Abstract
Thrombin-activable fibrinolysis inhibitor (TAFI) is a recently described carboxypeptidase that is potentially involved in the regulation of fibrinolysis by decreasing plasminogen binding to the fibrin surface. This role makes the TAFI gene a good candidate in atherothrombotic diseases. The great interindividual variability of plasma TAFI antigen levels is poorly explained by lifestyle characteristics, thus suggesting its genetic determination. To test this hypothesis, the promoter and the 3′-untranslated region of the TAFI gene were screened for polymorphisms, and their contribution to the variability of plasma TAFI antigen levels was evaluated. Seven new polymorphisms are described, 5 in the promoter (C-2599G, −2345 2G/1G, A-1690G, G-1102T, and G-438A) and 2 in the 3′UTR (C+1542G and T+1583A). All these polymorphisms were in strong linkage disequilibrium with each other and with the previously described Ala147Thr polymorphism. They generated 4 main haplotypes, accounting for 80% of all observed haplotypes. In univariate analyses, all polymorphisms were associated with plasma TAFI Ag levels and, individually, contributed to a large fraction of plasma TAFI Ag levels, ranging from 20% to 52%. In a stepwise regression analysis including all polymorphisms, several combinations remained significantly and independently associated with plasma TAFI Ag levels: C+1542G associated with Ala147Thr, T+1583A, or −2345 2G/1G explaining 61.6%, 60.2%, and 58.1% of the variance, respectively. These findings clearly demonstrate that circulating levels of TAFI are strongly determined by polymorphic variations in the promoter and the 3′UTR of theTAFI gene.
Introduction
Thrombin-activable fibrinolysis inhibitor (TAFI), also known as procarboxypeptidase B and procarboxypeptidase U, is a recently described plasma protein that can potentially inhibit fibrinolysis by removing carboxyterminal lysine residues from partially degraded fibrin, decreasing plasminogen binding on its surface.1-4 TAFI is secreted by the liver as a zymogen and can be efficiently activated by the thrombin–thrombomodulin complex in vitro. Because of its role in the balance between the coagulation and the fibrinolytic systems, the TAFI gene may be considered a potential candidate in atherothrombotic diseases. Studies in a rabbit jugular vein model of thrombolysis revealed that inhibition of activated TAFI activity increased therapeutic fibrinolysis.5,6 Moreover, it has been recently shown that high levels of TAFI antigen (Ag) could be a risk factor for venous thrombosis.7
A study conducted in consecutive patients attending a metabolic ward for primary prevention showed a great interindividual variability in plasma TAFI Ag levels, which appeared to be poorly explained by lifestyle characteristics (2% and 3% in men and women, respectively).8 This weak influence of environmental factors suggested that TAFI variations could be genetically determined.
The human TAFI gene is located on chromosome 13q14.11 and consists of 11 exons spanning approximately 48 kb. The sequence of its promoter and 3′-untranslated region (3′UTR) has been recently published.9 Until now, only 2 polymorphisms in the coding sequence have been described: one is a G-to-A substitution at nucleotide 505 on the cDNA sequence, leading to an Ala-to-Thr substitution at amino acid 147, and the other is a C-to-T substitution at nucleotide 678, resulting in a silent polymorphism.10In vitro studies did not show any effect of the Ala147Thr substitution on kinetics of activation of TAFI by thrombin–thrombomodulin complex. However, its effect on the protein level has not been assessed yet.
The aim of our study was to screen the promoter and the 3′UTR of theTAFI gene for polymorphisms and to evaluate their contribution to plasma TAFI Ag variability with the recently described Ala147thr polymorphism.
Materials and methods
Study population
The study population was composed of 121 men of Caucasian origin aged between 40 and 60 years living in the Marseilles area (France) and without history of cardiovascular disease. They were recruited on the occasion of a health check-up in a health care center. All the subjects gave written informed consent to participate in the study.
TAFI antigen determination
Blood samples were obtained from the antecubital vein between 8 and 10 am, after overnight fasting, collected on citrate (3.8% citrate, 0.129 M), and centrifuged (2500g for 30 minutes at 4°C). Platelet-poor plasma was kept frozen, below −80°C, until analysis. Antigen (Ag) determination of TAFI was performed with a commercially available kit from Milan Analytica (LaRoche, Switzerland) as previously described.8Results are expressed as percentage control pooled plasma from 30 healthy volunteers.
Search for novel polymorphisms in the TAFI gene by PCR-SSCP and sequencing
For the screening of the TAFI gene, we compared genomic DNA from 40 healthy subjects recruited from a systematic screening of a healthy population.11 Twenty subjects with high and 20 subjects with low plasma TAFI Ag levels (200% ± 24%; 60% ± 7%, respectively) were included. These sets of DNA were not part of the studied population.
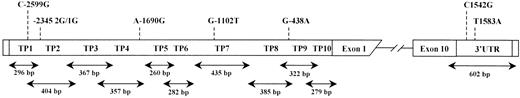
Genomic DNA was extracted from peripheral blood leukocytes by the salting-out method.12 The sequenced promoter region9 was divided into 10 overlapping fragments of approximately 300 bp maximum, as shown in Figure1. Amplified DNA fragments larger than this were digested with appropriate restriction endonucleases (Table1). Fragments were amplified and biotin-labeled during amplification. The 3′UTR was screened in one DNA fragment using the direct primer at the end of exon 10, just before the first TAA stop signal of the translation, and the reverse primer after the last signal of polyadenylation. The sequence of the 3′UTR was obtained by the addition of the cDNA sequence published by Eaton et al13 and of the genomic sequence published by Boffa et al.9 Primer sequences, lengths, locations of amplified fragments, and annealing temperatures are reported in Table 1. When a shift in the pattern of migration was observed, 2 or 3 sets of DNA corresponding to the different patterns were amplified and directly sequenced. Polymorphisms located in the promoter were numbered from the start of transcription according to the published sequence.9 Polymorphisms located in the 3′UTR were numbered from the first ATG located in position 19 on the cDNA sequence published in GenBank (accession number NM001872).
Screening strategy for TAFI gene polymorphism and localization of polymorphisms identified in the promoter and the 3′-untranslated region of the gene.
Ten overlapping fragments of TAFI promoter (TP1 to TP10) include the gene sequence from nucleotide −2699 to +25. 3′UTR includes the 3′-end of exon 10 and the untranslated part from nucleotide 1268 to 1880 on cDNA sequence. The length of each fragment is mentioned below the arrows.
Screening strategy for TAFI gene polymorphism and localization of polymorphisms identified in the promoter and the 3′-untranslated region of the gene.
Ten overlapping fragments of TAFI promoter (TP1 to TP10) include the gene sequence from nucleotide −2699 to +25. 3′UTR includes the 3′-end of exon 10 and the untranslated part from nucleotide 1268 to 1880 on cDNA sequence. The length of each fragment is mentioned below the arrows.
Sizes and primer sequences of PCR fragments for SSCP analysis of the promoter and the 3′UTR of the TAFI gene
| Primer . | Primer sequences . | DNA position* (5′ to 3′) . | PCR size (bp) . | Annealing temperature (°C) . | Enzyme used before SSCP . |
|---|---|---|---|---|---|
| Promoter | |||||
| T1a | CCTCATAAAGCAGTGCAAC | − 2676 | 296 | 58 | |
| T1b | GTATGCTGTATTCTGGGTG | − 2380 | |||
| T2a | CACCTCAACTGGACTATGT | − 2474 | 404 | 58 | RsaI |
| T2b | TGTTCCCTTGCAGTTTAGC | − 2070 | |||
| T3a | ATGGATCTGAGAAGCACC | − 2150 | 367 | 60 | TaqI |
| T3b | TTTCCTCTCTGGGGAAAC | − 1783 | |||
| T4a | CACCTGTAGACTTTTGC | − 1971 | 357 | 59 | |
| T4b | CACTGAAGGAGAGAAAG | − 1614 | |||
| T5a | TTTTCTTTCTCTCCTTCAGTG | − 1634 | 260 | 59 | |
| T5b | ATCAATGAGACTGGTTATCCA | − 1374 | |||
| T6a | ACTTCACCCAAAGAAGC | − 1467 | 282 | 59 | |
| T6b | CCTGGCCAGAAGAAAAT | − 1185 | |||
| T7a | ATTTTCTTCTGGCCAGG | − 1200 | 435 | 61 | Hinf1 |
| T7b | CCTACATTCCCATTGGT | − 765 | |||
| T8a | ACCAATGGGAATGTAGG | − 782 | 385 | 59 | MboI |
| T8b | CCTTGCCTCTGACTTTT | − 397 | |||
| T9a | TGGATTGGATTAGCCAG | − 476 | 322 | 59 | |
| T9b | TTGGCTTAGTTAGGCTG | − 154 | |||
| T10a | CCCAAGGAGTTACACAT | − 254 | 279 | 59 | |
| T10b | AAGCTTCATCCCAACAG | − 25 | |||
| 3′ part of exon 10 + 3′UTR | |||||
| 3′a | CTTGGCATGTCATTAGG | +1268 | 612 | 59 | BamHI |
| 3′b | TGCGGCATTTGTTGACA | + 1880 |
| Primer . | Primer sequences . | DNA position* (5′ to 3′) . | PCR size (bp) . | Annealing temperature (°C) . | Enzyme used before SSCP . |
|---|---|---|---|---|---|
| Promoter | |||||
| T1a | CCTCATAAAGCAGTGCAAC | − 2676 | 296 | 58 | |
| T1b | GTATGCTGTATTCTGGGTG | − 2380 | |||
| T2a | CACCTCAACTGGACTATGT | − 2474 | 404 | 58 | RsaI |
| T2b | TGTTCCCTTGCAGTTTAGC | − 2070 | |||
| T3a | ATGGATCTGAGAAGCACC | − 2150 | 367 | 60 | TaqI |
| T3b | TTTCCTCTCTGGGGAAAC | − 1783 | |||
| T4a | CACCTGTAGACTTTTGC | − 1971 | 357 | 59 | |
| T4b | CACTGAAGGAGAGAAAG | − 1614 | |||
| T5a | TTTTCTTTCTCTCCTTCAGTG | − 1634 | 260 | 59 | |
| T5b | ATCAATGAGACTGGTTATCCA | − 1374 | |||
| T6a | ACTTCACCCAAAGAAGC | − 1467 | 282 | 59 | |
| T6b | CCTGGCCAGAAGAAAAT | − 1185 | |||
| T7a | ATTTTCTTCTGGCCAGG | − 1200 | 435 | 61 | Hinf1 |
| T7b | CCTACATTCCCATTGGT | − 765 | |||
| T8a | ACCAATGGGAATGTAGG | − 782 | 385 | 59 | MboI |
| T8b | CCTTGCCTCTGACTTTT | − 397 | |||
| T9a | TGGATTGGATTAGCCAG | − 476 | 322 | 59 | |
| T9b | TTGGCTTAGTTAGGCTG | − 154 | |||
| T10a | CCCAAGGAGTTACACAT | − 254 | 279 | 59 | |
| T10b | AAGCTTCATCCCAACAG | − 25 | |||
| 3′ part of exon 10 + 3′UTR | |||||
| 3′a | CTTGGCATGTCATTAGG | +1268 | 612 | 59 | BamHI |
| 3′b | TGCGGCATTTGTTGACA | + 1880 |
DNA numbering of the TAFI promoter region is according to a proposed transcription start site (9). DNA numbering of the TAFI cDNA sequence is according to (13) and (9) for the end of the 3′UTR.
Analysis was performed with the procedure previously described.14 Briefly, each DNA was amplified with dUTP biotin incorporation, then, after a 5-minute denaturation at 95°C, they were loaded on 6% or 10% polyacrylamide gel with or without glycerol. A nondenaturing buffer (TBE, 0.5×) was used, and electrophoresis was performed at room temperature. Then DNA was transferred on a nylon membrane and revealed by the alkaline–phosphatase–streptavidin–NBT-BCIP procedure. All potential band shifts were analyzed by direct sequencing of a separately amplified fragment, using ABI Prism Bigdye Terminator and ABI Prism 377 sequencer (PE Biosystems, Foster City, CA).
Allele-specific polymerase chain reaction analyses
Genotyping of polymorphisms in the promoter and the 3′UTR was performed using allele-specific polymerase chain reactions (PCR). A PCR was performed for each allele determination according to the following conditions. Amplification was carried out in 25 μL in a Thermocycler 9600 Perkin Elmer (Applied Biosystems, Foster City, CA). Each sample contained 62 ng genomic DNA in 1× Taq polymerase buffer (3.5 mM MgCl2), 0.77 mM dNTP, 5 pmol each primer (forward and reverse primers in each case plus the allele-specific primer that corresponds to the analyzed genotype), and 0.38-U Taq polymerase (Biotaq; Quantum Bioprobe, Quebec Canada). A first denaturation at 95°C for 2 minutes was followed by 40 cycles for 1 minute at annealing temperature (determined for each reaction), at 72°C for 1 minute (extension), at 95°C for 45 seconds (denaturation), and then at 72°C for 5 minutes. All primer sequences and annealing temperatures are described in Table 2. Five microliters of each amplification sample was loaded on a 2% agarose gel stained with ethidium bromide.
Oligonucleotides and annealing temperatures used for genotyping polymorphisms located in the promoter and the 3′UTR of the TAFI gene by allele-specific PCR
| Polymorphism . | Primer sequences direct/reverse (D/R) . | PCR size (bp) . | Primer allele-specific Sequences . | PCR Allele-specific size (bp) . | Annealing temperature (°C) . |
|---|---|---|---|---|---|
| C-2599G | D:CCTCATAAAGCAGTGCAAC | 296 | C:CTAAAGGAATCATATCCA | 235 | 55 |
| R:GTATGCTGTATTCTGGGTG | G:CTAAAGGAATCATATCGA | ||||
| − 2345 2G/1G | D:CACCTCAACTGGACTATGT | 404 | 1G:AGTTTTTAAAACATGAAAGA | 293 | 54 |
| R:TGTTCCCTTGCAGTTTAGC | 2G:AGTTTTTAAAACATGAAAGGA | ||||
| A-1690G | D:CACCTGTAGACTTTTGC | 357 | A:TTAACTATTTTGACTGTTTA | 95 | 55 |
| R:CACTGAAGGAGAGAAAG | G:TTAACTATTTTGACTGTTTG | ||||
| G-1102T | D:CAGTGGAGGTCACTTTA | 311 | G:AGTTTGAGACCATCCAGG | 151 | 57 |
| R:ACTGCAACCTTCACTCT | T:AGTTTGAGACCATCCAGT | ||||
| G-438A | D:ACCAATGGGAATGTAGG | 698 | A:GAGGAACTCACTTTTTACAT | 301 | 60 |
| R:TTGGCTTAGTTAGGCTG | G:GAGGAACTCACTTTTTACGT | ||||
| Ala147Thr | A:GTTTCTGGAAAAGAACAAA | 105 | 58 | ||
| R:ATGGCCTATGAACCACAAGC | G:GTTTCTGGAAAAGAACAAG | 59 | |||
| C+1542G | D:CCAGCAAGACCAAATCA | 408 | C:AGTCAAACGTCGAAACT | 238 | 55 |
| R:ATTACCGTGGAGCAAAC | G:AGTCAAACGTCGAAAGT | ||||
| T+1583A | D:CTTGGCATGTCATTAGG | 612 | A:CTCAAGCAAGTTTAATCAAA | 294 | 57 |
| R:TGCGGCATTTGTTGACA | T:CTCAAGCAAGTTTAATCAAT | 55 |
| Polymorphism . | Primer sequences direct/reverse (D/R) . | PCR size (bp) . | Primer allele-specific Sequences . | PCR Allele-specific size (bp) . | Annealing temperature (°C) . |
|---|---|---|---|---|---|
| C-2599G | D:CCTCATAAAGCAGTGCAAC | 296 | C:CTAAAGGAATCATATCCA | 235 | 55 |
| R:GTATGCTGTATTCTGGGTG | G:CTAAAGGAATCATATCGA | ||||
| − 2345 2G/1G | D:CACCTCAACTGGACTATGT | 404 | 1G:AGTTTTTAAAACATGAAAGA | 293 | 54 |
| R:TGTTCCCTTGCAGTTTAGC | 2G:AGTTTTTAAAACATGAAAGGA | ||||
| A-1690G | D:CACCTGTAGACTTTTGC | 357 | A:TTAACTATTTTGACTGTTTA | 95 | 55 |
| R:CACTGAAGGAGAGAAAG | G:TTAACTATTTTGACTGTTTG | ||||
| G-1102T | D:CAGTGGAGGTCACTTTA | 311 | G:AGTTTGAGACCATCCAGG | 151 | 57 |
| R:ACTGCAACCTTCACTCT | T:AGTTTGAGACCATCCAGT | ||||
| G-438A | D:ACCAATGGGAATGTAGG | 698 | A:GAGGAACTCACTTTTTACAT | 301 | 60 |
| R:TTGGCTTAGTTAGGCTG | G:GAGGAACTCACTTTTTACGT | ||||
| Ala147Thr | A:GTTTCTGGAAAAGAACAAA | 105 | 58 | ||
| R:ATGGCCTATGAACCACAAGC | G:GTTTCTGGAAAAGAACAAG | 59 | |||
| C+1542G | D:CCAGCAAGACCAAATCA | 408 | C:AGTCAAACGTCGAAACT | 238 | 55 |
| R:ATTACCGTGGAGCAAAC | G:AGTCAAACGTCGAAAGT | ||||
| T+1583A | D:CTTGGCATGTCATTAGG | 612 | A:CTCAAGCAAGTTTAATCAAA | 294 | 57 |
| R:TGCGGCATTTGTTGACA | T:CTCAAGCAAGTTTAATCAAT | 55 |
For some polymorphisms, modifications were brought to this procedure. For the A-1690G polymorphism, we used 2.5 pmol forward primer to obtain better sensitivity. For the C+1542G polymorphism, the allele-specific primers were chosen in reverse sequence to obtain specific amplification.
Because the intronic sequence flanking the Ala147Thr polymorphism is unknown, genotyping of this polymorphism was performed by using only one allele-specific and one reverse primer. C+1542G direct and reverse primers were used concomitantly as an internal control of amplification.
Statistical analysis
Data were analyzed using the SAS software (SAS Institute, Cary, NC). Allele frequencies were estimated by gene counting. Hardy-Weinberg equilibrium was tested by χ2 analysis with 1 df. Pairwise linkage disequilibrium coefficients between polymorphisms were estimated by log-linear model analysis,15 and the extent of disequilibrium was expressed in terms of D' = D/Dmax or −D/Dmin. Haplotype frequencies were estimated and compared between TAFI tertiles using the Arlequin software (Arlequin [computer program]. Version 2000. Geneva, Switzerland: University of Geneva; 2000). Association of polymorphisms with TAFI Ag levels was tested by analysis of variance. The distribution of TAFI levels was log-transformed to remove positive skewness, and geometric means (95% confidence interval) were provided. Multivariate regression analysis was performed to assess the independent effect of several polymorphisms considered together. A codominant model assuming additive allele effects of polymorphisms on log(TAFI) was tested. Because this model was well fitted to the data for all the polymorphisms, it was adopted in all genotype–phenotype association analyses.
Results
Systematic search for polymorphisms of the TAFIgene promoter
We divided the sequenced promoter region into 10 overlapping fragments, as shown in Figure 1. These fragments were amplified and biotin labeled during amplification. The PCR-SSCP analysis was performed with 40 genomic DNA, 20 from subjects with a high TAFI antigen plasma level (200% ± 24%), and 20 subjects with a low TAFI antigen plasma level (60% ± 7%), in 2 experiments. When a shift in the pattern of migration was observed, 2 or 3 sets of DNA corresponding to the different patterns were amplified and direct sequenced. Five frequent polymorphisms were identified in the promoter: a C-to-G substitution at position −2599 upstream from the start of transcription, an insertion–deletion of one G nucleotide at position −2345, an A-to-G substitution at position −1690, a G-to-T substitution at position −1102, and a G-to-A substitution at position −438 (Figure 1).
3′UTR
For this analysis we chose a DNA fragment starting from the end of exon 10 just before the first TAA stop signal of the translation and ending at the last signal of polyadenylation. Two polymorphisms were identified in the 3′UTR: a C-to-G substitution at nucleotide +1542 of the cDNA sequence published by Eaton13 and a T-to-A substitution at position +1583 (Figure 1). None of the genomic DNA sequenced contained the sequence TCTTCTCCTTT, which spans nucleotides 1680 to 1690 of the published cDNA sequence. We found sequence TGCACG instead of TCAG between nucleotide 1403 to 1405 on all the sequenced genomic DNA.
Allele frequencies and linkage disequilibrium between polymorphisms
Genotype distribution did not deviate from Hardy-Weinberg expectations, except for the Ala147Thr polymorphism for which a slight excess of heterozygotes was observed (P < .02). All polymorphisms were common; frequency of the minor allele ranged from 0.24 to 0.49 (Table 3). All polymorphisms were in strong linkage disequilibrium with each other (Table 3). A-1690G, G-1102T, and G-438A were in almost complete concordance, and, consequently, only the G-438A polymorphism was considered in further analyses.
Allele frequencies and pairwise linkage disequilibrium coefficients between polymorphisms
| Polymorphism . | Allele . | Allele frequencies . | ± D′ . | ||||
|---|---|---|---|---|---|---|---|
| − 2345 . | − 438 . | aa147 . | + 1542 . | + 1583 . | |||
| −2599 | C, G | .51/.49 | − 1.00 | + 1.00 | − 0.85 | + 0.83 | − 0.78 |
| − 2345 | 2G, 1G | .69/.31 | − 1.00 | + 0.84 | − 0.75 | + 0.77 | |
| −438 | G, A | .71/.29 | − 1.00 | + 0.85 | − 0.75 | ||
| aa147 | Ala, Thr | .76/.24 | − 0.86 | + 0.95 | |||
| + 1542 | C, G | .70/.30 | − 0.92 | ||||
| + 1583 | T, A | .70/.30 | |||||
| Polymorphism . | Allele . | Allele frequencies . | ± D′ . | ||||
|---|---|---|---|---|---|---|---|
| − 2345 . | − 438 . | aa147 . | + 1542 . | + 1583 . | |||
| −2599 | C, G | .51/.49 | − 1.00 | + 1.00 | − 0.85 | + 0.83 | − 0.78 |
| − 2345 | 2G, 1G | .69/.31 | − 1.00 | + 0.84 | − 0.75 | + 0.77 | |
| −438 | G, A | .71/.29 | − 1.00 | + 0.85 | − 0.75 | ||
| aa147 | Ala, Thr | .76/.24 | − 0.86 | + 0.95 | |||
| + 1542 | C, G | .70/.30 | − 0.92 | ||||
| + 1583 | T, A | .70/.30 | |||||
All coefficients are significantly different from 0 (P < .001).
The A-1690G, G-1102T, and G-438A polymorphisms were in nearly complete concordance; thus, A-1690G and G-1102T are not reported in this table.
The 6 remaining polymorphisms generated 4 main haplotypes, accounting for more than 80% of all the haplotypes observed in the whole sample (Table 4). None of the other observed haplotypes had a frequency higher than 5%. Within the 4 main haplotypes, the −2345 2G/1G, the Ala147Thr, and the T+1583A polymorphisms were in complete association.
Frequency of the main haplotypes of the TAFIgene in the whole sample and the 1st and 3rdtertiles of the TAFI Ag distribution
| . | Haplotype . | Frequency . | |||||||
|---|---|---|---|---|---|---|---|---|---|
| − 2599 . | − 2345 . | − 438 . | aa147 . | + 1542 . | + 1583 . | Whole . | 1sttertile . | 3rd tertile . | |
| H1 | G | 2G | A | Ala | G | T | 0.26 | 0.50 | 0.01 |
| H2 | G | 2G | G | Ala | C | T | 0.16 | 0.13 | 0.15 |
| H3 | C | 2G | G | Ala | C | T | 0.19 | 0.17 | 0.23 |
| H4 | C | 1G | G | Thr | C | A | 0.21 | 0.08 | 0.36 |
| . | Haplotype . | Frequency . | |||||||
|---|---|---|---|---|---|---|---|---|---|
| − 2599 . | − 2345 . | − 438 . | aa147 . | + 1542 . | + 1583 . | Whole . | 1sttertile . | 3rd tertile . | |
| H1 | G | 2G | A | Ala | G | T | 0.26 | 0.50 | 0.01 |
| H2 | G | 2G | G | Ala | C | T | 0.16 | 0.13 | 0.15 |
| H3 | C | 2G | G | Ala | C | T | 0.19 | 0.17 | 0.23 |
| H4 | C | 1G | G | Thr | C | A | 0.21 | 0.08 | 0.36 |
Comparison of haplotype frequencies between the 1stand 3rd tertiles: P < 10−5.
Association of TAFI polymorphisms with plasma TAFI Ag levels
All the polymorphisms were strongly associated with TAFI Ag levels (P < 10−4). In all cases, the model was compatible with an additive allele effect on the log-transformed variable. Geometric means and 95% confidence intervals according to genotypes are shown in Table 5. The percentage of variance explained by genotypes varied from 52% for the C+1542G polymorphism to 20% for the −2345 2G/1G polymorphism.
Association between TAFI genotypes and plasma TAFI Ag levels
| Polymorphism . | n . | TAFI Ag level geometric mean (95% CI) . | R2 assuming a codominant model . |
|---|---|---|---|
| C-2599G | |||
| CC | 31 | 97.5 (56.3-168.8) | |
| CG | 62 | 68.7 (37.4-126.2) | 0.25 |
| GG | 27 | 58.0 (28.1-119.7) | |
| −2345 2G/1G | |||
| 2G/2G | 57 | 62.8 (33.5-117.6) | |
| 2G/1G | 51 | 77.5 (39.0-153.9) | 0.20 |
| 1G/1G | 12 | 106.7 (68.0-167.5) | |
| G-438A | |||
| GG | 59 | 90.0 (50.0-162.1) | |
| GA | 54 | 60.9 (37.3-99.5) | 0.41 |
| AA | 7 | 42.5 (20.6-87.8) | |
| Ala147Thr | |||
| Ala/Ala | 62 | 60.9 (31.9-116.4) | |
| Ala/Thr | 56 | 84.8 (48.0-149.7) | 0.23 |
| Thr/Thr | 2 | 100.5 (70.6-143.0) | |
| C+1542G | |||
| CC | 57 | 91.8 (55.2-152.9) | |
| CG | 55 | 62.8 (39.2-100.5) | 0.52 |
| GG | 8 | 35.2 (23.8-52.0) | |
| T+1583A | |||
| TT | 62 | 60.9 (31.9-116.4) | |
| TA | 44 | 81.5 (45.2-146.6) | 0.30 |
| AA | 14 | 107.8 (71.4-162.7) |
| Polymorphism . | n . | TAFI Ag level geometric mean (95% CI) . | R2 assuming a codominant model . |
|---|---|---|---|
| C-2599G | |||
| CC | 31 | 97.5 (56.3-168.8) | |
| CG | 62 | 68.7 (37.4-126.2) | 0.25 |
| GG | 27 | 58.0 (28.1-119.7) | |
| −2345 2G/1G | |||
| 2G/2G | 57 | 62.8 (33.5-117.6) | |
| 2G/1G | 51 | 77.5 (39.0-153.9) | 0.20 |
| 1G/1G | 12 | 106.7 (68.0-167.5) | |
| G-438A | |||
| GG | 59 | 90.0 (50.0-162.1) | |
| GA | 54 | 60.9 (37.3-99.5) | 0.41 |
| AA | 7 | 42.5 (20.6-87.8) | |
| Ala147Thr | |||
| Ala/Ala | 62 | 60.9 (31.9-116.4) | |
| Ala/Thr | 56 | 84.8 (48.0-149.7) | 0.23 |
| Thr/Thr | 2 | 100.5 (70.6-143.0) | |
| C+1542G | |||
| CC | 57 | 91.8 (55.2-152.9) | |
| CG | 55 | 62.8 (39.2-100.5) | 0.52 |
| GG | 8 | 35.2 (23.8-52.0) | |
| T+1583A | |||
| TT | 62 | 60.9 (31.9-116.4) | |
| TA | 44 | 81.5 (45.2-146.6) | 0.30 |
| AA | 14 | 107.8 (71.4-162.7) |
All polymorphisms were significantly associated with TAFI levels (P < 10−4).
We then investigated by multivariate analysis whether several polymorphisms were independently associated with TAFI levels. In a stepwise regression analysis including all the polymorphisms, 2 remained significantly and independently associated with plasma TAFI Ag levels, the C+1542G and the Ala147thr polymorphisms. Together, these 2 polymorphisms explained 61.6% of the variance. Two other pairs of polymorphisms explained nearly the same percentage of TAFI variance: the C+1542G polymorphism in combination either with the T+1583A polymorphism (60.2%) or with the −2345 2G/1G polymorphism (58.1%). However, as noted above, the Ala147Thr, the T+1583A, and the −2345 2G/1G polymorphisms are in strong association with each other, making it difficult to clearly differentiate the effect of these 3 polymorphisms on TAFI levels.
Frequencies of the 4 main haplotypes were compared between the 1st and 3rd tertiles of the TAFI distribution, and the difference was highly significant (P < 10−5, Table 4). The major source of difference was attributable to haplotype H1 (frequency: 0.50 vs 0.01 in the 2 tertiles, respectively). This haplotype differed from the 3 others by the C/G substitution at position +1542 and the G/A substitution at position −438. Among the 3 remaining haplotypes, the frequency of haplotype H4 strongly differed between the 2 tertiles (0.08 vs 0.36). Haplotype H4 differed from H2 and H3 by the Ala/Thr amino acid change, the T/A substitution at position +1583, and the insertion/deletion at position −2345.
Table 6 gives geometric TAFI means when the C+1542G and the Ala147Thr polymorphisms are combined. An additive effect of the 2 polymorphisms on TAFI levels was clearly shown (test on interaction between genotypes, P = .77). An almost 3-fold increase in TAFI Ag levels was observed between individuals combining the +1542GG and 147Ala/Ala genotypes and those combining the +1542CC and 147Ala/Thr genotypes (35.2 vs 98.5, respectively). The single individual carrying the combination +1542CC and 147Thr/Thr had an even higher TAFI level. However, it should be stressed again that the effect of the Ala147Thr polymorphism alone is not distinguishable from that of the haplotype combining the Ala147Thr, the T+1583A, and the −2345 2G/1G polymorphisms because of the strong association between the 3 polymorphisms.
Plasma TAFI Ag levels (geometric means, 95% CI) according to the C+1542G and the Ala147Thr genotypes
| Ala147Thr genotype . | C+1542G . | ||
|---|---|---|---|
| CC . | CG . | GG . | |
| Ala/Ala | 78.3 | 58.6 | 35.2 |
| (71.4-85.8) | (54.2-63.3) | (30.1-41.1) | |
| n = 23 | n = 31 | n = 8 | |
| Ala/Thr | 98.5 | 68.7 | |
| (91.1-106.5) | (62.8-75.3) | — | |
| n = 31 | n = 23 | ||
| Thr/Thr | 114.4 | 89.1 | |
| n = 1 | n = 1 | — | |
| Ala147Thr genotype . | C+1542G . | ||
|---|---|---|---|
| CC . | CG . | GG . | |
| Ala/Ala | 78.3 | 58.6 | 35.2 |
| (71.4-85.8) | (54.2-63.3) | (30.1-41.1) | |
| n = 23 | n = 31 | n = 8 | |
| Ala/Thr | 98.5 | 68.7 | |
| (91.1-106.5) | (62.8-75.3) | — | |
| n = 31 | n = 23 | ||
| Thr/Thr | 114.4 | 89.1 | |
| n = 1 | n = 1 | — | |
In a model assuming a codominant effect of polymorphisms on log (TAFI): C+1542G effect P < 10−4, Ala147Thr effect P < 10−4, interactionP = .77.
Discussion
TAFI is supposed to play a key role in the regulation of fibrinolysis through its potential to regulate plasminogen activation. Consequently, it has been suggested that high levels of TAFI could be a risk factor for atherothrombotic diseases. This hypothesis has been recently strengthened by a study showing a weak association between high TAFI levels and deep vein thrombosis.7 A great interindividual variability of plasma levels has been shown, weakly explained by environmental factors.8 This suggested a genetic determination of plasma TAFI concentrations. Here we screened the promoter and the 3′UTR of the human TAFI gene to identify polymorphisms linked to plasma TAFI levels.
We described 7 new polymorphisms: 5 in the promoter, 2 in the 3′UTR. All these polymorphisms were in strong linkage disequilibrium with each other and with the previously described Ala147Thr polymorphism. Because of the strong association between polymorphisms, they generated 4 main haplotypes that accounted for 80% of all haplotypes observed in the sample. In univariate analyses, all polymorphisms were associated with plasma TAFI Ag levels and, individually, contributed to a large fraction of plasma TAFI variability, ranging from 20% to 52%. However, the multivariate analysis suggested that at least 2 polymorphisms had independent and additive effects on TAFI levels. When combining these 2 polymorphisms, the percentage of variance explained reached more than 60%, indicating a strong influence of the TAFI gene on the determination of plasma TAFI levels. The polymorphisms that exhibited the highestR2 in multivariate analysis included the C+1542G and the Ala147Thr polymorphisms. Close R2 were also observed when the C+1542G polymorphism was combined either with the −2345 2G/1G or with the T+1583A polymorphism. Given the nearly complete association existing between the Ala147Thr, the −2345 2G/1G, and the T+1583A polymorphisms, it is not possible to determine whether their effect is caused by one of them or by their haplotype combination.
The results suggest that the regulation of plasma TAFI levels by theTAFI gene is complex and probably involves more than one single functional variant. The strong association observed between polymorphisms located within the promoter and the 3′UTR of theTAFI gene and TAFI Ag levels suggests that at least some of these polymorphisms are located within transcriptional regulatory regions. The role of the 2 polymorphisms within the 3′UTR in the mRNA stability can also be envisaged. However, we cannot exclude the possibility that the results observed in this study reflect the role of an unidentified functional polymorphism located elsewhere and that would be in linkage disequilibrium with all these polymorphisms. Further characterizations of sequence variations of the TAFIgene with in vitro experiments testing the functionality of the 5′ and 3′UTR polymorphisms are needed.
Our results must be compared to the study of the transcriptional activity of the 5′-flanking region of the TAFI gene in mammalian cells reported by Boffa et al.9 Transient transfection assays of reporter plasmids containing portions of the TAFI 5′-flanking region into mammalian cells identified an approximately 70-bp region between nucleotides −141 and −73 crucial for liver-specific transcription, whereas regions encompassing our identified polymorphisms did not seem to play a role in transcription. However, it is interesting to note that DNA used by Boffa et al9 in the studies of transcriptional activity contained all the alleles we have described to be associated with average or low levels of TAFI Ag levels. This may explain why no important effect on transcriptional activity was detected in these experiments. Effects on transcriptional level might have been observed with alleles linked to high levels of TAFI Ag.
In accordance with the results reported by Boffa et al,9none of the genomic DNA sequenced in our study contained the sequence TCTTCTCCTTT, which spans nucleotides 1680 to 1690 of the published cDNA sequence.13 Moreover, we found sequence TGCACG instead of TCAG between nucleotides 1403 and 1405 on all sequenced genomic DNA.
In conclusion, this study showed that interindividual variations of plasma TAFI Ag levels are strongly genetically determined. We described 7 new polymorphisms, 5 in the promoter and 2 in the 3′UTR of theTAFI gene, that accounted for a large part of this genetic variability. In vitro studies are needed to evaluate the functional importance of these polymorphisms. Moreover, it will be important to assess the role of these polymorphisms in relation to arterial or venous thromboembolic diseases.
We thank V. Nicaud and D. A. Trégouët (INSERM U525) for their help in statistical analyses and M. Billerey and S. Debroas for their skillful assistance.
Supported by INSERM, the Commission of the European Community, Hifmech Study contract BMH4-CT96-0272, the Fondation pour la Recherche Médicale, the Programme Hospitalier de Recherche Clinique (PHRC 1996), and Université de la Méditerranée. H.A. is recipient of a grant from Diagnostica Stago and Association Nationale de la Recherche Technique, France (Convention CIFRE 407/99).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Irène Juhan-Vague, Laboratoire d'Hématologie, CHU Timone, 13385 Marseilles cedex 5, France.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal