1 (3%) | 1 (4%) | — | — | — |
| nt 2363 G → A plus nt 2414 C → T | — | 16 (59%) | 7 (27%) | 10 (31%) | 12 (43%) | — | — | — |
| nt 2363 G → A | 15 (60%) | 3 (11%) | 6 (23%) | 5 (16%) | — | 10 (38%) | 11 (44%) | — |
| Wild-type sequence | 10 (40%) | 5 (19%) | 11 (42%) | 14 (44%) | 15 (53%) | 16 (62%) | 14 (56%) | 28 (100%) |
CN indicates congenital neutropenia; AML, acute myeloid leukemia; BMT, bone marrow transplantation; PB, peripheral blood; PMC, polymorphonuclear cell; MNC, mononuclear cell; nt, nucleotide position.
Before initiation of G-CSF therapy.
Nucleotide position refers to Fukunaga et al.5
In 1997, karyotype analysis for the patient reported here was still normal. At the time leukemia was diagnosed, 4 distinct variants of mutated G-CSFR genes were detectable: one variant with the mutation at nt 2363 alone, one with the mutation at nt 2414 alone, one with both mutations, and another with a newly diagnosed nonsense mutation at nt 2390, also introducing a stop codon (Figure 1 and Table1).
After induction of chemotherapy, the clones of leukemic cells expressing mutations at nt 2414 and nt 2390 and displaying monosomy 7 disappeared. However, cells harboring the mutation at nt 2363 were still detectable during clinical remission until BMT was performed in July 1998. After BMT, none of the mutations could be detected in the patient's hematopoietic cells (Table 1).
The increasing number of mutations reported here document the instability of the G-CSFR gene, which is proposed to be one of the reasons for malignant transformation.
Until now we detected 7 CN patients with multiple and 11 with single nonsense mutations in the critical region of theG-CSFR gene.3 To date, 5 patients with multiple mutations and 6 patients with single mutations developed secondary leukemia. Accordingly, we found a highly significant correlation between the occurrence of one or more G-CSFR gene mutations and leukemogenesis in CN patients rather than a correlation between the number of different mutations and the risk of developing leukemia. Furthermore, as CN patients with a single G-CSFR gene mutation as well as CN patients with multiple mutations developed secondary AML with a comparable incidence, multiple mutations may indicate a general and possibly increasing instability of theG-CSFR gene. The risk of developing AML in patients with CN seems to increase drastically with the occurrence of the first single mutation in the unstable G-CSFR gene as one early step in leukemogenesis. Other chromosomal aberrations are rather late events only detectable in the overt leukemic cells. Our data confirm that mutations of the G-CSFR gene are acquired during the course of disease and represent important initial steps from CN toward leukemia.
The contribution of the G-CSF therapy to the development ofG-CSFR gene mutations or leukemia remains unclear. However, data from the pre–G-CSF era already demonstrate that CN is a preleukemic disorder leading to leukemia in some patients. In addition, data of the Severe Chronic Neutropenia International Registry demonstrate that none of the patients with cyclic and idiopathic neutropenia who were also treated with G-CSF with similar dosages and for the same length of time developed either point mutations in the critical region of the G-CSFR gene or leukemia.1 4
Taking into consideration the time course of mutations in the CN patient reported here and the highly significant correlation between the occurrence of G-CSFR gene mutations and leukemogenesis in CN patients, we recommend a yearly evaluation of G-CSFRgene mutations in all CN patients to consider BMT options early if nonsense mutations in the critical region of the G-CSFR gene are identified.1,3,4 6
Supported by a grant from the Deutsche Krebshilfe (10-1548-We2).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Karl Welte, Medizinische Hochschule Hannover, Paediatrische Haematologie und Onkologie, Carl-Neuberg-Strasse 1, 30625 Hannover, Germany; e-mail:welte.karl@mh-hannover.de.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal