Abstract
In prior studies, it was demonstrated that the redox metabolism of doxorubicin leads to the formation of promutagenic oxidized DNA bases in human chromatin, suggesting a potential mechanism for doxorubicin-related second malignancies. To determine whether a similar type of DNA damage is produced in the clinic, peripheral blood mononuclear cell DNA from 15 women treated with infusional doxorubicin (165 mg/m2) as a single agent was examined for 14 modified bases by gas chromatography/mass spectrometry with selected ion monitoring. Prior to the 96-hour doxorubicin infusion, 13 different oxidized bases were present in all DNA samples examined. Chemotherapy, producing a steady-state level of 0.1 μM doxorubicin, increased DNA base oxidation up to 4-fold compared to baseline values for 9 of the 13 bases studied. Maximal base oxidation was observed 72 to 96 hours after doxorubicin treatment was begun; the greatest significant increases were found for Thy Gly (4.2-fold), 5-OH-Hyd (2.5-fold), FapyAde (2.4-fold), and 5-OH-MeUra (2.4-fold). The level of the promutagenic base FapyGua increased 1.6-fold (P < .02), whereas no change in 8-OH-Gua levels was observed in peripheral blood mononuclear cell DNA during the doxorubicin infusion. These results suggest that DNA base damage similar to that produced by ionizing radiation occurs under clinical conditions in hematopoietic cells after doxorubicin exposure. If doxorubicin-induced DNA base oxidation occurs in primitive hematopoietic precursors, these lesions could contribute to the mutagenic or toxic effects of the anthracyclines on the bone marrow.
Introduction
The anthracycline antibiotic doxorubicin plays an important role in the treatment of a wide variety of hematologic malignancies as well as breast cancer and osteogenic sarcoma.1 Although many competing hypotheses exist to explain the antineoplastic mechanism(s) of action of doxorubicin, there is little doubt that this drug interacts pleiotropically with DNA.2 In addition to DNA interactions that may be important for the therapeutic effects of the drug, doxorubicin is a well-characterized mutagen.3,4 When used clinically in combination with cyclophosphamide, doxorubicin is associated with a dramatically increased risk of second malignancy, particularly acute myelomonocytic leukemia.5,6 However, the specific DNA lesions underlying the carcinogenic effect of doxorubicin remain to be elucidated.7
Early investigations of the doxorubicin-DNA interaction characterized the ability of the planar anthracycline ring to intercalate into DNA8 with more recent studies demonstrating a special affinity of the drug for dGdC-rich regions flanked by A:T base pairs.9 Unfortunately, little evidence has been developed to demonstrate that intercalation of DNA by doxorubicin, per se, could explain the varied biochemical alterations (or mutagenicity) produced by this drug.10 Furthermore, because of the intercalative function of the anthracycline ring, preclinical studies of the doxorubicin-DNA interaction initially focused on the ability of the anthracycline to inhibit DNA and RNA synthesis as well as specific DNA polymerases.11-13 However, the doxorubicin concentrations required for inhibition of these enzymes were found to be in excess of those achievable clinically, which may explain the lack of correlation between doxorubicin-related inhibition of bulk DNA or RNA synthesis and tumor cell killing.14 15
More recent studies of the doxorubicin-DNA interaction have focused on the effect of the anthracycline on the nuclear matrix-associated enzyme topoisomerase II.16 Doxorubicin inhibits topoisomerase II through the formation of DNA strand passage intermediates that can be detected as protein-associated DNA single- and double-strand breaks linked to the enzyme; intercalation of DNA by anthracyclines is not required for inhibition of topoisomerase II.17 However, in certain cell lines the formation and disappearance of doxorubicin-related DNA breaks has not correlated with tumor cell killing; in others, DNA single-strand cleavage is modest and double-strand breaks are essentially undetectable at supralethal drug concentrations.18,19 Because doxorubicin also exerts more cytotoxicity than expected per DNA break,20 it is likely that mechanisms of DNA damage other than those related to topoisomerase II operate in parallel with the formation of the doxorubicin–DNA–topoisomerase II ternary complex.
Continued interest in the nature of the interaction of doxorubicin with DNA has recently provided new insights into potential mechanisms of anthracycline-induced mutagenicity. It now seems clear that redox activation of doxorubicin by H2O2, dithiothreitol and Fe3+, or formaldehyde produces a unique alkylating species capable of covalently binding DNA.21-23The discovery of this novel drug adduct, although not yet demonstrated in clinical material, provides the first evidence for a long sought after, potentially lethal, covalently bound doxorubicin species that could provide a critical link in our understanding of the doxorubicin-DNA interaction. Because previous studies have demonstrated that flavin dehydrogenase–mediated redox cycling of doxorubicin leads to reactive oxygen metabolism, including the formation of H2O2, in mammalian nuclei as well as intact cells,24 25 it is reasonable to suggest that doxorubicin-induced oxidative modifications in DNA might also play an important role in either the therapeutic or carcinogenic properties of this agent.
In a previous study, we examined whether the flavoenzyme-related, one-electron reduction of doxorubicin induced DNA base modifications in isolated human chromatin.26 Redox cycling of doxorubicin catalyzed by NADH dehydrogenase generated a wide range of promutagenic oxidized DNA bases, including Thy Gly3 and 8-OH-Gua, typical of base modifications produced by a chemical species with the characteristics of the hydroxyl radical (·OH).27-29 Furthermore, base oxidation was significantly increased by Fe3+, and significantly decreased by substitution of the non–redox cycling anthracycline 5-iminodaunorubicin for doxorubicin.
Our in vitro experiments, as well as recent interest in the mechanism of anthracycline-related second malignancies, stimulated a further examination of free radical metabolism in patients treated with doxorubicin. In the study presented here, we evaluated the potential of high-dose infusional doxorubicin to produce base oxidation in peripheral blood mononuclear cell (PBMC) DNA. We found that doxorubicin administered for 96 hours as the first agent of a multiagent conditioning regimen for women undergoing high-dose chemotherapy with bone marrow stem cell support30 significantly increased levels of 9 different oxidized bases detectable in PBMC DNA by gas chromatography/mass spectrometry (GC/MS) with selected ion monitoring. The pattern of base modification was similar, in part, to our previous in vitro study with human chromatin and suggests the involvement of ·OH in the oxidation of PBMC DNA after doxorubicin exposure in the clinic.
Patients, materials, and methods
Materials
Doxorubicin hydrochloride was purchased from Pharmacia & Upjohn (Kalamazoo, MI); Tris base, proteinase K, and sodium dodecyl sulfate (SDS) were obtained from Boehringer Mannheim (Indianapolis, IN). EDTA and sodium chloride were from Mallinckrodt (Paris, KY). Tergitol type NP-40, RNAse A, 2-amino-6,8-dihydroxypurine (8-OH-Gua), azaT, formic acid, and Histopaque-1077 were purchased from Sigma Chemical (St Louis, MO). Ethyl alcohol [200 proof] was from Quantum Chemical (Tuscola, IL). Spectra/Por dialysis membranes with a molecular weight cutoff of 3500 were purchased from Spectrum Companies (Gardena, CA). 2-OH-Ade was from ACROS Organic (Fairlawn, NJ). BSTFA and acetonitrile were purchased from Pierce (Rockford, IL). Pure samples of 5-OH-5-MeHyd, 5-OH-Ura, 5-OH-6H-Thy, 5-OH-Cyt, 5-OH-MeUra, Thy Gly, 5,6diOH-Ura, FapyAde, 8-OH-Ade, Xan, and FapyGua were kindly provided by Dr Miral Dizdaroglu of the National Institute of Standards and Technology, Gaithersburg, MD. Dulbecco phosphate-buffered saline solution (PBS) without calcium and magnesium was purchased from Irvine Scientific (Santa Ana, CA). Only deionized, glass-distilled water was used in this study.
Clinical treatment program, pharmacokinetic determinations, and PBMC sampling
The PBMCs for determination of DNA base modification and plasma for doxorubicin pharmacokinetics were obtained from patients entered in a clinical trial of high-dose infusional doxorubicin, cyclophosphamide, and paclitaxel with autologous peripheral blood progenitor cell support for women with high-risk primary and chemotherapeutically responsive metastatic breast cancer. The clinical trial and blood sampling studies were approved by the Institutional Review Board of the City of Hope National Medical Center. All patients gave their voluntary, written informed consent for participation in the investigational treatment program. Blood from 15 patients of median age 47 years (range, 33-63 years) treated between December 1996 and February 1998 was examined for modified DNA bases; an evaluation of doxorubicin pharmacokinetics was performed for 6 patients.
The preliminary clinical results of the high-dose chemotherapy regimen used for this study have recently been reported.30 The initial therapeutic component involved a 96-hour, continuous intravenous infusion of doxorubicin at a dose of 165 mg/m2, followed by high-dose cyclophosphamide 100 mg/kg, and escalating doses of paclitaxel delivered over 24 hours. Peripheral blood progenitor cells (at least 8 × 108 mononuclear cells/kg) were administered after the completion of high-dose chemotherapy. To examine the pharmacokinetics of doxorubicin administered at this dose and schedule, as well as oxidized DNA base levels, 20-mL blood samples were collected in EDTA-coated tubes from a peripheral intravenous line contralateral to the site of treatment immediately prior to the start of the infusion and 48, 72, and 96 hours after the initiation of doxorubicin. Plasma was separated, and doxorubicin concentrations were determined by high-performance liquid chromatography (HPLC) with fluorescence detection using daunorubicin as the internal standard.31 PBMCs were isolated from whole blood obtained at these time points by Ficoll-Hypaque gradient. Briefly, each 20-mL sample of anticoagulated whole blood was diluted 1:3 in PBS and layered onto Histopaque-1077. Following centrifugation at 1500g for 20 minutes, the PBMC-containing interface was transferred to a 15-mL conical centrifuge tube and washed once in ice-cold PBS. The viability of the PBMC was always more than 90% as assessed by exclusion of trypan blue dye. After the wash, the cell pellet was resuspended in 0.5 mL TEN buffer (10 mM Tris HCl, pH 7.8, 1 mM EDTA, 0.15 M NaCl) and stored at −20°C until all samples from each patient were collected. Prior to further study, cells were defrosted and all samples from each patient were simultaneously processed further for DNA isolation, hydrolysis, derivatization, and GC/MS analysis.
DNA preparation
DNA from PBMCs was prepared with no exposure to phenol according to the modified procedure of Nicolaides and Stoeckert.32 Ten percent Tergitol NP-40 was added to a final concentration 0.6%. Samples were gently shaken for 2 minutes on ice; nuclei were obtained by centrifugation at 1400g for 10 minutes at 4°C. Collected nuclei were resuspended in nuclear lysis buffer (10 mM Tris HCl, pH 8.0, 2 mM EDTA, 0.4 M NaCl) and digested overnight at 37°C with gentle shaking after the additions of 10% SDS (final concentration 0.6%) and proteinase K solution (2 mg/mL in 1% SDS and 2 mM EDTA). The final proteinase K concentration was 0.27 mg/mL. The next day solubilized nuclei were incubated with RNAse (final concentration 0.10 mg/mL) at 50°C for 2 hours. Proteins were precipitated from nuclei by adding 6 M NaCl (saturated solution) to a final concentration of 1.5 M, shaking vigorously for 30 seconds; samples were then centrifuged at 10 000g for 10 minutes at room temperature. Supernatants were removed, and DNA was precipitated with 2 volumes of ethanol kept at −20°C for at least 1 hour and then centrifuged at 10 000g. DNA precipitate was rinsed with 70% ethanol and centrifuged again at 10 000g. Dry DNA was dissolved in water at room temperature. After centrifuging again to remove undissolved debris, DNA samples were dialyzed overnight at room temperature against water. DNA concentration was estimated by measuring the optical density at 260 nm using a Milton Roy Spectronic 1201 Spectrophotometer.
Measurement of oxidative DNA base modifications by GC/MS
Each set of patient DNA samples obtained before or at specific times during the doxorubicin infusion contained the same amount of DNA by estimation of OD260. A known amount of azaT was added to each sample as an internal standard for quantitative determination of modified DNA bases. For GC/MS measurements, all samples in a series were lyophilized, hydrolyzed with 66% formic acid at 140°C for 45 minutes, lyophilized again, and derivatized with a mixture of BSTFA/acetonitrile (4:1) at 130°C for 45 minutes. For measurements of the relative molar response factors (RMRFs) needed for quantitative calculations of the modified bases in DNA samples with azaT as an internal standard, samples containing known amounts of 13 DNA modified bases monitored in these studies and the known amount of azaT were prepared and kept frozen. 5-OH-Hyd was not included because it was not available. Periodically, during the course of this work, we lyophilized and derivatized these samples in the same manner as the patient DNA samples, and performed GC/MS analysis to calculate RMRFs for all modified bases. These averages of the RMRFs were used for calculations of the modified bases in the tested DNA samples from PBMCs. Each set of patient samples was examined by GC/MS in 1 day with the same instrument tuning. A Hewlett-Packard (Palo Alto, CA) model 5890A gas chromatograph with a model 5970B mass spectral detector was used for these studies. The injector port was kept at 250°C, interface at 280°C, and oven temperature was kept at 150°C for 2 minutes followed by a gradient of 8°C/min up to 260°C; then the oven was kept at 260°C for an additional 2 minutes. A Hewlett-Packard Ultra 2 (cross-linked methyl-siloxane) column (12 meters, 0.20 mm ID, film thickness 0.33 m) was used. The column head pressure was 50 kPa with a 20:1 split. Using these conditions, 14 oxidative products of DNA bases and azaT were detected simultaneously in one chromatogram.
To compare changes in the content of modified bases in our DNA samples quantitatively, azaT was used as an internal standard; the areas of the peaks of the ions of interest were estimated using the integration software package of the Hewlett-Packard GC/MS. The RMRF for each compound with respect to the internal standard is described by the following equation33:
×
The average value for the RMRF of each modified DNA base measured in this trial is shown in Table1. These values were obtained from GC/MS measurements performed during the same time period in which modified DNA bases were quantitated in the patient samples for this study. Although an authentic sample of 5-OH-Hyd was not available for determination of its RMRF, because 5-OH-Hyd was detected in the PBMC DNA of the patients treated with doxorubicin, we assumed the RMRF for this compound to be the same as that calculated for 5-OH-5-Me-Hyd due to the similarity of their MS spectra. Having established the RMRFs for these bases, the amount of each modified base in our DNA samples, with azaT as the internal standard, was calculated using the following equation:
The amount of each modified base in each extracted DNA sample was calculated per milligram of DNA sample. Our calibration samples did not contain additional DNA or any other “dummy mass” to offset destruction or alteration of bases during formic acid hydrolysis and derivatization. It has been assumed that the destructive effect of hydrolysis on the modified DNA bases may, in part, be quenched by the bulk of the DNA in our patient samples. Furthermore, additional DNA was not added to the calibration samples because there is no unambiguous method to ensure that the effects of hydrolysis are identical on modified and unmodified bases. The DNA amount was determined by measuring the optical density of each DNA sample at 260 nm (OD260 = 1 for a DNA concentration of 50 μg/mL).
Experimentally derived relative molar response factors and oxidized DNA base content in peripheral blood mononuclear cells of patients prior to doxorubicin
| DNA base . | Ion used . | RMRF* . | Oxidized DNA base level (nmol/mg DNA)* . | Oxidized DNA base level (oxidized bases/ 106 bases)† . |
|---|---|---|---|---|
| Thy Gly | 259 | 0.49 ± 0.13 | 5.98 ± 6.99 | 1914 ± 2237 |
| 5-OH-Hyd | 317 | — | 0.81 ± 1.25 | 259 ± 400 |
| FapyAde | 354 | 1.66 ± 0.55 | 0.64 ± 0.64 | 205 ± 205 |
| 5-OHMe-Ura | 358 | 0.31 ± 0.07 | 0.60 ± 0.85 | 192 ± 272 |
| 5,6diOH-Ura | 417 | 2.35 ± 0.87 | 1.20 ± 0.83 | 384 ± 266 |
| 5-OH-5-MeHyd | 331 | 0.40 ± 0.10 | 1.64 ± 2.66 | 525 ± 851 |
| 5-OH-Ura | 329 | 0.72 ± 0.51 | 0.60 ± 0.54 | 192 ± 173 |
| FapyGua | 442 | 8.20 ± 6.1 | 6.62 ± 6.24 | 2118 ± 1997 |
| Xan | 353 | 0.45 ± 0.30 | 4.71 ± 3.97 | 1507 ± 1270 |
| 2-OH-Ade | 352 | 0.25 ± 0.13 | 0.33 ± 0.77 | 106 ± 246 |
| 5-OH-Cyt | 328 | 0.46 ± 0.11 | 0.73 ± 0.72 | 234 ± 230 |
| 8-OH-Ade | 352 | 0.32 ± 0.12 | 1.52 ± 0.73 | 486 ± 237 |
| 8-OH-Gua | 440 | 4.66 ± 2.76 | 13.30 ± 9.76 | 4256 ± 3123 |
| DNA base . | Ion used . | RMRF* . | Oxidized DNA base level (nmol/mg DNA)* . | Oxidized DNA base level (oxidized bases/ 106 bases)† . |
|---|---|---|---|---|
| Thy Gly | 259 | 0.49 ± 0.13 | 5.98 ± 6.99 | 1914 ± 2237 |
| 5-OH-Hyd | 317 | — | 0.81 ± 1.25 | 259 ± 400 |
| FapyAde | 354 | 1.66 ± 0.55 | 0.64 ± 0.64 | 205 ± 205 |
| 5-OHMe-Ura | 358 | 0.31 ± 0.07 | 0.60 ± 0.85 | 192 ± 272 |
| 5,6diOH-Ura | 417 | 2.35 ± 0.87 | 1.20 ± 0.83 | 384 ± 266 |
| 5-OH-5-MeHyd | 331 | 0.40 ± 0.10 | 1.64 ± 2.66 | 525 ± 851 |
| 5-OH-Ura | 329 | 0.72 ± 0.51 | 0.60 ± 0.54 | 192 ± 173 |
| FapyGua | 442 | 8.20 ± 6.1 | 6.62 ± 6.24 | 2118 ± 1997 |
| Xan | 353 | 0.45 ± 0.30 | 4.71 ± 3.97 | 1507 ± 1270 |
| 2-OH-Ade | 352 | 0.25 ± 0.13 | 0.33 ± 0.77 | 106 ± 246 |
| 5-OH-Cyt | 328 | 0.46 ± 0.11 | 0.73 ± 0.72 | 234 ± 230 |
| 8-OH-Ade | 352 | 0.32 ± 0.12 | 1.52 ± 0.73 | 486 ± 237 |
| 8-OH-Gua | 440 | 4.66 ± 2.76 | 13.30 ± 9.76 | 4256 ± 3123 |
RMRF indicates relative molar response factor.
Mean ± SD.
1 nmol oxidized base/mg DNA is 32 molecules of the modified base/105 DNA bases.
Statistical methods
All samples obtained from a single patient were analyzed by GC/MS on the same day to minimize day-to-day assay variability. The levels of modified bases were normalized to pretreatment values determined in each individual. A 2-tailed, paired t test was used, after consultation with Dr Jeff Longmate of the City of Hope Department of Biostatistics, to compare the fold change in each modified base after doxorubicin treatment to its pretreatment value using the entire (n = 15) patient sample; NS is not significant,P > .05.
Results
Identification of modified DNA bases by GC/MS
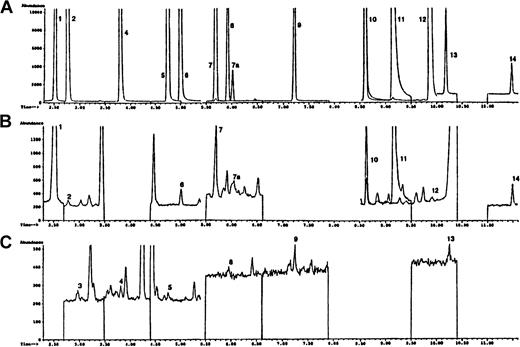
The chromatogram shown in Figure 1A using known standards is representative of 12 modified DNA bases monitored in this study. These compounds are: 5-OH-5MeHyd, 5-OH-Ura, 5-OH-Cyt, 5-OHMe-Ura, cis and trans Thy Gly, 5,6diOH-Ura, FapyAde, 8-OH-Ade, 2-OH-Ade, Xan, FapyGua, and 8-OH-Gua. Not included in this chromatogram are 5-OH-6-H-Thy, which was also monitored but was not detected in our DNA samples, and 5-OH-Hyd, a pure sample of which was not available. However, 5-OH-Hyd (monitored ion 317) was identified in all of the DNA samples we studied and was found to elute after 5-OH-5-MeHyd. The internal standard azaT (monitored ion 256) elutes before 5-OH-5-MeHyd. Panels B and C of Figure 1 demonstrate typical chromatograms from PBMCs of a patient immediately before she received high-dose, infusional doxorubicin.
Selected ion chromatograms obtained during the GC/MS analysis of PBMC DNA.
(A) Represents a mixture of trimethylsilylated standards. (B) The chromatogram demonstrates the higher abundance ions and the chromatogram in panel C shows the lower abundance ions of a trimethylsilylated hydrolysate of DNA isolated from the PBMCs of a representative patient entered into this trial before initiation of the doxorubicin infusion. Experimental details are given in “Patients, materials, and methods.” Peak indicates DNA base (ion monitored): 1, 6-azathymine (m/z 256) (internal standard); 2, 5-OH-5-MeHyd (m/z 331); 3, 5-OH-Hyd (m/z 317); 4, 5-OH-Ura (m/z 329); 5, 5-OHMeUra (m/z 358); 6, 5-OH-Cyt (m/z 328); 7, 7a, cis and transThy glycol (m/z 259); 8, 5,6-dihydroxyuracil (m/z 417); 9, FapyAde (m/z 354); 10, 8-OH-Ade (m/z 352); 11, Xan (m/z 353); 12, 2-OH-Ade (m/z 352); 13, FapyGua (m/z 442); 14, 8-OH-Gua (m/z 440).
Selected ion chromatograms obtained during the GC/MS analysis of PBMC DNA.
(A) Represents a mixture of trimethylsilylated standards. (B) The chromatogram demonstrates the higher abundance ions and the chromatogram in panel C shows the lower abundance ions of a trimethylsilylated hydrolysate of DNA isolated from the PBMCs of a representative patient entered into this trial before initiation of the doxorubicin infusion. Experimental details are given in “Patients, materials, and methods.” Peak indicates DNA base (ion monitored): 1, 6-azathymine (m/z 256) (internal standard); 2, 5-OH-5-MeHyd (m/z 331); 3, 5-OH-Hyd (m/z 317); 4, 5-OH-Ura (m/z 329); 5, 5-OHMeUra (m/z 358); 6, 5-OH-Cyt (m/z 328); 7, 7a, cis and transThy glycol (m/z 259); 8, 5,6-dihydroxyuracil (m/z 417); 9, FapyAde (m/z 354); 10, 8-OH-Ade (m/z 352); 11, Xan (m/z 353); 12, 2-OH-Ade (m/z 352); 13, FapyGua (m/z 442); 14, 8-OH-Gua (m/z 440).
Quantitative measurement of modified bases in DNA from PBMCs of doxorubicin-treated patients
The content (mean ± SD) of 13 modified bases found in PBMC DNA prior to anthracycline chemotherapy is shown in Table 1. The relative amounts of the individual modified bases varied by over 20-fold prior to doxorubicin treatment. Following initiation of the doxorubicin infusion, an increase in oxidized base content was detected in PBMC DNA in both a time-dependent and base-specific fashion. As shown in Table 2, significant increases in modified DNA base content were observed primarily at the completion of the 96-hour anthracycline infusion; however, a significant rise in Thy Gly, 5-OH-Hyd, and 5-OH-Ura levels was also found 72 hours after drug treatment was begun. The average fold increase over baseline values was highest for Thy Gly followed by 5-OH-Hyd, FapyAde, 5-OHMe-Ura, 5,6-diOH-Ura, and 5-OH-5-MeHyd. Smaller, but significant increases were also observed for 5-OH-Ura and FapyGua (Table 2). No change in the levels of 3 modified bases, 5-OH-Cyt, 8-OH-Ade, and 8-OH-Gua, was observed during the 96 hours of doxorubicin treatment. Although oxidized DNA base levels varied between patients (as can be appreciated by review of the SDs for pretreatment values seen in Table1), the baseline for individual patients was quite stable. As shown in Table 2, for the most part no significant alterations in oxidized DNA base levels were observed for the first 48 hours after the doxorubicin infusion had begun.
Oxidized DNA base content in peripheral blood mononuclear cells of patients receiving a 96-hour infusion of 165 mg/m2 doxorubicin
| DNA base . | 0 h . | 48 h . | 72 h . | 96 h . |
|---|---|---|---|---|
| Thy Gly | 1 | 1.5 ± 1.2 | 1.9 ± 1.1* | 4.2 ± 4.6* |
| 5-OH-Hyd | 1 | 1.3 ± 0.7 | 1.9 ± 1.1* | 2.5 ± 1.3* |
| FapyAde | 1 | 1.9 ± 2.2 | 1.6 ± 1.5 | 2.4 ± 1.9* |
| 5-OHMe-Ura | 1 | 0.9 ± 0.4 | 1.4 ± 1.1 | 2.4 ± 2.1† |
| 5,6diOH-Ura | 1 | 1.2 ± 0.7 | 1.4 ± 1.0 | 2.0 ± 1.8† |
| 5-OH-5-MeHyd | 1 | 1.4 ± 1.3 | 2.2 ± 2.6 | 1.8 ± 1.0* |
| 5-OH-Ura | 1 | 1.7 ± 1.7 | 1.5 ± 0.8† | 1.8 ± 1.3* |
| FapyGua | 1 | 1.9 ± 1.5 | 1.6 ± 1.2 | 1.6 ± 0.8* |
| Xan | 1 | 1.4 ± 0.9 | 1.4 ± 0.9 | 1.4 ± 0.7 |
| 2-OH-Ade | 1 | 1.6 ± 0.8† | 2.1 ± 2.6 | 1.4 ± 1.0 |
| 5-OH-Cyt | 1 | 1.0 ± 0.5 | 0.9 ± 0.4 | 1.1 ± 0.9 |
| 8-OH-Ade | 1 | 1.0 ± 0.7 | 1.0 ± 0.7 | 1.0 ± 0.8 |
| 8-OH-Gua | 1 | 1.1 ± 0.9 | 1.0 ± 0.9 | 0.7 ± 0.5 |
| DNA base . | 0 h . | 48 h . | 72 h . | 96 h . |
|---|---|---|---|---|
| Thy Gly | 1 | 1.5 ± 1.2 | 1.9 ± 1.1* | 4.2 ± 4.6* |
| 5-OH-Hyd | 1 | 1.3 ± 0.7 | 1.9 ± 1.1* | 2.5 ± 1.3* |
| FapyAde | 1 | 1.9 ± 2.2 | 1.6 ± 1.5 | 2.4 ± 1.9* |
| 5-OHMe-Ura | 1 | 0.9 ± 0.4 | 1.4 ± 1.1 | 2.4 ± 2.1† |
| 5,6diOH-Ura | 1 | 1.2 ± 0.7 | 1.4 ± 1.0 | 2.0 ± 1.8† |
| 5-OH-5-MeHyd | 1 | 1.4 ± 1.3 | 2.2 ± 2.6 | 1.8 ± 1.0* |
| 5-OH-Ura | 1 | 1.7 ± 1.7 | 1.5 ± 0.8† | 1.8 ± 1.3* |
| FapyGua | 1 | 1.9 ± 1.5 | 1.6 ± 1.2 | 1.6 ± 0.8* |
| Xan | 1 | 1.4 ± 0.9 | 1.4 ± 0.9 | 1.4 ± 0.7 |
| 2-OH-Ade | 1 | 1.6 ± 0.8† | 2.1 ± 2.6 | 1.4 ± 1.0 |
| 5-OH-Cyt | 1 | 1.0 ± 0.5 | 0.9 ± 0.4 | 1.1 ± 0.9 |
| 8-OH-Ade | 1 | 1.0 ± 0.7 | 1.0 ± 0.7 | 1.0 ± 0.8 |
| 8-OH-Gua | 1 | 1.1 ± 0.9 | 1.0 ± 0.9 | 0.7 ± 0.5 |
N = 15. Data represent the fold increase (± SD) in oxidized DNA base levels normalized to pretreatment values for the entire patient population 48, 72, and 96 h after the initiation of infusional doxorubicin.
PBMC indicates peripheral blood mononuclear cell.
P < 0.02 versus PBMC DNA from patients before doxorubicin infusion began.
P < 0.05 versus PBMC DNA from patients before doxorubicin infusion began.
Doxorubicin levels
The concentration of doxorubicin in plasma after initiation of a 96-hour continuous intravenous infusion rises quickly to a steady-state value of 60 ng/mL (0.1 μM) that is maintained for the duration of treatment.
Discussion
The oxidative metabolism of doxorubicin, which generates reactive oxygen species during cycles of reduction and oxidation of the anthracycline quinone, has generally been accepted to play an important role in the cardiac toxicity of this drug.34 However, the importance of drug-induced free radical formation for doxorubicin toxicity in other tissues has been questioned. To determine whether by-products of the reductive activation of doxorubicin could be observed under clinical conditions in patients receiving anthracycline therapy, and to provide potential insights into the mechanism by which doxorubicin increases the rate of second malignancies of the hematopoietic system, we evaluated PBMC DNA from 15 women receiving doxorubicin as a 96-hour intravenous infusion for the presence of oxidized base lesions known to be promutagenic.
Methodologic issues related to oxidized DNA base quantitation
The methodology used in this study for the calculation of doxorubicin-induced oxidative DNA base damage has been described previously.35,36 The calculated number of nanomoles of each modified base per milligram DNA is a function of (1) the RMRF for this base, (2) how the amount of DNA in the sample was estimated, and (3) whether a constant fraction of DNA-bound modified bases was released during hydrolysis. However, RMRFs can be determined in several different ways, including the method described in this study, with values that may vary substantially.37 Furthermore, the ion peak response of most modified DNA bases can change with the tuning of the GC/MS. This means that RMRFs may differ significantly if they are calculated from multiple injections of mixtures of known amounts of modified DNA bases and an internal standard performed at different times before and after the GC/MS has been tuned. However, this last possibility produced only a small degree of error in the studies presented here. We examined the same set of standards with different instrument tuning during a period of 5 months and found the calculated RMRFs to vary by less than 10%. Finally, for the purposes of this study, the DNA content in each tested sample was estimated by OD260, and a constant fraction of modified bases was assumed to have been released from the DNA backbone during hydrolysis in all samples processed and hydrolyzed in identical fashion.36
Oxidative base modifications in PBMC DNA during infusional doxorubicin therapy
The objective of this work was to determine whether doxorubicin produced oxidative damage to PBMC DNA of patients undergoing a 96-hour doxorubicin infusion. In our study, doxorubicin was delivered as a prolonged intravenous infusion to diminish its cardiotoxic potential,38 while retaining therapeutic activity, in the setting of high-dose chemotherapy for women with responsive metastatic and high-risk primary breast cancer.30 Under these conditions, the steady-state doxorubicin plasma level was 0.1 μM when the drug was infused for 96 hours at a total dose of 165 mg/m2; this level can be compared to the typical peak doxorubicin concentration of 0.6 to 0.9 μM, which rapidly decays below 0.05 μM after bolus administration of a 60 mg/m2dose.39 In the major clinical studies of doxorubicin- and epirubicin-related leukemias, the anthracycline antibiotics were delivered as an intravenous bolus at doses that ranged from 60 to 70 mg/m2 per administration.5
We monitored the content of 14 modified bases in DNA isolated from PBMCs of 15 patients before and during high-dose infusional doxorubicin treatment and found 13 modified bases present in all DNA samples examined. Infusional doxorubicin significantly increased DNA base oxidation (up to 4-fold) over baseline levels for 9 of the 13 species monitored beginning, for the most part, 72 hours after anthracycline treatment was initiated. Maximal base oxidation was observed at the end of the 96-hour infusion. Unfortunately, because of the design of our clinical trial, repair of doxorubicin-induced DNA base oxidation could not usefully be examined. This was due to the possibility that alkylation-related DNA damage from high-dose cyclophosphamide (100 mg/kg) administered immediately following completion of the doxorubicin infusion could have substantially altered interpretation of DNA base oxidation results subsequent to the 96-hour time point.
Two previous studies have examined the production of oxidized DNA base lesions in patients treated with an anthracycline.40,41 In the work of Faure and colleagues, levels of 5-(hydroxymethyl)uracil (HMUra) and 8-oxo-deoxyguanine (8-oxo-dGuo) were determined in 24-hour urine collections following doxorubicin-containing combination chemotherapy in patients treated for a wide variety of malignancies. A significant, 25% increase in urinary HMUra was found in the 24 hours following chemotherapy; no change in 8-oxo-dGuo concentration was observed. Olinski and associates examined oxidative DNA base damage by GC/MS in lymphocytes of cancer patients receiving single-agent chemotherapy with the doxorubicin analogue epirubicin.41Treatment was administered as a short intravenous bolus of epirubicin at a dose of 70 mg/m2; DNA base damage was evaluated 1 and 24 hours after therapy. In concert with the results presented in the current report, the greatest increase (2- to 2.5-fold) for any single oxidized base after treatment with epirubicin was shown for Thy Gly. Significant increases in 5-OH-Ura and 2-OH-Ade, as well as significant interpatient variability in overall DNA base oxidation, were also found in both series. However, comparison of Table 2 in the present study with the data of Olinski and associates also demonstrates substantive differences. Other than for Thy Gly, DNA base oxidation observed following bolus epirubicin was substantially smaller (< 1.5-fold) compared to that following infusional doxorubicin. Furthermore, we observed increases in other modified bases, namely, 5-OH-Hyd, FapyAde, 5-OHMe-Ura, 5,6diOH-Ura, 5-OH-5-MeHyd, FapyGua, and Xan during the 96-hour infusion of doxorubicin not reported in the other study. On the other hand, we did not observe changes in 5-OH-Cyt, 8-OH-Ade, or 8-OH-Gua levels following infusional doxorubicin. However, it must be recognized that because of substantial variations in the pretreatment levels of these 3 bases, as well as the potential for artifactual alterations in 8-OH-Gua levels due to our hydrolysis conditions,42 a definitive conclusion regarding the effect of doxorubicin on the production of these 3 oxidized bases in PBMC may not be possible in the context of our clinical trial.36
The explanation for the greater degree and wider array of oxidized DNA base damage following infusional doxorubicin remains to be determined but could be related to the substantially greater systemic exposure provided by the 96-hour dosing schedule,43 leading to the consumption of intracellular antioxidants, as well as the possibility that longer-term drug treatment may transiently overwhelm the DNA repair capacity of the hematopoietic compartment. In fact, recent in vitro studies from our laboratory suggest that DNA repair enzyme messenger RNAs may be down-regulated following continuous doxorubicin exposure at precisely the time when increased DNA base oxidation begins to be quantifiable (data not shown). In any case, the importance of these observations is related to the increased frequency of infusional doxorubicin usage in the clinic, especially during the treatment of both hematologic malignancies and solid tumors in the pediatric age group, as part of a strategy to decrease the potentially devastating long-term side effects of doxorubicin cardiac toxicity in the young.44
Mechanisms of doxorubicin-induced PBMC DNA base oxidation
The spectrum of oxidized DNA base damage observed in this study is similar to that produced by ionizing radiation (and thus, presumably, is due to a species with the chemical reactivity of ·OH).33,45 It is also similar to our previous study in which doxorubicin-related ·OH formation oxidized DNA bases in vitro in human chromatin.26 Only Xan has not routinely been observed in DNA after exposure to systems generating ·OH; however, Xan has been detected in DNA extracted from human tumors at levels greater than that in surrounding normal tissues.46 It has been suggested that Xan could be formed by attack of ·OH at the C-2 of guanine, or by deamination of guanine by other species.475-OH-6-H-Thy was not detected in any of our DNA samples. This compound is formed from thymine where, in the first step, ·OH reacts with the thymine molecule to form the 5-hydroxy-6-yl radical. Further oxidation forms Thy Gly, and reduction forms 5-OH-6H-Thy.27 Apparently, in PBMC DNA exposed to doxorubicin, the formation of Thy Gly is favored over 5-OH-6-H-Thy.
Potential role of DNA base oxidation in the mechanism of action and mutagenicity of doxorubicin
In addition to supporting the possibility that the oxidative metabolism of doxorubicin occurs under clinically relevant conditions, this study provides insight into potential mechanisms of doxorubicin toxicity and mutagenicity for hematopoietic cells. Reactive oxygen species are known to produce a wide range of DNA lesions including apurinic sites, strand breaks, and DNA-protein cross-links, in addition to base oxidation. However, the promutagenic effects of several oxidized bases have been very well characterized.48 Thy Gly, FapyGua, 5-OH-Ura, and 5-OHMe-Ura, which were increased 1.6- to 4.2-fold after doxorubicin treatment in this study, have all been shown to be potentially mutagenic under specific experimental conditions.28,49 In a previous study,50 we demonstrated that 1.5- to 4-fold increases in the levels of FapyGua, Thy Gly, FapyAde, and 5-OH-Hyd observed following exposure of a reporter plasmid to an ·OH-generating system in vitro (similar to the changes found in the current trial) were associated with a significantly increased mutation frequency when the oxidized plasmid was transfected into monkey kidney CV-1 cells. The predominant lesions observed in that report occurred at C-G base pairs. It is also known that FapyGua may lead to GC→CG transversions,51 2-OH-Ade pairs with adenine and guanine,52 5-OH-Ura leads to GC→AT transitions and GC→CG transversions,53 and 5-OH-5-Me-Ura is known to be mutagenic.54,55 Furthermore, ring-fragmented DNA bases (such as FapyGua), as well as Thy Gly, have also been shown to block DNA replication or increase reading error frequency by DNA polymerase, leading either to potentially mutagenic or cytotoxic DNA lesions.48,56 57 Similar inefficiencies in DNA polymerase function may be produced by conformational changes in DNA that occur as a consequence of DNA base oxidation. Thus, if doxorubicin-related DNA base oxidation occurs clinically either in primitive hematopoietic precursors or in tumor cells (in addition to PBMC), there is substantial experimental precedent for the contribution of these lesions to the toxic effects of the anthracycline.
In conclusion, we observed a significant increase in 9 oxidatively modified DNA bases in the PBMC of cancer patients undergoing a 96-hour doxorubicin infusion. The largest increases found were for Thy Gly, 5-OH-Hyd, FapyAde, 5-OHMe-Ura, as well as FapyGua. These results strongly suggest that doxorubicin-enhanced reactive oxygen metabolism occurs in the clinic and produces potentially mutagenic DNA base lesions that can be detected in PBMCs. However, further studies will be required to define the precise mechanisms of oxidatively modified DNA base mutagenesis or toxicity, and the kinetics of doxorubicin-induced oxidized DNA base formation and repair, in both normal and malignant hematopoietic precursors.
We wish to thank Lynn Baltazar for her expert secretarial assistance in the preparation of this manuscript.
Abbreviations: thymine glycol, Thy Gly; 8-hydroxyguanine, 8-OH-Gua; 6-azathymine, azaT; 2-hydroxy-6-aminopurine, 2-OH-Ade; N, O-bis(trimethylsilyl) trifluoroacetamide, BSTFA; 5-hydroxy-5-methylhydantoin, 5-OH-5-MeHyd; 5-hydroxyuracil, 5-OH-Ura; 5-hydroxy-6-hydrothymine, 5-OH-6H-Thy; 5-hydroxycytosine, 5-OH-Cyt; 5-(hydroxymethyl)uracil, 5-OH-MeUra; 5,6-dihydroxyuracil, 5,6diOH-Ura; 4,6-diamino-5-formamidopyrimidine, FapyAde; 8-hydroxyadenine, 8-OH-Ade; Xanthine, Xan; 2,6-diamino-4-hydroxy-5-formamidopyrimidine, FapyGua; 5-hydroxyhydantoin, 5-OH-Hyd.
Supported by grants CA 33572, CA 63265, and CA 62505 from the National Cancer Institute.
These results were reported, in part, in the Proceedings of the American Association for Cancer Research. 1998;39:489.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
James H. Doroshow, Department of Medical Oncology, City of Hope National Medical Center, 1500 E Duarte Rd, Duarte, CA 91010; e-mail: jdoroshow@coh.org.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal