Abstract
Although hematopoiesis is known to proceed from stem cells through a graded series of multipotent, oligopotent, and unipotent precursor cells, it has been difficult to resolve these cells physically one from another. There is, therefore, corresponding uncertainty about the exact distribution and timing of the expression of genes known to be important in hematopoietic differentiation. In earlier work, the generation of a set of amplified complementary DNAs (cDNAs) from single precursor cells was described, whose biologic potential was determined by the outcome of cultured sibling cells. In this study, the new acquisition of cDNA from multipotent myeloid precursor cells is described, as is the mapping of RNA-level expression of 17 distinct cytokine receptors (c-kit, Flk-1, Flk-2/Flt-3, c-fms, gp130, erythropoietin receptor, GM-CSFRα, G-CSFR, TNFR1, IL-1RI, IL-1RII, IL-2Rβ, IL-3-specific β receptor, IL-4R, IL-6Rα, IL-7Rα, and IL-11Rα) to the enlarged sample set, spanning stages from pentapotent precursors through oligopotent intermediates to committed and maturing cells in the myeloid and lymphoid lineages. Although the enhanced scope and resolving power of the analysis yielded previously unreported observations, there was overall agreement with known biologic responsiveness at individual stages, and major contradictions did not arise. Moreover, each precursor category displayed a unique overall pattern of hybridization to the matrix of 17 receptor probes, supporting the notion that each sample pool indeed reflected a unique precursor stage. Collectively, the results provide supportive evidence for the validity of the cDNA assignments to particular stages, the depth of the information captured, and the unique capacity of the sample matrix to resolve individual stages in the hematopoietic hierarchy.
Introduction
Maintenance of the various hematopoietic lineages is achieved by the continuous growth and differentiation of a hierarchy of precursor cells in the blood-forming tissues. Positioned at the top are rare, multipotent stem cells possessing a unique capacity for extensive, if not permanent, self-renewal.1 Stem cell progeny progress through a series of pluripotent and eventually bipotent intermediates of diminishing self-renewal ability and differentiative repertoire.2-5 These yield, in turn, to precursors restricted to development along single hematopoietic lineages.
The genes and mechanisms that specify lineage divergence are still only dimly understood. Cell purification has been generally considered a crucial step to eventual identification of the genes whose differential expression would account for key differentiation decisions. However, presently known markers have not cleanly resolved the variety of differentiation intermediates known to exist on the basis of clonal analyses. Although multipotent cell lines have frequently been assumed to offer a solution to the problem of heterogeneity,6 the evidence clearly indicates heterogeneity comparable to normal hematopoietic tissue and that only infrequent cells within the lines actually remain multipotential.7-10
A different approach was recently described4 that allows isolation of precursor subsets in homogeneous form and supports analysis of global patterns of gene expression. The method is based on the synchronous nature of differentiation observed in nascent colonies or “starts” consisting of 4 to 8 cells. Single precursor cells are drawn from such starts, and their transcripts are amplified by global reverse transcription–polymerase chain reaction (RT-PCR). The remaining siblings are cultured individually in conditions supportive of multilineage development, and their clonal outcomes are determined. Identical differentiative outcomes of each sibling reporter allow tentative assignment of the same potential to the cells processed for complementary DNA (cDNA).
In our earlier study, we described construction in this way of a set of cDNA samples from tripotent, bipotent, and unipotent hematopoietic precursor cells derived from murine bone marrow.4 Hybridization analysis of the sample set with a small number of probes for lineage-specific transcripts provided confirmation of the lineage assignments, particularly of the more mature elements of the set. However, there was at that time only a limited representation of pluripotent elements. Moreover, validation of pluripotential assignments was less likely to be achievable with probes characteristic of maturing cells, leaving the challenge of identifying probes that might have known specificity for individual pluripotent intermediates.
Hematopoiesis is known to be driven by a variety of cytokines, some of which act with distinct differential specificity on pluripotential precursors at particular stages or on committed precursors in particular lineages.11 Such response patterns indicate that the timed initiation of expression of the cognate factor receptors is likely to play a central role in the sequence of events leading to lineage divergence and establishment in the hematopoietic system. An understanding of the earliest steps of the process will, therefore, require a description of where and when expression of the key receptors begins, particularly in the precursor intermediates that are presently unresolved by physical techniques. Such an analysis is now possible based on our single-cell cDNA approach. In addition to its importance in resolving receptor expression, such an analysis, if congruent with known patterns of differential responsiveness of precursor subsets to particular cytokines, would add substantially to confidence in the validity and specificity of our cDNA sample matrix.
In this study, we describe the acquisition of new cDNA samples from multipotent precursor cells from adult marrow, including pentapotent and tetrapotent instances, and report on the distribution of expression of transcripts for 17 distinct cytokine receptors using the expanded cDNA sample matrix. The resultant data are the first to resolve expression of these transcripts among hematopoietic precursors at different commitment stages, and they provide essential validation of the integrity and fidelity of our hierarchy sample set.
Materials and methods
The cDNA samples characterized in this study were generated in 2 sets. Collection and characterization of the initial set was described in detail.4 It comprised 69 individual cDNA samples from hematopoietic cells ranging from tripotent precursors to terminally maturing cells in erythroid, myeloid, and lymphoid lineages. In this study we describe the generation of 86 additional samples. The new instances significantly augmented the sample sizes of precursor stages already included in the original set (tripotent E/Meg/Mac, bipotent E/Meg, committed Meg and Mac precursors) and of terminally maturing cells. Of greatest interest, the work extended representation of the sample archive to more primitive tetrapotent and pentapotent cells.
Cells and cultures
Marrow cells from femurs of adult (12- to 16-week-old) CBA/J mice were used without purification. The rhodamine 123loLy6A/E (Sca-1)hi fraction was obtained by FACS from adult C57BL6/J marrow as detailed.12 13
Terminally maturing hematopoietic cells were sampled from single lineage colonies growing in methylcellulose cultures containing IL-1, IL-3, and erythropoietin.4 Mast cells were generated from marrow cells cultured 4 to 7 weeks in medium containing IL-3 and concanavalin A.4 Mature T cells were sampled from ConA-stimulated, 2- to 4-day cultures of CD4+ or CD8+ T cells isolated from adult C57BL/6J spleens. NIH3T3 and 95/1.7 fibroblast monolayers14 were grown in serum-containing medium.
For sampling of precursor cell cDNA and for assessment of developmental potential of sibling cells, colony starts from marrow cell precursors were initiated in sparsely seeded methyl cellulose cultures4,13 containing IL-1, c-kit ligand, IL-3, IL-11, 5637 bladder carcinoma cell-conditioned medium, and erythropoietin. The differentiation potential of sister cells was tested at 4 days by micromanipulating single siblings into gridded secondary methylcellulose plates and culturing them in the same cytokines. Small numbers of cells were drawn at intervals from growing secondary colonies and were stained on glass slides with May-Grünwald-Giemsa for morphologic assessment.4Megakaryocytic lineage cells were identified by histochemical staining for acetylcholinesterase.15 For detailed assessment of lineage content of large, actively growing secondary colonies, cells were dispersed and subcultured at 7 to 9 days into tertiary methylcellulose plates where erythroid and other myeloid growth could more readily be identified. They were also subcultured into liquid medium containing IL-3 and ConA for 4 to 7 weeks, after which cells were stained with Alcian blue for specific detection of mast cells.16
Amplified cDNA
cDNA was generated from single precursor cells or from terminally differentiating cells in unilineage colonies or cultures in which individual samples comprised 50 to 100 cells. The method for generating globally amplified cDNA has been described in detail elsewhere.17,18 Briefly, cell samples were lysed in a 4-μL volume containing NP-40, and first-strand cDNA primed by oligo(dT) was generated to a length of 150 to 400 bases on the liberated mRNA templates by reverse transcriptase. After addition of a 3′ poly(dA) tail with terminal transferase, the resultant cDNA mixture was amplified by PCR using a single oligo(dT)-containing primer. The amplified product and subsequent re-amplifications using the same primer have repeatedly been observed to preserve qualitatively the relative abundance relations in the original mRNA population.4,17 19 Nevertheless, care was taken to ensure that hybridization analyses were performed on cDNA that had not been re-amplified through more than 2 rounds from the originally archived samples.
The individual cDNA samples generated in this study were hybridized with cDNA probes for constitutive housekeeping transcripts (L32) and lineage-specific transcripts α- and β-globin, lysozyme, and immunoglobulin to confirm the integrity of the new samples and conformance with our earlier observed patterns of expression.4
For hybridization analysis of cytokine receptor probes, amplified cDNA samples from cells at similar positions in the hematopoietic differentiation hierarchy were pooled (0.5 μL per sample) and re-amplified. Seventeen such pools, each representative of a distinct stage, were prepared, and 20 μL each was slot-blotted onto Hybond N+ nylon membranes (Amersham, Uppsala, Sweden). Each blot also included a “blank” control consisting of a similar quantity of DNA amplified in a global RT-PCR reaction performed in the absence of a cellular template and determined by sequencing to consist of primer concatamers.
For secondary target-specific PCR, globally amplified cDNA was diluted 1:10 000 into PCR buffer (10 mM Tris, pH 8.3, 50 mM KCl, 1.5 mM Mg++, 2 mM dNTPs, 1 μM specific upstream and downstream primers) and amplified with Taq polymerase through 35 or 40 cycles (94°C at 15 seconds/60°C at 30 seconds/72°C at 60 seconds). The amplified product was visualized after electrophoresis on 1.8% agarose gels containing ethidium bromide. Gels were then blotted onto Hybond N+ nylon membranes, and the membranes were probed with the corresponding radiolabeled specific amplification product.
Probes
Specific probes, detailed in Table1, were designed to target terminal 3′ untranslated sequences close to the polyadenylation sites. Probes prepared as PCR fragments were amplified with the indicated primer pairs enclosing terminal 3′ sequences, using globally amplified cDNA from appropriate cell lines or primary cell sources as templates. For the design of PCR primers, extra care was taken to ensure proximity to the true 3′ termini. From posted GenBank cDNA sequences, the 3′ termini were used to probe the public murine EST databases to confirm termination at the same positions in ESTs prepared with oligo(dT) primers. For c-kit, IL-2Rβ, and IL-7Rα, which lacked classical polyadenylation signals in the GenBank sequences, EST sequence clusters were found that extended the posted sequences (for c-kit, by 19 bases; IL-2Rβ, 660 bases; IL-7Rα, 250 and 650 bases, to alternative polyadenylation sites) and contained appropriate polyadenylation signals (Table 1 footnote). These revisions have been posted as annotations to the NCBI reference sequences listed in the footnote to Table 1. Once primers directed against extreme 3′ tracts were designed, the enclosed sequences of the intended probes were screened against the GenBank nonredundant nucleotide database to ensure unique specificity for the target transcripts. All amplified fragments were validated by size determination and sequencing.
Probes and primers for cytokine receptors
| Probe . | Size (b) . | Source . | Offset* . | Primers† . | Reference‡ . | Donor . |
|---|---|---|---|---|---|---|
| IL-1RI | 545 | PCR | 37 | 5′-ggagagcagtgacagagaa+ | 81 | |
| 5′tgatcaataatatgcactgt− | ||||||
| IL-1RII | 763 | 3′-EcoRI-XbaI | 0 | 82 | U. Gubler | |
| IL-2Rβ1-153 | 1700 | 3′ fragment of mIL2Rb-12 | 660 | 83 | T. Taniguchi | |
| 235 | PCR | 47 | 5′ tttatgtgttcttgggcgat+ | |||
| 5′ acttttcccagatgttgcat− | ||||||
| IL-3Rβ | 980 | 3′BamHI of A1C2A-32 | 0 | 84 | A. Miyajima | |
| IL-4Rα | 1300 | 3′ XhoI of pEV40-mIL4R | 200 | 85 | A. Miyajima | |
| IL-6Rα | 990 | 3′EcoRI of pMR301I | 0 | 86 | T. Kishimoto | |
| gp130 | 1100 | 3′ EcoRI of pBSSkm130 | 0 | 70 | T. Kishimoto | |
| IL-7Rα1-153 | 269 | PCR | 270/665 | 5′ ggcataccatgaccatgtgt+ | 87 | C. Paige |
| 5′ cacacataggaggactcaca− | ||||||
| Upstream | 317 | PCR | 19 | 5′ gccaatctacaggagttcaa+ | ||
| 5′ cacccaaccacagtatcttt− | ||||||
| Downstream | 249 | 0 | 5′ ctgccaattttcctcttggt+ | |||
| 5′ ccagaaaatagcgcatgctt− | ||||||
| IL-11Rα | 332 | PCR | 56 | 5′ cttctcttgccttggcctgg+ | 88 | |
| 5′ gaccgcacacactctccaat− | ||||||
| c-fms | 427 | 3′ApaLI-EcoRI | 0 | 22 | L. Rohrschneider | |
| 217 | Progenitor cell PCR clone | 0 | Iscove, unpublished | |||
| c-kit1-153 A | 315 | PCR (blot) | 25 | 5′ gtcttggatattcttgaaag+ | 4 | |
| oligo(dT)− | ||||||
| B | 248 | PCR (secondary) | 12 | 5′ tacatcagatgtcagatgtt+ | 89 | |
| 5′ tagaattcttcagaactgtc− | ||||||
| EpoR | 465 | 3′BanI-XhoI of pER190-133 | 0 | 90 | A. D'Andrea | |
| flk-11-155 | 247 | PCR | 13 | 5′ ttctctgtcaagtggcggta+ | 91 | |
| 5′ aggcagaaaccagtagacat− | ||||||
| flk-2 | 265 | 76 | 5′ aagattagcctcgcctctga+ | 92 | ||
| 5′ cccttagcaaaaaacgggtt | ||||||
| G-CSFR1-154 | 260 | PCR | 200 | 5′ cttctcaggctataccctga+ | 93 | |
| 5′ tggatctcactatgtaggct− | ||||||
| GM-CSFRα | 346 | pBSmGMRa71 | 21 | 59 | L. S. Park | |
| TNFR-I | 860 | 3′BamHI-EcoRI | 248 | 94 | T. Mak |
| Probe . | Size (b) . | Source . | Offset* . | Primers† . | Reference‡ . | Donor . |
|---|---|---|---|---|---|---|
| IL-1RI | 545 | PCR | 37 | 5′-ggagagcagtgacagagaa+ | 81 | |
| 5′tgatcaataatatgcactgt− | ||||||
| IL-1RII | 763 | 3′-EcoRI-XbaI | 0 | 82 | U. Gubler | |
| IL-2Rβ1-153 | 1700 | 3′ fragment of mIL2Rb-12 | 660 | 83 | T. Taniguchi | |
| 235 | PCR | 47 | 5′ tttatgtgttcttgggcgat+ | |||
| 5′ acttttcccagatgttgcat− | ||||||
| IL-3Rβ | 980 | 3′BamHI of A1C2A-32 | 0 | 84 | A. Miyajima | |
| IL-4Rα | 1300 | 3′ XhoI of pEV40-mIL4R | 200 | 85 | A. Miyajima | |
| IL-6Rα | 990 | 3′EcoRI of pMR301I | 0 | 86 | T. Kishimoto | |
| gp130 | 1100 | 3′ EcoRI of pBSSkm130 | 0 | 70 | T. Kishimoto | |
| IL-7Rα1-153 | 269 | PCR | 270/665 | 5′ ggcataccatgaccatgtgt+ | 87 | C. Paige |
| 5′ cacacataggaggactcaca− | ||||||
| Upstream | 317 | PCR | 19 | 5′ gccaatctacaggagttcaa+ | ||
| 5′ cacccaaccacagtatcttt− | ||||||
| Downstream | 249 | 0 | 5′ ctgccaattttcctcttggt+ | |||
| 5′ ccagaaaatagcgcatgctt− | ||||||
| IL-11Rα | 332 | PCR | 56 | 5′ cttctcttgccttggcctgg+ | 88 | |
| 5′ gaccgcacacactctccaat− | ||||||
| c-fms | 427 | 3′ApaLI-EcoRI | 0 | 22 | L. Rohrschneider | |
| 217 | Progenitor cell PCR clone | 0 | Iscove, unpublished | |||
| c-kit1-153 A | 315 | PCR (blot) | 25 | 5′ gtcttggatattcttgaaag+ | 4 | |
| oligo(dT)− | ||||||
| B | 248 | PCR (secondary) | 12 | 5′ tacatcagatgtcagatgtt+ | 89 | |
| 5′ tagaattcttcagaactgtc− | ||||||
| EpoR | 465 | 3′BanI-XhoI of pER190-133 | 0 | 90 | A. D'Andrea | |
| flk-11-155 | 247 | PCR | 13 | 5′ ttctctgtcaagtggcggta+ | 91 | |
| 5′ aggcagaaaccagtagacat− | ||||||
| flk-2 | 265 | 76 | 5′ aagattagcctcgcctctga+ | 92 | ||
| 5′ cccttagcaaaaaacgggtt | ||||||
| G-CSFR1-154 | 260 | PCR | 200 | 5′ cttctcaggctataccctga+ | 93 | |
| 5′ tggatctcactatgtaggct− | ||||||
| GM-CSFRα | 346 | pBSmGMRa71 | 21 | 59 | L. S. Park | |
| TNFR-I | 860 | 3′BamHI-EcoRI | 248 | 94 | T. Mak |
Offset (b) of 3′ end of probe from polyA tail.
Primers: (+) upstream, sense; (−) downstream, antisense.
Defining report.
Sequence extended to 3′ terminus by ESTs, as annotated in NCBI RefSeqs: c-kit-RefSeq NM_021099; IL-2Rβ-RefSeq NM_008368; IL-7Rα-RefSeq NM_008372.
Atypical polyadenylation signal ATAAAA, 3′ terminus confirmed by ESTs.
G-CSFR probe excludes a 3′ terminal motif spanning nucleotides 3114 to 3245 (GenBank accession #M58288.1) common to numerous GenBank entries.
After labeling of the probes with 32P by random priming (oligolabeling kit; Pharmacia, Uppsala, Sweden), blots were hybridized with 10 mL probe containing at least 106 cpm/mL in buffer containing 1% sodium dodecyl sulfate, 1M NaCl, 10% dextran sulfate, and 100 μgm/mL salmon sperm DNA, at 70°C (c-kit, 60°C) for 16 hours. Blots were washed twice for 1 hour in 0.2 × SSC and 0.1% sodium dodecyl sulfate at 70°C. Autoradiographs were exposed at −70°C with intensifying screens.
Results
Sampling of cDNA from individual multipotent hematopoietic precursor cells
Genes with roles in self-renewal and lineage commitment decisions are likely to be expressed in multipotent cells that possess these capacities, and a subset of such genes might be expected to be expressed with greatest intensity in the earliest precursor stages. This study describes a crucial step in establishing a system able to detect genes that display strongly stage-specific patterns of expression.
Our earlier work4 established that when precursor cells in adult marrow initiate colony growth in culture, individual cells replated from a given 4- to 8-cell colony start usually generate colonies of identical composition. When colonies are grown from unseparated murine marrow cells, the great majority contain only a single differentiating lineage, and, not surprisingly, individual cells from such colony starts display identical unilineage potential (Figure1, top panels).
Numbers of lineages generated by sibling cells taken from 4 to 8 cell colony starts.
Colony starts were initiated from unseparated marrow (BM; total, 15 starts), marrow cells after overnight selection in thymidine (TdR; total, 28 starts), or the rhodamine 123loLy6A/Ehi marrow cell fraction (total, 54 starts). Left-hand panels indicate the relative frequencies at which colony starts displayed single or multiple lineage potential. For example, after thymidine selection, 58% of the colony starts displayed the potential to differentiate into 2 lineages. Right-hand panels indicate the relative proportions of starts whose siblings all yielded the same kind of colony, as a function of the total number of lineages found in the colonies. For example, after thymidine selection, daughter cells from 63% of bipotent colony starts had identical bilineage outcomes after individual culture, whereas 37% of starts yielded secondary colonies that did not have identical outcomes. In the lower-right panel, lightly shaded bars represent secondary colonies that were identical. A lower level of stringency, in which one sibling subclone with a disparate outcome was allowed, is represented by darkly shaded bars.
Numbers of lineages generated by sibling cells taken from 4 to 8 cell colony starts.
Colony starts were initiated from unseparated marrow (BM; total, 15 starts), marrow cells after overnight selection in thymidine (TdR; total, 28 starts), or the rhodamine 123loLy6A/Ehi marrow cell fraction (total, 54 starts). Left-hand panels indicate the relative frequencies at which colony starts displayed single or multiple lineage potential. For example, after thymidine selection, 58% of the colony starts displayed the potential to differentiate into 2 lineages. Right-hand panels indicate the relative proportions of starts whose siblings all yielded the same kind of colony, as a function of the total number of lineages found in the colonies. For example, after thymidine selection, daughter cells from 63% of bipotent colony starts had identical bilineage outcomes after individual culture, whereas 37% of starts yielded secondary colonies that did not have identical outcomes. In the lower-right panel, lightly shaded bars represent secondary colonies that were identical. A lower level of stringency, in which one sibling subclone with a disparate outcome was allowed, is represented by darkly shaded bars.
To enhance the proportion of colony starts developing from pluripotential precursors in our earlier study, we eliminated the most rapidly cycling precursors from marrow in vitro by cold thymidine arrest overnight4 before plating in semisolid medium. The middle panels in Figure 1 summarize a small result set obtained with the same technique used in the present work, showing the enhanced proportion of clonal starts displaying potential for generating progeny in 2 and occasionally 3 lineages. Of all starts with bilineage outcomes, 60% yielded a uniform set of bilineage colonies from the subcultured siblings (middle right panel). As in our original study, colony starts displaying potential to generate cells in more than 3 myeloid lineages were rare.
To improve further the potential for sampling colony starts from uncommitted precursor subsets, we tested marrow cells that had been enriched by cell sorting for the stem cells measured by long-term in vivo reconstitution. The RholoLy6Ahisubfraction of adult marrow contains 85% of the long-term reconstituting activity of marrow while excluding 99% of the more advanced clonogenic cells.12,13 When colony starts were initiated from this fraction, half the starts with dispersed morphology13 contained cells able to differentiate into 3 to 5 distinct myeloid lineages (Figure 1, bottom panels). Mast cell potential was monitored specifically by testing the ability of sibling subcolonies to spawn 6- to 12-week IL-3–dependent liquid cultures of Alcian blue-positive mast cells. The proportion of multiple (3-4) lineage starts yielding uniformly multilineage subclones was 45%. Even in the case of pentapotent starts, half yielded subclones that uniformly contained at least 4 lineages (erythroid, megakaryocyte, macrophage, and neutrophil), though some subclones failed to spawn long-term mast cell cultures (Figure 1, lower right panel). Thus, approximately half of the multipotent clonogenic cells obtained from the stem cell fraction were able to undergo 2 to 4 serial divisions without restricting their myeloid differentiation potential.
In view of these results, we concentrated on dispersed morphology starts generated from the stem cell-enriched fraction. From starts consisting of 4 to 16 cells, one or more cells were individually processed to global cDNA, while their siblings were subcultured to determine their lineage potential. Eight starts were identified with potential to differentiate into at least erythroid, megakaryocyte, monocyte, and neutrophil lineages. From these, 15 single-cell cDNA samples were harvested. Sibling cells from 5 of the 8 starts generated tetralineage colonies in methylcellulose. In 3 of these 5, all siblings generated tetralineage colonies, whereas in 2 others some siblings generated tetralineage colonies and others yielded colonies containing 3 or fewer lineages. In the remaining 3 of 8 multilineage starts, most of the colonies from sibling cells also yielded long-term liquid cultures of mast cells. The 4 associated cDNA samples from these “pentapotent” instances were segregated for separate analysis (Figure 1). Because these numbers are small, it was not anticipated that hybridization differences between the tetrapotent and pentapotent sets could be considered significant.
Additional cDNA samples were harvested from tripotent, bipotent, and unipotent precursor cells—again, as determined by sibling outcomes. Such samples were entered into our archive only if sibling outcomes were identical. New samples were also obtained from terminally differentiating myeloid and T lymphocytic cells. Relative to the sample collection described originally,4 the present effort enlarged the number of cDNA entries to 155 from the original 69. Of greatest interest, representation was now expanded to include multipotent cells able to differentiate into at least 4 or 5 myeloid lineages. An overview of the sample matrix is shown in Figure2, indicating numbers of samples of each type and a schematic representation of their hierarchical relationships.
Slot blots of amplified cDNA from various positions in the hematopoietic precursor hierarchy.
On the left are shown 2 alternative layouts (A and B) for the blots shown in subsequent figures. Layouts indicate the lineage potential of each stage, and the number (in parentheses) of independent amplified cDNA samples pooled before application to the blot. E, erythroid; Mg or Meg, megakaryocyte; Mc or Mac, monocyte/macrophage; Mst, mast cell; N, neutrophil; BFU-E, precursors of large erythroid-only 6- to 8-day colonies; CFU-E, precursors of small erythroid 2 day colonies; p, unipotent precursors of 7-day colonies. The left column of each blot contains pentapotent (E/Mg/Mc/N/Mst), tetrapotent, tripotent, bipotent, and unipotent precursor cell cDNAs. Middle and right columns contain unipotent precursor cell cDNAs (pMac) and samples from terminally maturing cells (E, N, Mast, etc). The sample indicated by “?” was of uncertain origin but probably mature macrophage. On the top right is an autoradiogram of a type A layout blot hybridized with a radiolabeled probe for L32. Below is a hierarchy tree representative of the various stages arrayed on the blots. The topmost level represents pentapotent precursors, the next levels tetrapotent, tripotent, bipotent, and unipotent precursors, while the bottom row represents terminally maturing cells in the indicated lineages. The ghosted circle represents a committed mast cell precursor stage, assumed to exist but not actually sampled. Radiographic densities from the autoradiogram were excised, squared, and superimposed on the corresponding positions in the hierarchy.
Slot blots of amplified cDNA from various positions in the hematopoietic precursor hierarchy.
On the left are shown 2 alternative layouts (A and B) for the blots shown in subsequent figures. Layouts indicate the lineage potential of each stage, and the number (in parentheses) of independent amplified cDNA samples pooled before application to the blot. E, erythroid; Mg or Meg, megakaryocyte; Mc or Mac, monocyte/macrophage; Mst, mast cell; N, neutrophil; BFU-E, precursors of large erythroid-only 6- to 8-day colonies; CFU-E, precursors of small erythroid 2 day colonies; p, unipotent precursors of 7-day colonies. The left column of each blot contains pentapotent (E/Mg/Mc/N/Mst), tetrapotent, tripotent, bipotent, and unipotent precursor cell cDNAs. Middle and right columns contain unipotent precursor cell cDNAs (pMac) and samples from terminally maturing cells (E, N, Mast, etc). The sample indicated by “?” was of uncertain origin but probably mature macrophage. On the top right is an autoradiogram of a type A layout blot hybridized with a radiolabeled probe for L32. Below is a hierarchy tree representative of the various stages arrayed on the blots. The topmost level represents pentapotent precursors, the next levels tetrapotent, tripotent, bipotent, and unipotent precursors, while the bottom row represents terminally maturing cells in the indicated lineages. The ghosted circle represents a committed mast cell precursor stage, assumed to exist but not actually sampled. Radiographic densities from the autoradiogram were excised, squared, and superimposed on the corresponding positions in the hierarchy.
For investigation of stage-specific gene expression, single-cell cDNA samples from each precursor or mature cell type were pooled, and each pool was transferred as a single slot onto hierarchy blots. Pooling was intended not only to simplify the generation of blots but also to average out the marked variation in transcript expression observed at the single cell but not population levels.4 20 Two alternative blot layouts are shown in Figure 2, indicating the numbers of samples pooled for application to each slot. Also shown is the result obtained when a blot with layout A was hybridized with a probe for L32, a constitutively expressed “housekeeping” transcript encoding a ribosomal subunit protein. The transcript was detected as expected in all cellular samples. A background control (“blank”), consisting of an equivalent amount of primer elongation artifact amplified in the global RT-PCR reaction in the absence of cell-derived mRNA template, did not bind the probe. In the lower right panel of Figure 2, the autoradiographic images from each individual slot were redimensioned and positioned over the corresponding locations in the lineage tree. This arrangement allows rapid assessment of differential hybridization intensities—in this example, it illustrates a general expression pattern for L32, with somewhat lesser hybridization to the sample from maturing neutrophils and none to the blank.
Hybridization of the L32 probe to the pools prepared from pentapotent and tetrapotent cells was frequently weaker than for most of the other pools, though the difference was not marked in the exposure shown in Figure 2. When the single-cell cDNA components of each pool were tested individually, each of the 4 pentapotent samples hybridized weakly, whereas 6 of the 11 tetrapotent samples hybridized more strongly. To investigate the basis for the difference, the pentapotent pool and a pool of the strongly hybridizing tetrapotent subset were cloned separately into plasmid libraries. A random sampling of 31 plasmid inserts from the pentapotent pool revealed 84% to consist of primer artifact rather than bona fide cDNA. In contrast, all 39 inserts from the strongly hybridizing tetrapotent subset were cell-derived.
Hierarchy blot analysis of cytokine receptor transcript expression
To assess expression of cytokine receptor transcripts, hierarchy blots were hybridized with radiolabeled receptor cDNA probes. Because the global RT-PCR procedure begins with oligo(dT) priming, efficient detection of transcripts in the amplified product requires probes that include 3′ terminal sequence. Two levels of analysis were performed. An initial screen was carried out with probes prepared simply by isolating nominal 3′ termini from supplied plasmid probes. In most instances, the resultant hybridization signals were convincingly above background, and in those instances further experimentation was not performed. However, a number of probes failed to hybridize above background even though cells at particular stages were known to be responsive to the corresponding cytokines. These negative results prompted a second level of analysis. The first step was to ensure that the probes were indeed inclusive of the extreme 3′ ends of their target transcripts. In 3 instances (c-kit, IL-2Rβ, IL-7Rα), the available GenBank mRNA sequences were incomplete. These were successfully extended by identification of contiguous EST clusters containing polyadenylation signals followed by polyA tails (detailed in “Materials and methods”). Suitable primer pairs enclosing terminal sequence were devised and used to amplify the corresponding fragments from globally amplified cDNA, as indicated. Once the sequence of the amplified fragments was confirmed, they were radiolabeled and used as probes on hierarchy blots. Additionally, the primers were used in secondary PCR reactions applied to each global cDNA pool represented on the hierarchy blots.
Cytokine receptor transcripts expected to demonstrate lineage-restricted expression.
Once a matrix of cDNA samples was assembled that had an expanded representation of uncommitted stages, it became feasible to compare RNA-level expression patterns of receptors for cytokines acting before and after lineage commitment to the corresponding biologic response profiles. For heteromultimeric receptors, we chose to probe only for subunits associated with ligand specificity. Subunits of 17 distinct cytokine receptors were analyzed.
Most hematopoietic cytokines have been described as influencing cellular targets at multiple stages and frequently in more than a single lineage. Three cytokines most associated with dominant action in a single lineage provided an opportunity to compare expression both across lineages and through transitions from multiple to single potentiality.
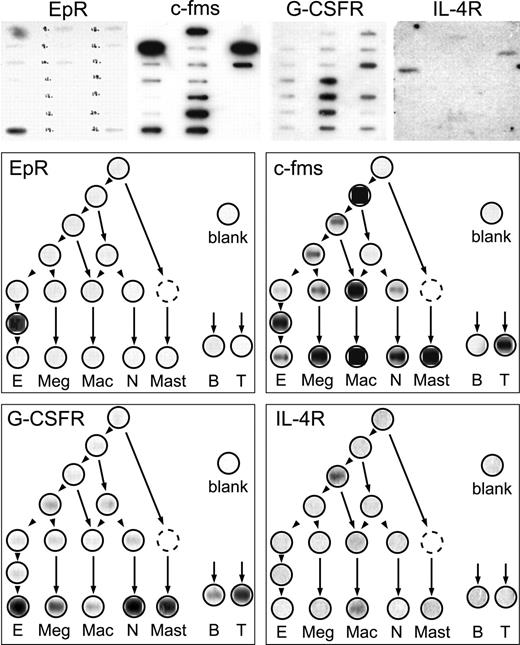
Erythropoietin is the archetypal lineage-specific cytokine whose biologic effects are considered confined mainly to the erythroid lineage. Moreover, its stage of action within that lineage is well defined, commencing only after unique commitment has occurred and centering on precursor cells called colony-forming unit (CFU-E) that immediately precede the onset of globin synthesis.21 As shown in Figure 3, hybridization of an EpR probe was decidedly strongest over cDNA from CFU-E, exactly in accord with the biologic response profile.
Hybridization patterns of probes for receptor transcripts expected to show lineage-restricted expression.
Blots (layout A) hybridized with the indicated probes are shown across the top. Below, the autoradiographic signals are arranged over the corresponding positions in the hierarchy trees, as explained in the legend to Figure 2.
Hybridization patterns of probes for receptor transcripts expected to show lineage-restricted expression.
Blots (layout A) hybridized with the indicated probes are shown across the top. Below, the autoradiographic signals are arranged over the corresponding positions in the hierarchy trees, as explained in the legend to Figure 2.
The receptor for the macrophage growth factor M-CSF is c-fms22; its biologic activity and physical binding are associated with committed cells in the macrophage lineage.23,24 Genetic disruption of c-fms leads mainly to a failure of development of osteoclasts from their macrophage precursors.25 Figure 3 confirms the expression of c-fms transcripts in committed macrophage precursors and in terminally maturing monocytes–macrophages. Distinct hybridization was also observed in maturing populations of other hemopoietic lineage cells. Because these samples of terminally maturing cells were derived from populations that could have contained a small macrophage component, the significance of these signals is not yet clear. Very strong hybridization was also detected in single tetrapotent precursor cells but was absent in downstream precursors that had dual macrophage–neutrophil potential.
Ambient granulocyte–colony-stimulating factor (G-CSF) levels limit neutrophil production in vivo,26 and G-CSF stimulates predominantly committed neutrophil precursor growth in cultures of murine marrow.27 Although it mobilizes multipotent precursors from marrow in vivo and effects on earlier precursors have been described in bulk cultures, compelling evidence for direct action on uncommitted cells is sparse. Binding studies localized receptors to neutrophilic and monocytic lineages, with receptor numbers increasing with maturation.28 29 Expression of transcripts for G-CSF receptor was, therefore, anticipated to show a corresponding pattern. Using a 241-bp probe extending to 50 bases upstream of the poly(dA) tail, we observed strong hybridization throughout the precursor hierarchy (not shown). To determine whether this diffuse hybridization could reflect the presence of a repetitive motif, the probe was compared against the GenBank database and revealed the presence of a tract (positions 3114-3245, accession #M58288), 25 b upstream of the polyadenylation signal, that is represented frequently in the mouse genome. Accordingly, we used a new 3′ probe that excluded this motif (Table 1 footnote) and observed strongest hybridization to cDNA from maturing cells in most lineages, including neutrophils, and weaker signals in bipotent and committed neutrophilic precursors (Figure 3). Fibroblasts were also distinctly positive.
IL-4 is best known as a regulator of growth and immunoglobulin switching during B lymphocyte development, but it also has pleiotropic effects on various kinds of myeloid colony formation in culture.30 However, its best-established primary myeloid targets are mast cells31 and monocytic lineage cells32 that respond by deactivation,33cytokine down-regulation,34 and dendritic cell conversion.35 In general correspondence with this pattern of biologic activity, strongest hybridization of the IL-4Rα probe was detected in maturing macrophage populations and more weakly in their immediate committed precursors. Hybridization was also detected distinctly in tripotent precursors and just above background in mast cell cDNA.
Receptor transcripts expected to be expressed in very primitive hematopoietic precursors.
C-kit is expressed on reconstituting stem cells,36-38 and its ligand promotes their survival and growth in culture.13 It is also required for transit of early stages of erythroid commitment upstream of CFU-E39and for mast cell differentiation and growth in vivo.40Although hybridization of the 3′ UTR probe (A probe, Table 1) was expected not only to cDNA from multipotent stages but also to committed erythroid precursors and mast cells, we obtained significant hybridization only to pentapotent cell cDNA (Figure4). To extend the sensitivity of detection, we subjected each globally amplified cDNA sample pool to secondary c-kit–specific amplification using the B primers indicated in Table 1. A single fragment of the expected size was amplified from the pentapotent cDNA sample (Figure 4) and confirmed as c-kit by sequencing. The same fragment was also amplified from cDNA corresponding to tripotent and bipotent E/Meg and Neut/Mac precursors and to committed erythroid precursors (BFU-E), but not from any of the other committed precursor cDNA pools and not from long-term mast cell cDNA.
Expression of receptor transcripts expected to be found in primitive hematopoietic precursors.
Autoradiographs for layout A hierarchy blots probed with c-kit (A, Table 1) and flk-1 are shown above left, and the corresponding signals are arranged on hierarchy trees below left. (right) Global cDNAs from the indicated sources were subjected to target-specific secondary PCR, and the products were run out on agarose gels containing ethidium bromide. Each gel contained molecular weight markers corresponding to the number of bases shown at right. Positive PCR controls are shown at right for flk-1 (globally amplified cDNA from murine aorta) and for flk-2 (globally amplified cDNA from sorted rhodamine 123loLy6A/Ehi murine marrow cells). C-kit, B primers (Table 1), 35 cycles, 60°C annealing, target fragment size 248 bases; flk-1, 35 cycles, 65°C, target fragment size 248 bases; flk-2/flt-3, 41 cycles, 65°C, target fragment size 265 bases. Below each ethidium gel is the corresponding gel blot hybridized with the corresponding radiolabeled probe.
Expression of receptor transcripts expected to be found in primitive hematopoietic precursors.
Autoradiographs for layout A hierarchy blots probed with c-kit (A, Table 1) and flk-1 are shown above left, and the corresponding signals are arranged on hierarchy trees below left. (right) Global cDNAs from the indicated sources were subjected to target-specific secondary PCR, and the products were run out on agarose gels containing ethidium bromide. Each gel contained molecular weight markers corresponding to the number of bases shown at right. Positive PCR controls are shown at right for flk-1 (globally amplified cDNA from murine aorta) and for flk-2 (globally amplified cDNA from sorted rhodamine 123loLy6A/Ehi murine marrow cells). C-kit, B primers (Table 1), 35 cycles, 60°C annealing, target fragment size 248 bases; flk-1, 35 cycles, 65°C, target fragment size 248 bases; flk-2/flt-3, 41 cycles, 65°C, target fragment size 265 bases. Below each ethidium gel is the corresponding gel blot hybridized with the corresponding radiolabeled probe.
Flk-1 encodes the receptor for vascular endothelial growth factor. It is required for emergence of endothelium and blood cells in early embryogenesis41 and for survival and growth of a common endothelial and hematopoietic progenitor differentiating from embryonic stem cells in culture.42 The receptor protein has been reported on primitive human hematopoietic precursors,43and transcripts have been detected in human hematopoietic precursors and in megakaryocytes.44 On hierarchy blots we detected a robust hybridization signal only in maturing megakaryocyte cDNA. Secondary Flk-1–specific PCR additionally detected transcripts in maturing macrophages but not in any other lineages or precursor stages (Figure 4). These results suggest possible secondary effects of vascular endothelial growth factor, the cognate ligand of Flk-1, on hematopoiesis through interaction with megakaryocytes or monocytes.
Flk-2/Flt-3 is required for the generation of B lymphoid precursor cells in vivo,45 and its ligand enhances survival and renewal of primitive myeloid precursor cells in culture.46-49 We were unable to detect significant probe hybridization on hierarchy blots (not shown). PCR primers directed against the 3′ terminus successfully amplified their target from cDNA globally amplified from a reconstituting stem cell fraction purified from adult mouse marrow (Figure 4), a result confirmed by sequencing of the amplified fragment. However, specific PCR failed to detect transcripts in any of the globally amplified hierarchy cDNAs. The result suggested that although transcripts may be expressed in resting reconstituting stem cells, they are down-regulated in more advanced, though still multipotent, myeloid progeny growing in culture.
Transcripts expected to be expressed with distinctive specificities in pluripotent precursors.
The receptors in this group recognize ligands known to affect precursors in more than one lineage and, in most instances, also precursors that have more than one lineage potential. Despite this common feature, the associated agonists differ from one another in thespectrum of targets each affects. The group was considered to provide an informative overall test of the fidelity of the staging and lineage assignments of our single-cell cDNA samples. Hybridization results for the group are shown in Figure5.
Receptor transcripts expected to differ among pluripotent precursors of different kinds.
Autoradiographs from type A (IL-7Rα, IL-6Rα, IL-11Rα) and type B blots are shown (top left), and the corresponding arrangements in hierarchy trees are shown beneath them. (right) Results of target-specific secondary PCR amplification, as detailed in the legend to Figure 4. Primers were directed at alternative upstream or downstream IL-7Rα transcript termini, as explained in the footnote to Table 1. Upstream primers, 35 cycles, 67°C annealing, followed by 5 cycles, 60°C annealing, target fragment size 317 bases. Downstream primers, 35 cycles, 67°C, target fragment size 249 bases.
Receptor transcripts expected to differ among pluripotent precursors of different kinds.
Autoradiographs from type A (IL-7Rα, IL-6Rα, IL-11Rα) and type B blots are shown (top left), and the corresponding arrangements in hierarchy trees are shown beneath them. (right) Results of target-specific secondary PCR amplification, as detailed in the legend to Figure 4. Primers were directed at alternative upstream or downstream IL-7Rα transcript termini, as explained in the footnote to Table 1. Upstream primers, 35 cycles, 67°C annealing, followed by 5 cycles, 60°C annealing, target fragment size 317 bases. Downstream primers, 35 cycles, 67°C, target fragment size 249 bases.
IL-3 is known to target committed precursors in the erythroid, megakaryocyte, neutrophil, and mast cell lineages and their pluripotent precursors.50-52 However, in the mouse there is little evidence that direct responsiveness extends further upstream to the level of long-term reconstituting cells. The IL-3-specific β receptor subunit, present in the mouse but lacking in humans, confers high ligand affinity to the α subunit53,54 and is required for maximum affinity of the receptors on hematopoietic cells in vivo.55 As anticipated, moderate to strong hybridization of the β-specific probe was seen in all erythroid–myeloid lineages and in bipotent and tripotent precursor stages, whereas distinctly less was observed to tetrapotent and pentapotent samples.
Granulocyte macrophage–colony-stimulating factor (GM-CSF) stimulates almost exclusively neutrophil and macrophage lineages and their bipotent precursors in murine marrow cultures,56 and it particularly differs from IL-3 in not stimulating the growth of erythroid colonies. Receptors have been detected on macrophage, neutrophil, and eosinophil lineage cells.57 The α receptor subunit is specific for the GM-CSF ligand.58 59The pattern of hybridization of a GM-CSFRα probe differed distinctly from that of the IL-3Rβ–specific probe: strong hybridization was obtained to tetrapotent cDNA, but high-level expression was maintained as expected into macrophage and neutrophil, but not erythroid or megakaryocyte, lineages.
IL-7 is a growth factor required by committed B and T lymphocyte precursors.60,61 It also enhances the growth of macrophage and neutrophil lineage cells in culture provided other cytokines are present, including c-kit ligand, or the classical colony-stimulating factors GM-CSF, M-CSF, or IL-3.62,63The α receptor subunit has been demonstrated by FACS analysis on macrophages and their precursors and on B and T lymphocytes,64 and monocytes are directly responsive.65 A 3′ probe prepared on the basis of the available GenBank sequence (accession #M29697.1) for the IL-7 α receptor sequence yielded little hybridization to a hierarchy blot (not shown). As referenced in the footnote to Table 1, we extended the mRNA sequence several hundred bases downstream to 2 potential alternative polyadenylation sites by identifying EST clusters in the NCBI murine EST database. Primer pairs specific to each of the alternative 3′ ends were used to amplify products from our globally amplified B cell cDNA pool. Products of the predicted sizes were obtained (Figure 5, “upstream” and “downstream”), and their identities were confirmed by sequencing. The upstream α receptor probe hybridized most clearly to cDNA from macrophages and B cells on the hierarchy blot (Figure 5). To obtain additional sensitivity, both primer pairs were used to probe our cDNA sample matrix by specific secondary PCR amplification. As also shown in Figure 5, the upstream primers detected transcripts in maturing B cell and macrophage cDNA, while the downstream primers detected targets in the same maturing samples and in mast cell, T cell, and, at a lower level, tetrapotent cDNAs. Overall, IL-7Rα transcripts were most abundant in cDNA from maturing macrophages and growing B cells, but they were also detected in multipotent cells and in mast- and T-cell populations. The results reinforce the evidence that macrophages are a likely target of IL-7. Because macrophages are potential sources of G-CSF and GM-CSF, the findings also suggest a possible indirect mechanism for reported effects of IL-7 on myeloid colony formation.
Tumor necrosis factor (TNF)-α and -β exert inhibitory action on the growth of hemopoietic precursor cells in bulk cultures, with macrophage precursor growth the most affected,66 and on the growth of neutrophil–macrophage precursors in single-cell cultures.67 The TNF receptor subtype 1 is the main mediator of these responses, as shown by loss of the inhibitory effect in cultures of marrow from TNFR1−/− mice.67Our TNFR1 probe hybridized predominantly to cDNA from bipotent neutrophil–macrophage precursor cDNA and from committed and maturing cells in the macrophage lineage (Figure 5). A strong signal was also seen on cDNA from NIH3T3 fibroblasts.
Although the effects of IL-6 have been reported in bulk cultures on various hematopoietic stages and lineages, binding of labeled IL-6 was more narrowly distributed on macrophage and megakaryocyte lineage cells and morphologically undifferentiated precursor cells.68 In good agreement, we observed hybridization of our IL-6Rα probe predominantly to cDNA from macrophages and their committed precursors and on upstream bipotent neutrophil–macrophage and tetrapotent precursor cells.
IL-11, when combined with c-kit ligand and other cytokines in cultures of murine marrow, strikingly promotes the growth of in vivo short-term erythroid reconstituting cells13 and multilineage and committed erythroid and megakaryocyte progenitors.69 These effects on pluripotent cells in culture are more pronounced than those of IL-6. Little information is available on differential binding of IL-11 to murine marrow cells in different lineages. As shown in Figure 5, the actions on more primitive cells are paralleled by distinct hybridization of an IL-11Rα probe to cDNA from bipotent and tripotent precursors upstream of the erythroid and megakaryocyte lineages, samples that showed significantly less signal with the IL-6Rα probe. Committed neutrophil precursors and ConA-activated T cells also hybridized the IL-11Rα probe strongly, but little signal was detected in megakaryocytes or their committed precursors.
Receptors with little prior indication of myeloid stage- or lineage- specificity of expression.
IL-6, IL-11, LIF, and Oncostatin M have distinct cognate receptors that incorporate gp130 as a common signaling subunit. Because the gp130 receptor subunit is ubiquitously expressed in various tissues,70 it was expected to show a wider distribution of expression than the IL-6 and IL-11 receptors individually. In agreement, our probe hybridized more widely than either IL-6Rα or IL-11Rα to cDNA from pluripotent and committed precursors, with distinctly less signal in maturing cells in all lineages (Figure6).
Receptor transcripts expected to be broadly expressed.
Autoradiographs from type A blots are shown (top left), and corresponding hierarchy trees are shown beneath. (right) Results of target-specific secondary PCR amplification for IL-2Rβ transcripts in hierarchy cDNA samples; 35 cycles, 60°C annealing, target fragment size 235 bases.
Receptor transcripts expected to be broadly expressed.
Autoradiographs from type A blots are shown (top left), and corresponding hierarchy trees are shown beneath. (right) Results of target-specific secondary PCR amplification for IL-2Rβ transcripts in hierarchy cDNA samples; 35 cycles, 60°C annealing, target fragment size 235 bases.
IL-1 shows a variety of enhancing and inhibitory effects in bulk cultures of myeloid and lymphoid cells in the presence of other primary agonists.52,71 Cells in murine marrow fractions enriched for either primitive stem cells or more advanced and committed progenitors bind IL-1 specifically,38 and mature neutrophils and monocytes are also primary responders,72suggesting widespread receptor expression in the hematopoietic hierarchy. The type 1 receptor is the principal signaling element responsible for biologic responses to IL-1, while the evidence suggests that type 2 receptors play modulating rather than primary roles and have a more restricted pattern of expression.72 73 As seen in Figure 6, type 1 receptor transcripts were detected ubiquitously in the marrow hierarchy, whereas type 2 transcript expression was found predominantly in maturing macrophages. These patterns thus also conform to expectations.
Although IL-2 plays its dominant biologic role in regulating lymphocyte growth downstream of T cell activation, granulocyte and macrophage lineage cells are reported to express receptor components for IL-2 and respond to it.74-76 The β receptor subunit is essential for the formation of a high-affinity receptor complex and transduction of a proliferative signal.77 Human monocytes are variably reported to express or not express the β subunit protein,75,77 while in mice it (CD122) is expressed on maturing cells in the neutrophil series.76 An IL-2 receptor β plasmid probe (Table 1) initially showed little hybridization on our hierarchy blots (not shown) and lacked an obvious polyadenylation signal. As described earlier for IL-7, it was possible to identify by EST cluster analysis a likely 3′ polyadenylation site located 660 bases downstream of the 3′ end of the available murine sequence in the GenBank database (accession #M28052.1). Using a primer pair directed against the identified terminus, we were able to amplify the correct target sequence from our globally amplified T cell cDNA, and the resultant probe hybridized robustly to cDNA from T cells but not from any of our other hierarchy samples (Figure 6). Target-specific secondary PCR additionally detected transcripts in tetrapotent cDNA but not in cDNA from more advanced cells, including the neutrophilic or monocytic lineages.
Discussion
The single-cell approach described in this study was designed to resolve gene expression patterns to individual stages during lineage divergence from multipotent stem cells. Generation of nearly homogeneous samples of cells at various individual stages is beyond the range of current cell-sorting strategies. However, we showed in an earlier study that homogeneity at specific stages could be achieved by sampling individual cells and described the generation and probing of a set of stage-specific cDNAs that included unipotent, bipotent, and a small number of tripotent precursors of various myeloid lineages.4
The present study describes the acquisition of new cDNA samples from single cells that were tetrapotent and pentapotent for the generation of progeny in erythroid, megakaryocyte, macrophage, neutrophil, and mast cell lineages. Multipotent cells were obtained in 2 stages, first by FACS enrichment of marrow for the fraction containing the long-term reconstituting stem cells, and then by selecting the disperse morphology colony starts generated from this fraction in culture. After 4 days in culture, only 1 in 60 of such starts had the capacity to reconstitute recipient mice.13 However, approximately half of them yielded multilineage differentiation, and approximately half of those yielded subclones that were uniformly multilineage. Thus, the approach allows access to small homogeneous populations of cells that are downstream of the long-term reconstituting cells but are still multipotent. In most instances, precursors were tetrapotent for erythroid, megakaryocyte, macrophage, and neutrophil differentiation. Where sibling cells exhibited the additional potential for spawning long-term mast cell cultures, the associated cDNA samples were segregated from the rest with the designation “pentapotent.” The numbers of samples were, however, too small for differences in hybridization behavior between tetrapotent and pentapotent cDNAs to be considered significant.
As determined by sibling outcomes, numerous cDNA samples were also obtained from downstream stages representing tripotent, bipotent, and unipotent hematopoietic precursor cells. In all these instances, samples were entered into the archive only if sibling outcomes were uniform. Overall, the new entries enlarged our sample set from 69 in our original study to 168, of which 120 derived from single growth-competent precursor cells and the rest derived from terminally maturing populations.
Abundance of regulated, but not of constitutive, transcripts varies widely from cell to cell, and there is evidence that much of the variability originates in the processed cells themselves rather than in the RT-PCR procedure.4 20 We analyzed sample pools rather than the entire array of single-cell cDNA samples, a tactic that allowed us to average the results from multiple samples at a particular stage without compromising the homogeneity assured by their single-cell origin.
Many of our receptor probes hybridized robustly to the hierarchy blots, yielding coherent patterns of differential expression. However, other probes were more problematic, either failing to hybridize above background to any of the pools (IL-2Rβ, IL-7Rα, flk-2/flt-3) or failing to hybridize to some stages expected to be positive on the basis of known responsiveness (c-kit) or hybridizing more broadly than expected (G-CSFR). Extra effort was expended on ensuring that probes were indeed representative of the 3′ termini of the corresponding transcripts and that they did not contain nonspecific motifs that could cause promiscuous hybridization. This effort led to identification of previously undocumented 3′ termini for murine IL-2Rβ, IL-7Rα, and c-kit transcripts and to the refinement of our G-CSFR probe to exclude a commonly occurring motif at the 3′ end. The second-generation G-CSFR probe showed a more restricted hybridization pattern confirmatory of previously reported results, and the new probes for IL-2Rβ and IL-7Rα directly hybridized to T and B lymphocyte cDNAs as expected.
The global RT-PCR procedure was designed to amplify all transcripts without bias. We and others have provided evidence that the procedure preserves relative abundance relationships in the original mRNA population.17 19 The amplification product thus differs substantially from that obtained by target-specific RT-PCR, by which, given enough cycles, even a few transcripts can amplify to yield a strong band. It was anticipated that receptor transcripts in precursors at very low abundance would be more difficult to detect by hybridization to our cDNA samples than those in higher abundance. We indeed found that our second-generation probes for c-kit, IL-2Rβ, and IL-7Rα still did not hybridize as widely as anticipated, whereas flk-2/flt-3 did not hybridize to any of our hierarchy samples. When secondary, target-specific PCR was performed on the globally amplified cDNAs, low levels of IL-2Rβ, IL-7Rα, c-kit, and Flk-1 transcripts were revealed that were not evident from simple hybridization analysis. The results reinforce the evidence that relative abundance relationships are maintained in globally amplified cDNA and establish that the amplified samples contain more information than is accessible by simple probe hybridization.
In the multipotent sample set comprising pentapotent and tetrapotent cDNAs, we detected transcripts for c-kit, gp130, IL-6Rα, GM-CSFRα, c-fms, and IL-1RI by direct hybridization, and IL-7Rα and IL-2Rβ only by target-specific PCR. In contrast, readily detectable signals for IL-3Rβ-specific and IL-11Rα only commenced in downstream tripotent and bipotent cDNAs, while even secondary target-specific PCR failed to detect flk-1 or flk-2/flt-3 in multipotent cDNAs. Hybridization of EpR, IL-2Rβ, GCSFR, and flk-1 probes was clearest in terminally differentiating cells of specific lineages.
Transcripts for Flk-1 and Flk-2/Flt-3 were not detected in multipotent cell cDNA. Because these transcripts were present as expected in megakaryocytes and blood vessel cDNA, respectively, the observation reinforces our view that most in vitro colony starts, generated as described here, consist of cells that are more advanced in differentiation than the earliest cells capable of long-term reconstitution in vivo. Our findings that transcripts for IL-1RI, IL-6Rα, and gp130, but not G-CSFR, are readily detectable in multipotent cells are concordant with published mRNA expression78 and cytokine-binding38 results on marrow fractions enriched by cell separation for primitive hematopoietic stem cells. However, our observations differ in detecting clear hybridization of c-fms and GM-CSFRα but little hybridization of IL-3Rβ-specific or erythropoietin receptor probes to multipotent cell cDNA. These discrepancies may be explainable by the different experimental approaches taken. Orlic et al used target-specific PCR and detected transcripts at a higher level of sensitivity than by direct hybridization. In addition, our findings are specific to cells operationally established to have multipotency, whereas the earlier work was conducted on populations that were likely to be operationally heterogeneous but that were not directly characterized. Further, we analyzed cells taken from tissue cultures rather than directly from marrow and without the long delays involved in cell sorting that could lead to changes in relative transcript abundance.
Unexpectedly, c-kit transcripts, detectable readily in multipotent and bipotent precursor cDNAs, could not be detected in cDNA from 6- to 8-week-old primary mast cell cultures. The result could reflect the down-regulation of c-kit expression by IL-379 80 or ConA or other details of the culture conditions.
The enhanced scope and resolving power of the analysis yielded a number of previously unreported observations. Although c-fms is understood to play its role in the monocyte lineage, we detected transcripts not only in that lineage but also in multipotent cells and committed precursors in the erythroid, megakaryocyte, and neutrophil lineages. The evidence for a role of G-CSF receptors in growth and regulation of neutrophil lineage cells is similarly clear. It was therefore unexpected that transcripts would be most abundant in terminally maturing cells in all hematopoietic lineages rather than in neutrophil or more primitive precursors. Conversely, though IL-7 and IL-2 receptor transcripts were detected as expected in B and T cells, respectively, we report the first indication of their expression in multipotent precursors. We also confirm the expression of Flk-1 transcripts in megakaryocytes and report the first indication that they are also expressed in macrophages.
Despite the emergence of unanticipated observations, an important motivation for undertaking this analysis was to assess the extent ofagreement between known biologic response patterns of hematopoietic precursors to specific cytokines and hybridization profiles obtained from our sample set to cytokine receptor probes. A number of reservations apply to our description of RNA-level expression. First, the view provided here represents a snapshot of RNA-level abundance under arbitrary conditions of culture and cytokine environments that may or may not be representative of environments in vivo. Further, receptor mRNA abundance may not necessarily reflect corresponding levels of functional protein. Finally, even if corresponding and functional protein is present, it cannot be assumed without further direct analysis that partners and downstream proteins are in place to couple receptors to cellular responses. Nevertheless, our results in aggregate provide a unique overview of precisely timed and stage-specific patterns of expression of receptor transcripts. The level of resolution achieved with this analysis adds significant new detail to our view of the sequence of activation of specific genes during the divergence and subsequent maturation of the hematopoietic lineages.
The level of agreement we observed with known biologic responsiveness at individual stages was satisfying, and major contradictions did not arise. Our results correctly distinguished between cytokine receptor transcripts expressed at highest levels only after lineage commitment (EpR, G-CSFR, IL-7Rα, IL-2Rβ, and IL-1RII) and many of the others expressed at moderate to high levels in multipotent and bipotent cells, including c-kit, the gp130-associated receptors, IL-3Rβ-specific, and GM-CSFRα. In addition, the experiments correctly distinguished differing patterns of expression within the precommitted cDNA samples. IL-1RI and gp130 probes hybridized essentially ubiquitously to early cell cDNAs. Probes for other receptor transcripts showed more specific patterns. GM-CSFRα and TNFR1 probes hybridized, as expected, mainly to bipotent and committed neutrophil–macrophage precursor cDNA, contrasting with the IL-3-specific β receptor probe that hybridized not only to these but also, as expected, to cDNA from mast cells and precursors upstream of the erythroid and megakaryocyte lineages. Whereas the IL-6Rα probe hybridized mainly to cells upstream of the monocytic pathway, IL-11Rα transcripts were also detected, as expected, in precursors immediately upstream of the erythroid and megakaryocyte lineages. In addition to the observed agreement with biologic responsiveness, each precursor category displayed a unique overall pattern of hybridization to the matrix of 17 receptor probes, supporting the notion that each sample pool indeed reflected a unique precursor stage. Collectively, the results provide supportive evidence for the validity of the cDNA assignments to particular stages and for the unique capacity of our sample matrix to resolve individual stages in the hematopoietic hierarchy.
We thank D. Hyam for technical assistance, P. Benveniste for purified marrow stem cells, G. Keller and S. Robertson for aorta cDNA, and the authors named in Table 1 for plasmid probes.
Supported by operating grants to N.N.I. from the National Cancer Institute of Canada and the Canadian Institutes of Health Research.
Correspondence:Norman N. Iscove, Department of Medical Biophysics, University of Toronto, 610 University Avenue, Toronto, Ontario, Canada M5G 2M9; e-mail: iscove@uhnres.utoronto.ca.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.






This feature is available to Subscribers Only
Sign In or Create an Account Close Modal