Hypoxia-inducible factor (HIF) mediates a large number of transcriptional responses to hypoxia and has an important role in processes that include angiogenesis and erythropoiesis. The HIF DNA binding complex consists of 2 basic-helix-loop-helix PAS proteins designated α and β subunits. Regulation occurs principally through the α subunits, which are stabilized and activated in hypoxia. Although substantial evidence implicates reactive oxygen species (ROS) in the regulatory process, the precise mechanisms remain unclear. Mitochondria are an important source of ROS, and in one model it has been proposed that hypoxia increases the generation of ROS at complex III in the mitochondrion and that this signal acts through a transduction pathway to stabilize HIF-1α and to activate HIF. To test this model the induction of the HIF-1α subunit and the HIF target gene, glucose-transporter-1, was examined in a variety of mutant cells that lacked mitochondrial DNA (ρ0) or had other genetic defects in mitochondrial respiration. HIF induction by hypoxia was essentially normal in all cells tested. Hydrogen peroxide production was measured by the luminol/peroxidase method and found to be reduced in ρ0 versus wild-type cells and reduced by hypoxia in both ρ0 and wild-type cells. Furthermore, concentrations of rotenone that maximally inhibited respiration did not affect HIF activation by hypoxia. These data do not support the model outlined above and indicate that a functional respiratory chain is not necessary for the regulation of HIF by oxygen.
Introduction
Considerable progress has been made in understanding mechanisms that underlie the regulation of mammalian gene expression by oxygen. A heterodimeric DNA binding complex, hypoxia-inducible factor (HIF), consisting of 2 basic-helix-loop-helix PAS proteins, termed HIF-α and HIF-β, plays a critical role in a widespread transcriptional response induced by hypoxia.1-3 First discovered in the context of erythropoietin regulation,4 it is now recognized that HIF is central to the regulation of genes involved in a diverse range of functions in mammalian physiology, including angiogenesis, glucose and energy metabolism, iron metabolism, vasomotor regulation, and regulation of apoptosis and cell proliferation (for review, see5,6). The regulation of HIF itself occurs mainly through modifications of its α subunit, whereas the β subunit is a constitutive nuclear protein. The HIF-α subunit is rapidly degraded in normoxia by a ubiquitin-proteasome pathway, involving the von Hippel–Lindau tumor suppressor protein.7-11 In hypoxia this pathway is inhibited and HIF-α is stabilized. Nuclear translocation, coactivator recruitment, dimerization, and DNA binding follow in a multistep process that results in transactivation of target genes12,13 (for review, see14). A substantial body of evidence has implicated partially reduced reactive oxygen species (ROS) in the sensing and transduction mechanism,7 15-19 but knowledge of the source, precise species, and mode of interaction of such molecules with the transcriptional system are limited, and the experimental findings are conflicting.
The mitochondrion is the major oxygen-consuming organelle and, at least in some circumstances, is a major producer of oxygen radical species.20,21 As such it might be expected to play a central role in oxygen-sensitive processes, and evidence has suggested such a role in HIF regulation.18,19 Chandel et al18,19 found impairment of HIF activation by hypoxia in cells treated with pharmacologic inhibitors of complex I of the electron chain and in cells lacking mitochondrial DNA. They proposed that complex III of the mitochondrial respiratory chain acts as an oxygen sensor and is a source of increased ROS in hypoxia that have a stabilizing effect on HIF-1α. In this model, complex I inhibitors were proposed to act by blocking the electron flow proximally, so as to ablate such a signal. Mitochondrial function has also been implicated in other oxygen-sensing systems. For instance, Baysal et al22 recently demonstrated that germline mutations in the gene that encodes the small subunit of cytochrome b (cybS) in complex II predisposes to hereditary paraganglioma of the carotid body. Chronic hypoxia predisposes to similar tumors, leading those researchers to suggest that the genetic defect might mimic the normal hypoxic signal and that cybS is a critical component of the oxygen-sensing system of paraganglionic tissue. However, no specific model was proposed, and despite implicating the mitochondrion, the findings are not easy to reconcile with the model proposed by Chandel et al.19
Against a role for mitochondria in oxygen sensing are findings that cyanide has only modest or absent effects on hypoxia-inducible gene expression or HIF in a variety of systems.23-25 Further conflicting data comes from the observations on the effects of hydrogen peroxide. Although Chandel et al19 have noted that application of hydrogen peroxide to cells may activate HIF and HIF target genes, supporting their model that increased mitochondrial ROS in hypoxia might activate HIF, several other studies have described the opposite effect.7,15 16 Thus, hydrogen peroxide has been found to destabilize HIF even in hypoxia and to prevent the induction of HIF target genes by hypoxia. This has led to the converse proposal that ROS are produced in normoxia and might trigger HIF-1 degradation in oxygenated cells.
We have taken primarily a genetic approach to address the question as to what role a functional mitochondrial respiratory chain may play in the regulation of HIF. We have examined several series of mutant cell lines with specific genetic defects in the electron transport chain and also 2 established cell lines that lack mitochondrial DNA (mtDNA). Regulation of HIF-1α and HIF target genes was preserved in all these cell lines. Furthermore, pharmacologic inhibition of the respiratory chain by the complex I inhibitor rotenone did not impair HIF regulation at doses that fully inhibit electron transport. These data indicate that a functional respiratory chain is not required for the regulation of HIF by oxygen.
Materials and methods
Cell lines and culture conditions
The human osteosarcoma-derived cell line 143B(TK−) and its mtDNA-less derivative, 206ρ0, and the human lung carcinoma–derived cell line A549 and its mtDNA-less derivative, B2ρ0, were as described by the originators.26,27 The B2ρ0 cybrid cell line, B2.3243 0%, repopulated with normal human mitochondria and restored to respiratory competence, is described elsewhere.28 The respiration-deficient Chinese hamster fibroblast cell lines were provided by the originators and are described elsewhere.29-33 Their characteristics and derivation are summarized in Table 1.
Source and characteristics of respiration-deficient Chinese hamster fibroblast cell lines
| Cell line . | Derived from . | Defects . | Reference . |
|---|---|---|---|
| CCLI6-B2 | Chinese hamster CCLI6 (DON) | Mutation in X-linked NDUFA1* gene complex I activity 1%-2% of WT† | 29 |
| CCLI6-B9 | Chinese hamster CCLI6 (DON) | Mutation in small integral membrane anchor protein CII-3 of complex II | 30 |
| Complex II activity less than 5% of WT | |||
| Gal32 | Chinese hamster V79 | Mutation in nuclear gene | 31 |
| Complex IV activity 1%-2% of WT | |||
| Complex I activity 1%-2% of WT | |||
| Defective mitochondrial protein synthesis | |||
| V79-G4 | Chinese hamster V79 | Mutation in nuclear gene on X chromosome | 32 |
| Complex I activity 1%-2% of WT | |||
| Different complementation group to CCLI6-B9 | |||
| V79-G7 | Chinese hamster V79 | Mutation in nuclear gene | 33 |
| Complex IV activity 6.6% of WT | |||
| Complex V activity 11.6% of WT | |||
| Defective mitochondrial protein synthesis |
| Cell line . | Derived from . | Defects . | Reference . |
|---|---|---|---|
| CCLI6-B2 | Chinese hamster CCLI6 (DON) | Mutation in X-linked NDUFA1* gene complex I activity 1%-2% of WT† | 29 |
| CCLI6-B9 | Chinese hamster CCLI6 (DON) | Mutation in small integral membrane anchor protein CII-3 of complex II | 30 |
| Complex II activity less than 5% of WT | |||
| Gal32 | Chinese hamster V79 | Mutation in nuclear gene | 31 |
| Complex IV activity 1%-2% of WT | |||
| Complex I activity 1%-2% of WT | |||
| Defective mitochondrial protein synthesis | |||
| V79-G4 | Chinese hamster V79 | Mutation in nuclear gene on X chromosome | 32 |
| Complex I activity 1%-2% of WT | |||
| Different complementation group to CCLI6-B9 | |||
| V79-G7 | Chinese hamster V79 | Mutation in nuclear gene | 33 |
| Complex IV activity 6.6% of WT | |||
| Complex V activity 11.6% of WT | |||
| Defective mitochondrial protein synthesis |
Human NDUFA1 on the X chromosome encodes the MWFE polypeptide of mammalian complex I.
Wild type.
Chinese hamster fibroblast lines (both wild-type and respiration-deficient mutant lines), 143B, A549, and cybrid cell lines were grown in Dulbecco modified Eagle medium (DMEM; Sigma, Poole, United Kingdom). This medium contains 4.5 g/L glucose and 110 μg/mL sodium pyruvate and was supplemented with 10% fetal calf serum (Globepharm, Esher, United Kingdom), L-glutamine (8 mM), penicillin (20 IU/mL), streptomycin sulfate (50 μg/mL), and nonessential amino acids (×100; Sigma). The ρ0 cell lines were grown in DMEM, supplemented as above with the further addition of uridine (50 μg/mL). Cells were plated as appropriate 24 hours before experiments such that they were approaching confluence at the time of the experimental procedure. Culture medium was replaced at the start of each experiment. Hypoxic exposure was in a NAPCO 7001 incubator (Precision Scientific, Chicago, IL) with 0.1% oxygen or 1% oxygen, 0 5% carbon dioxide, balance nitrogen. Exposure to experimental conditions was for 4 hours unless otherwise stated.
The human hepatoma Hep3B cell line was from American Tissue Culture Center (Manassas, VA) and was grown in αMEM (Sigma) supplemented with 10% fetal calf serum, penicillin (100 IU/mL), L-glutamine (8 mM), and streptomycin sulfate (100 μg/mL). Hep3Bρ0 cells were depleted of mitochondrial DNA by incubating wild-type cells in ethidium bromide (50 ng/mL; Sigma) in medium supplemented with sodium pyruvate (110 μg/mL) and uridine (50 μg/mL) for periods of 3, 4, 6, and 8 weeks. Some ρ0 cells were additionally selected in medium containing rotenone (1 μg/mL).
Polymerase chain reaction determination of mitochondrial DNA and test for uridine auxotrophy
The ρ0 status was confirmed by undetectable mitochondrial DNA as assessed by failure of polymerase chain reaction (PCR) amplification of a 546–base pair (bp) fragment (residues 15 875 to 16 421) using primers designed from published Cambridge reference sequence, 5′-CAGGAAACAGCTATGACCTTGATTTCACGGAGGATGGTG-3′ and 5′-TGTAAA- ACGACGGCCAGTCTCAAATGGGCCTGTCCTTG-3′, data not shown), absent cyanide inhibitable respiration (Table2), and uridine auxotrophy, determined at regular intervals by failure of growth of ρ0 cells in the absence of uridine in the growth medium.
Measurement of oxygen consumption
| Cell line . | Oxygen consumption [fmol/(min × cell)] . | |
|---|---|---|
| Without KCN . | With KCN (2 mM) . | |
| 143B | 7.2 ± 2.7 | 2.7 ± 1.1 |
| 206ρ0 | 0.9 ± 0.3 | 0.8 ± 0.2 |
| A549 | 3.8 ± 0.5 | 0.4 ± 0.02 |
| B2ρ0 | 0.5 ± 0.1 | 0.5 ± 0.05 |
| B2ρ0 cybrid | 3.5 ± 0.9 | 0.6 ± 0.06 |
| Cell line . | Oxygen consumption [fmol/(min × cell)] . | |
|---|---|---|
| Without KCN . | With KCN (2 mM) . | |
| 143B | 7.2 ± 2.7 | 2.7 ± 1.1 |
| 206ρ0 | 0.9 ± 0.3 | 0.8 ± 0.2 |
| A549 | 3.8 ± 0.5 | 0.4 ± 0.02 |
| B2ρ0 | 0.5 ± 0.1 | 0.5 ± 0.05 |
| B2ρ0 cybrid | 3.5 ± 0.9 | 0.6 ± 0.06 |
Polarographic measurements of oxygen consumption in 206ρ0, B2ρ0, and B2ρ0 cybrid cell lines with and without exposure to cyanide (KCN) (2 mM). All measurements were done in triplicate, and data are shown as mean ± 1 SD.
Assay of HIF-1α levels and HIF target gene activation
Whole cell extracts were made as described previously25 and were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to Immobilon-P membranes. HIF-1α was detected using a monoclonal immunoglobulin G2b (clone H1α67) antibody (Novus Biologicals, Littleton, CO) at 1:1000 dilution. Detection was with a peroxidase-conjugated goat anti–mouse immunoglobulin (DAKO, Ely, United Kingdom) at 1:2000 dilution and enhanced chemiluminescence (ECL Plus; Amersham). After analysis, membranes were stained with Coomassie blue to verify equal protein loading and transfer. RNA was extracted by a modified acid/guanidinium thiocyanate/phenol/chloroform method (RNAzol B; Cinna/Biotec Laboratories, Houston, TX), dissolved in hybridization buffer (80% formamide; 40 mM PIPES, pH 6.5; 400 mM sodium chloride; and 1 mM EDTA, pH8), and analyzed by ribonuclease protection assays as described elsewhere.2 Quantification of the protected species was performed using a PhosphorImager (Molecular Dynamics, Sunnyvale, CA) and was related to an internal control assay for a constitutively expressed U6 small nuclear RNA. Fully homologous riboprobe templates for the Chinese hamster and human glucose transporter-1 (Glut-1) genes were generated by the PCR using oligonucleotides based on published sequences. 32P-labeled riboprobes were generated using SP6 or T7 RNA polymerase. The riboprobes used yielded protected fragments as follows: 227 bp for hamster Glut-1 (nucleotides 1310-1537; Accession No. LO7300), 136 bp for human Glut-1 (nucleotides 1063-1198; No. KO3195), 106 bp for U6 (nucleotides 1 to 107; No. X01366).
Oxygen consumption assay
Cells were cultured in 75-cm2 flasks. Following trypsinization, cells were centrifuged for 5 minutes and suspended in cell culture medium lacking fetal calf serum. Approximately 2.5 × 106 cells were suspended in 1 mL medium and then transferred to a water-jacketed reaction chamber (Hansatech Instruments, King's Lynn, United Kingdom). Temperature was maintained at 37°C. The cell suspension was stirred magnetically. The oxygen concentration was measured polarographically and recorded once per second. After the experiment, cells were counted (Coulter Counter; Coulter Electronics, Fullerton, CA), and the respiration rate was calculated from the decrease of the oxygen concentration in the chamber. All experiments were done in 3 or 4 independent cultures. Cyanide (2 mM) was added directly to cells in the chamber. Rotenone (Sigma) was added to cells just prior to trypsinization and then maintained at similar concentrations in all solutions used during the assays.
Measurement of hydrogen peroxide
To assess cellular production of hydrogen peroxide, concentrations were measured in the extracellular fluid using horseradish peroxidase (HRP)-enhanced luminol chemiluminescence.15 16 Cells were plated such that they approached confluence at the time of experimental procedure. Cell culture media was replaced by 250 μL Tyrode Salt solution (Sigma), and cells were incubated in a 20% oxygen atmosphere or at 1% oxygen in an Invivo2 400 Hypoxia Workstation (Ruskin Technology, Leeds, United Kingdom). Samples were taken after 2 hours. A volume of 50 μL was then mixed with 10 μL HRP solution (Sigma) in a disposable cuvette and placed in a luminometer (TD20/20; Turner Designs, Sunnyvale, CA). Luminol (Sigma) was injected automatically. Final concentrations were 1 μg/mL HRP and 250 μM luminol. Chemiluminescence was integrated for 5 seconds. A calibration curve was obtained by measuring the luminescence of external standards of hydrogen peroxide diluted in Tyrode Salts solution. A concentration of 0.5 μM was clearly distinguishable from baseline. Hydrogen peroxide concentrations in the experimental samples were calculated by reference to the calibration curve derived from external standards using an exponential growth regression (Sigma Plot; SPSS Science, Birmingham, United Kingdom).
Results
Action of rotenone on the regulation of HIF-1α by hypoxia
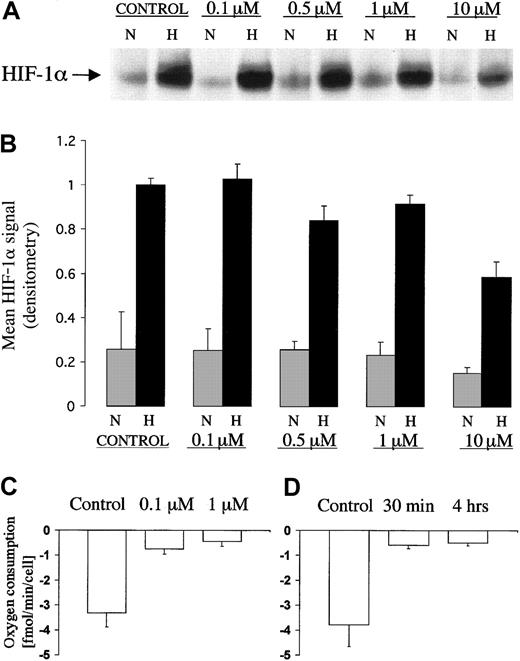
Pharmacologic inhibition of the respiratory chain has been used previously to test the involvement of mitochondrial electron transport in the regulation of HIF. Of particular interest are the reported effects of complex I inhibitor rotenone that at doses of 1 to 3 μg/mL (approximately 2.5 to 7.5 μM) was found to inhibit the induction of HIF by hypoxia.18 19 To investigate this further we compared the dose-response characteristic for the effect of rotenone on HIF induction by hypoxia, with that for the effect of rotenone on mitochondrial oxygen consumption in the Chinese hamster ovary cell line, CHO-K1 (Figure 1A). The results of 3 separate dose-response experiments that examine effects on HIF-1α protein induction by hypoxia are shown in Figure 1B. Only at the highest doses of rotenone (10 μM) was a consistent suppressive effect on the induction of HIF-1α protein levels observed. The effects of rotenone on oxygen consumption in the same cell line are shown in Figure 1C. Oxygen consumption was sensitive to much lower doses of rotenone and was maximally inhibited at a dose of 0.1 μM. To ensure that the duration of inhibition of the mitochondrial electron chain was sufficient to ensure complete inhibition during the entire 4 hour exposure that we had used to induce HIF-1α, we checked the time course of effects on oxygen consumption under the conditions of these experiments. The onset of inhibition of oxygen consumption by 0.1 μM rotenone in the CHO cell line was rapid (within 30 minutes) and was sustained throughout a period of at least 4 hours (Figure 1D). Thus, we found that, although high doses of rotenone reduced induction of HIF-1α by hypoxia, doses of rotenone that are sufficient for maximal inhibition of respiration in CHO-K1 cells had no effect on HIF-1α regulation.
HIF-1α protein expression and measurement of oxygen consumption in CHOK1 cell line exposed to different doses of rotenone.
(A) Representative immunoblot of HIF-1α showing dose response characteristic for the effect of rotenone on induction by hypoxia. Levels of HIF-1α in extracts of CHO-K1 cells after 4 hour exposure to normoxia (21% O2, N) or hypoxia (0.1% O2, H) at different doses of rotenone (0 to 10 μM). (B) Data are HIF-1α signals (mean ± 1 SD; n = 3) normalized to the HIF-1α signal from control cells in hypoxia. (C,D) Polarographic measurements of oxygen consumption in CHO-K1 cells exposed to rotenone. (C) Dose response for rotenone (0, 0.1 μM, 1 μM). Cells treated for 4 hours in normoxia (21% O2). (D) Time course. Cells treated with 0.1 μM rotenone for 30 minutes or 4 hours in normoxia (21% O2). Measurements were performed in triplicate. Data are mean ± 1 SD.
HIF-1α protein expression and measurement of oxygen consumption in CHOK1 cell line exposed to different doses of rotenone.
(A) Representative immunoblot of HIF-1α showing dose response characteristic for the effect of rotenone on induction by hypoxia. Levels of HIF-1α in extracts of CHO-K1 cells after 4 hour exposure to normoxia (21% O2, N) or hypoxia (0.1% O2, H) at different doses of rotenone (0 to 10 μM). (B) Data are HIF-1α signals (mean ± 1 SD; n = 3) normalized to the HIF-1α signal from control cells in hypoxia. (C,D) Polarographic measurements of oxygen consumption in CHO-K1 cells exposed to rotenone. (C) Dose response for rotenone (0, 0.1 μM, 1 μM). Cells treated for 4 hours in normoxia (21% O2). (D) Time course. Cells treated with 0.1 μM rotenone for 30 minutes or 4 hours in normoxia (21% O2). Measurements were performed in triplicate. Data are mean ± 1 SD.
Regulation of HIF-1α in stable cell lines with mitochondrial defects
To consider the possibility that other perturbations of mitochondrial metabolism might affect regulation of the HIF system more significantly, we next analyzed several different series of mutant cell lines with a range of genetic defects all of which have major effects on the operation of the mitochondrial electron chain.
First we tested several different series of previously described respiration-deficient Chinese hamster fibroblasts. Table 1 summarizes the origin and characteristics of these cells. In each case the phenotype is known to be stable in culture, and the presence of a respiration defect was confirmed by testing for metabolic sensitivity to removal of glucose from the medium or replacement of glucose by galactose as detailed in previous descriptions of these cells.31 34 Induction of HIF-1α protein during exposure to hypoxia (0.1% or 1% oxygen) was tested by immunoblotting. The results of typical experiments are illustrated in Figure2 (A,C). The respiration-deficient mutants CCL16-B2 and CCL16-B9 demonstrated HIF-1α induction that was indistinguishable from the wild-type line, CCL16, whereas the mutants Gal32, V79-G4, and V79-G7 were indistinguishable from the parental line, V79. As a further test of activation of the HIF system, we examined effects on expression of messenger RNA (mRNA) for the HIF target gene, Glut-1. Induction of Glut-1 mRNA was normal in all cell lines (Figure 2B, and data not shown).
HIF-1α protein and HIF target gene expression in Chinese hamster fibroblast mutant cell lines.
(A,C) Immunoblots showing levels of HIF-1α protein in cell extracts from respiration-deficient Chinese hamster fibroblast mutant cell lines. Cells were cultured in parallel for 4 hours in normoxia (21% O2, N) or hypoxia (0.1% O2, H). (A) Chinese hamster lung fibroblast wild-type CCL16 cells and the derived CCL16-B2 (B2) and CCL16-B9 (B9) cell lines. (B) RNAse protection assay showing expression of Glut-1 mRNA in wild-type CCL16 and CCL16-B2 (B2) cells. Cells were cultured in parallel for 16 hours in normoxia (21% O2, N) or hypoxia (0.1% O2, H). Signals from Glut-1 mRNA and U6 small nuclear RNA, as an internal control, are indicated. (C) Chinese hamster lung fibroblast wild-type V79 cells and the derived Gal32, V79-G4 (G4), and V79-G7 (G7) cell lines.
HIF-1α protein and HIF target gene expression in Chinese hamster fibroblast mutant cell lines.
(A,C) Immunoblots showing levels of HIF-1α protein in cell extracts from respiration-deficient Chinese hamster fibroblast mutant cell lines. Cells were cultured in parallel for 4 hours in normoxia (21% O2, N) or hypoxia (0.1% O2, H). (A) Chinese hamster lung fibroblast wild-type CCL16 cells and the derived CCL16-B2 (B2) and CCL16-B9 (B9) cell lines. (B) RNAse protection assay showing expression of Glut-1 mRNA in wild-type CCL16 and CCL16-B2 (B2) cells. Cells were cultured in parallel for 16 hours in normoxia (21% O2, N) or hypoxia (0.1% O2, H). Signals from Glut-1 mRNA and U6 small nuclear RNA, as an internal control, are indicated. (C) Chinese hamster lung fibroblast wild-type V79 cells and the derived Gal32, V79-G4 (G4), and V79-G7 (G7) cell lines.
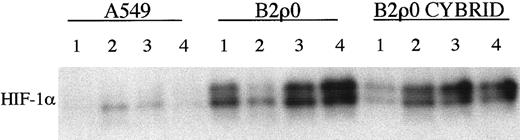
Next we tested a series of previously described stable ρ0 cell lines that had a complete defect in mitochondrial electron transport because of loss of mitochondrial DNA.26 27 The ρ0 status was confirmed by undetectable mitochondrial DNA as assessed by PCR, absent cyanide inhibitable respiration (Table 2), and metabolic sensitivity to growth in the absence of uridine. HIF-1α induction was analyzed by immunoblotting. For one set of cells, the human osteosarcoma-derived cell line 143B and its mtDNA-less derivative, 206ρ0, HIF-1α induction was essentially identical. Representative responses (in this case to 4 hour exposure to approximately 0.1% oxygen) are shown in Figure 3A. However, for the second set of cells, the human lung carcinoma–derived cell line A549 and its mtDNA-less derivative, B2ρ0, somewhat different results were obtained (Figure 3C). The clearest difference was in the level of HIF1-α observed under normoxic growth conditions. Although the results were quantitatively variable in normoxic culture, HIF-1α levels were consistently and sometimes quite markedly higher in the B2ρ0 line than in its parental control (Figure 3D). HIF-1α mRNA levels were similar between these cell lines (data not shown). When levels of mRNA for Glut-1 were examined, similar results were obtained. Induction of Glut-1 mRNA by hypoxia in the 206ρ0 cells was indistinguishable from parental 143B cells, whereas in B2ρ0 cells, levels of Glut-1 mRNA were higher than in the parental A549 cells (Figure 3B,E). Thus, among 2 cell lines analyzed that had complete loss of mitochondrial electron transport, one showed essentially normal regulation of HIF, whereas the other showed an increase in the normoxic level of HIF-1α. This finding might indicate the existence of a mitochondrial signal affecting HIF regulation that was redundant in one but not in the other cell background. Alternatively, the abnormal HIF expression in B2ρ0 cells might be due to some nonmitochondrial mechanism of metabolic adaptation that had occurred in one but not in the other cell line. To address this we analyzed a cybrid of the B2ρ0 cell line that had been repopulated with normal mitochondria. Figure4 shows the results of a series of assays of HIF-1α levels in normoxic parental A549 cells, B2ρ0 cells, and the cybrid line derived from B2ρ0 that had regained essentially normal levels of oxygen consumption (Table 2). Whereas HIF-1α levels were clearly elevated in B2ρ0 versus parental A549 cells, the repopulated cybrid showed a similar elevation, indicating that up-regulation of HIF-α was not directly related to the mitochondrial defect.
HIF-1α protein and HIF target gene expression in ρ0 cell lines.
(A,C,D) Immunoblots showing levels of HIF-1α in cell extracts from wild-type and mtDNA-less cell lines. Cells were cultured in parallel for 4 hours in normoxia (21% O2, N) or hypoxia (0.1% O2, H). (A) Human osteosarcoma 143B and the derived mtDNA-less 206ρ0 cell line. (B) Human osteosarcoma 143B and the derived 206ρ0 cell line. (C) Human lung cancer cell line A549 and the derived mtDNA-less B2ρ0 cell line. (D) Further extracts of normoxic A549 and B2ρ0 cells. βTubulin acts as an internal control. (B,E) Expression of Glut-1 mRNA. Cells were cultured in parallel for 16 hours in normoxia (21% O2, N) or hypoxia (0.1% O2, H). RNAse protection assays of total RNA was performed for Glut-1 and U6 small nuclear RNA as an internal control. (E) A549 and the derived B2ρ0 cell line.
HIF-1α protein and HIF target gene expression in ρ0 cell lines.
(A,C,D) Immunoblots showing levels of HIF-1α in cell extracts from wild-type and mtDNA-less cell lines. Cells were cultured in parallel for 4 hours in normoxia (21% O2, N) or hypoxia (0.1% O2, H). (A) Human osteosarcoma 143B and the derived mtDNA-less 206ρ0 cell line. (B) Human osteosarcoma 143B and the derived 206ρ0 cell line. (C) Human lung cancer cell line A549 and the derived mtDNA-less B2ρ0 cell line. (D) Further extracts of normoxic A549 and B2ρ0 cells. βTubulin acts as an internal control. (B,E) Expression of Glut-1 mRNA. Cells were cultured in parallel for 16 hours in normoxia (21% O2, N) or hypoxia (0.1% O2, H). RNAse protection assays of total RNA was performed for Glut-1 and U6 small nuclear RNA as an internal control. (E) A549 and the derived B2ρ0 cell line.
HIF-1α protein expression in B2ρ0 cybrid cell line.
Immunoblot of HIF-1α protein levels in normoxic A549, B2ρ0, and B2ρ0 cybrid cell lines. Cybrid cell line harbors mitochondria with 100% wild-type mtDNA and is restored to respiratory competence. Four independent sets of whole cell extracts 1 to 4 were assayed.
HIF-1α protein expression in B2ρ0 cybrid cell line.
Immunoblot of HIF-1α protein levels in normoxic A549, B2ρ0, and B2ρ0 cybrid cell lines. Cybrid cell line harbors mitochondria with 100% wild-type mtDNA and is restored to respiratory competence. Four independent sets of whole cell extracts 1 to 4 were assayed.
Taken together, these results indicate that in several different cell backgrounds a functional mitochondrial chain is not necessary for generation of the oxygen-sensitive signal that regulates the HIF system. Previous reports have described failure of HIF activation in Hep3Bρ0 cells that lacked mitochondrial DNA following a 3-week period of exposure to ethidium bromide but not maintained in long-term tissue culture.18 We considered that perturbation of HIF activation by mitochondrial depletion might be specific to the cell type or mode of depletion. To pursue this consideration we depleted Hep3B cells of mitochondria by exposure to ethidium bromide for periods ranging from 3 to 8 weeks and confirmed ρ0 status by the absence of detectable mitochondrial DNA by PCR amplification and uridine auxotrophy. When these cells were exposed to hypoxia, we again observed normal induction of HIF-1α (Figure 5A).
Immunoblots of HIF-1α protein in normoxic and hypoxic Hep3B cells under different conditions.
(A) Untreated Hep3B cells (lanes 1 and 2), Hep3B cells after 3 weeks' culture in medium containing ethidium bromide (EB) (50 ng/mL) to deplete mtDNA to ρ0 status (lanes 5 and 6), or for 3 weeks in the presence of ethidium bromide (50 ng/mL) followed by 1 week in the presence of rotenone (ROT) (1 μg/mL) (lanes 3 and 4). (B) Wild-type Hep3B cells grown under normal conditions or in the presence of rotenone (1 μg/mL) for 1 week. Cells were cultured in parallel for 4 hours in normoxia (21% O2, N) or hypoxia (0.1% O2, H).
Immunoblots of HIF-1α protein in normoxic and hypoxic Hep3B cells under different conditions.
(A) Untreated Hep3B cells (lanes 1 and 2), Hep3B cells after 3 weeks' culture in medium containing ethidium bromide (EB) (50 ng/mL) to deplete mtDNA to ρ0 status (lanes 5 and 6), or for 3 weeks in the presence of ethidium bromide (50 ng/mL) followed by 1 week in the presence of rotenone (ROT) (1 μg/mL) (lanes 3 and 4). (B) Wild-type Hep3B cells grown under normal conditions or in the presence of rotenone (1 μg/mL) for 1 week. Cells were cultured in parallel for 4 hours in normoxia (21% O2, N) or hypoxia (0.1% O2, H).
Because some protocols have described selection of ρ0 cells in high-dose rotenone after growth in ethidium bromide,18cells were grown in the presence of ethidium bromide for 4 weeks with the addition of rotenone (1 μg/mL) in the last week. HIF activation in hypoxia was assessed in the continued presence of rotenone and ethidium bromide. Exposure to rotenone for 1 week reduced HIF activation by hypoxia in both Hep3Bρ0 cells and wild-type Hep3B cells (Figure 5A,B).
Measurement of hydrogen peroxide in normoxia and hypoxia
Mitochondrial metabolism is an important source of ROS that includes hydrogen peroxide.20,21 Because hydrogen peroxide has been reported in different models to mediate the oxygen-sensitive signal that regulates HIF,7 15-19 we wished to determine whether, and in what way, hydrogen peroxide production was altered in the ρ0 cell lines. To provide an assay of production that avoids potentially confounding influences of intracellular redox active markers, we measured the extracellular accumulation of hydrogen peroxide by the luminol/peroxidase method in both 206ρ0 and B2ρ0 cell lines and their respective wild-type parental lines. Results are summarized in Figure 6. Hydrogen peroxide production was markedly reduced by hypoxia in both parental cell lines. The 206ρ0 cells showed a substantial reduction in hydrogen peroxide accumulation in normoxia that was further suppressed by hypoxia, whereas the B2ρ0 cells manifest an apparently more complete defect in hydrogen peroxide production.
Production of hydrogen peroxide by wild-type and ρ0 cells.
Cells were grown in 24-well plates. After 2 hours of incubation in normoxia (20% O2, N) or hypoxia (1% O2, H), an aliquot of extracellular medium was removed in which the hydrogen peroxide (H2O2) concentration was measured (as described in “Materials and methods”). Cells were counted after the experiments: 143B cultures contained 218 000 ± 3000 cells/well (mean ± 1 SD), 206ρ0 171 000 ± 13 000 cells/well, A549 576 000 ± 85 000 cells/well, and B2ρ0 235 000 ± 24 000 cells/well.
Production of hydrogen peroxide by wild-type and ρ0 cells.
Cells were grown in 24-well plates. After 2 hours of incubation in normoxia (20% O2, N) or hypoxia (1% O2, H), an aliquot of extracellular medium was removed in which the hydrogen peroxide (H2O2) concentration was measured (as described in “Materials and methods”). Cells were counted after the experiments: 143B cultures contained 218 000 ± 3000 cells/well (mean ± 1 SD), 206ρ0 171 000 ± 13 000 cells/well, A549 576 000 ± 85 000 cells/well, and B2ρ0 235 000 ± 24 000 cells/well.
Discussion
In this work we have demonstrated a normal pattern of HIF induction by hypoxia in the presence of major defects in mitochondrial respiration. HIF induction was assessed both by accumulation of the HIF-1α subunit in hypoxia and by induction of the mRNA encoding the HIF target gene, Glut-1. In each case similar results were obtained. The genetic deficiencies in mitochondrial metabolism that were tested included complete lack of mitochondrial DNA in several cell backgrounds and a series of known and unknown mutations that disrupt the electron chain partially or completely at different sites in Chinese hamster fibroblasts. In the pharmacologic studies, we found that high concentrations of rotenone, in the range 1 to 10 μM, could inhibit induction of HIF by hypoxia. However, the concentrations of rotenone required to inhibit electron transport were much lower, and we observed maximal inhibition of respiration using 0.1 μM rotenone. These concentrations fit well with other dose-response studies of rotenone. For instance, Barrientos and Moraes35 found that in the human osteosarcoma–derived 143B cell line, complex I activity was inhibited by approximately 50% at a rotenone concentration of 5 to 10 nM and that inhibition was complete with 0.1 μM. In this cell line, concentrations in the range 0.5 μM to 1 μM cause a disorganization of the microtubular structure. Because this effect was also seen in 206ρ0 cells, it was considered to be a secondary effect of rotenone unrelated to its action on complex I.35 Thus, the dose-response characteristics that we found for HIF activation are suggestive of secondary effects unrelated to the action of rotenone on complex I. Taken together these findings indicate that in a variety of cell backgrounds HIF regulation by oxygen appears to be independent of changes in overall mitochondrial electron transport. The results therefore provide evidence against the proposed model in which electron flow in the proximal part of the chain results in increased generation of ROS at complex III in hypoxia as a result of limitation in electron flow distal to that site.18 19
Our results also appear to be at odds with some of the experimental findings taken to support that model. Several possibilities may be considered, although we have at present no clear explanation for all the differences.
First, although loss of mitochondrial DNA in human hepatoma Hep3B and human kidney 293 cells has been reported to result in failure of hypoxic induction of HIF,18,19 we found entirely normal induction of HIF in 206ρ0 cells that were originally derived from a human osteosarcoma cell line. We considered that dependence of HIF regulation on mitochondrial electron transport might be cell-type specific. However, we were also unable to perturb HIF regulation by mitochondrial DNA depletion of Hep3B to ρ0 status. Another possible explanation is that mitochondrial DNA depletion might be expected to have widespread indirect effects on intermediary metabolism that could differ between one line and another and be responsible for effects on HIF regulation. Such processes might also explain conflicting data on the effects of mitochondrial inhibitors on HIF regulation. In recent work we, and others, have shown the critical involvement of a novel prolyl hydroxylase that modifies HIF-1α so as to allow recognition by pVHL.38,39 Such enzymes employ the Krebs cycle intermediate α-ketoglutarate as cosubstrate, so that indirect effects on the availability of this molecule might affect HIF-1α regulation. Interestingly, B2ρ0 cells showed up-regulation of HIF in normoxia and were clearly different in this respect from their parental line A549. However, the abnormality was not reversed by repopulation with normal mitochondria in a derived cybrid line. Thus, it seems that the abnormality in HIF regulation in B2ρ0 cell was either coincidental or more likely the consequence of selection of cells that bore some compensatory nonmitochondrial metabolic abnormality that was not reversed when functional mitochondria were reintroduced. Clearly, up-regulation of HIF in normoxia is not the same as the previously reported failure of induction by hypoxia,18 19 but the experiments do demonstrate a potential difficulty in assigning functional effects to mitochondrial DNA depletion itself.
Second, in some of the previously reported experiments on Hep3Bρ0 cells, the cells were exposed to high concentrations of rotenone as part of the ρ0 selection process.18 We found that similar concentrations of rotenone were able to abrogate HIF activation in both wild-type and ρ0 cells, suggesting that a secondary nonmitochondrial action of rotenone might account for effects on HIF activation in such cells.
Third, we found that hydrogen peroxide production by wild-type cells was greatly reduced both by loss of mitochondrial DNA and by hypoxia. In 206ρ0 cells, some production remained that was also reduced by hypoxia. This finding is consistent with previous findings that established mitochondrial metabolism as a major source of ROS20 21 and suggests that overall production of hydrogen peroxide by both mitochondrial and nonmitochondrial sources is decreased, rather than increased by hypoxia.
Overall, our results indicate that mitochondrial electron flow is not linked in any simple way to the regulation of HIF nor do they support the proposal that increases in ROS during hypoxia activate HIF.18 19 Although B2ρ0 cells showed a low rate of production of hydrogen peroxide and had high normoxic HIF-1α expression, these findings may not be causally connected as we have not observed any clear correlation between levels of hydrogen peroxide production and levels of HIF-1α in surveys of other cells. Whether and in what way reduced ROS generation in hypoxia might be linked to HIF activation remains an open question. In this respect, further analyses of HIF regulation and ROS output in cells bearing well-characterized mutations affecting ROS metabolism should be of interest.
Although our findings provide no support for current models of mitochondrial function in the activation of HIF by hypoxia, it is important to recognize that they do not rule out the mitochondrion as a potential contributor of signals that regulate HIF in other ways. This is particularly the case as we have not yet explained apparent differences from some reported findings. In work reported very recently, much reduced, though not absent, induction of HIF by hypoxia was reported in cybrid lines constructed from 206ρ0 cells repopulated by mitochondria from gorilla, chimpanzee, and pigmy chimp sources, although no experiments were reported on 206ρ0 cells themselves.36 These cybrid cells contain a partial defect in respiration that is mainly, but not entirely, due to a defect in complex I, most probably arising from mismatching of the mitochondrial-encoded hominoid proteins with the nuclear-encoded human proteins.35 The effects on HIF were interpreted as supporting the model in which ROS generated at complex III activate HIF, with this signal being inhibited by limitation of proximal electron flow arising from complex I deficiency.36 Because we did not observe similar effects either in 206ρ0 cells themselves, or in the Chinese hamster lines CCLI6-B2, CCLI6-B9, V79-G4, V79-G7, and Gal32, all of which have severe defects in complex I,29-33 this explanation seems unlikely. However, it remains possible that the defect created in the primate xenomitochondrial cybrids interferes in some different way with a mitochondrial signal that is unaffected by the other genetic defects in mitochondrial metabolism that we have examined. Interestingly, measurements of ROS production in these xenomitochondrial cybrids have demonstrated increased production, at least in normoxic culture conditions.35
Similar arguments apply to a different model that was postulated following the finding that defects in the mitochondrial complex II protein, cybS, predispose, like hypoxia, to paraganglioma.22 Although they did not examine HIF activation directly, those researchers postulated that cybS was critical for an oxygen-sensing system in paraganglionic tissue and that the cybS defect mimicked a hypoxic signal. On the basis of the current work it is unlikely that any effects of cybS mutations on HIF might arise simply as a consequence of an overall reduction in electron flow, mimicking that observed in hypoxia. However, it again remains possible that the cybS defect could in some other way perturb the signaling process, this time to activate, rather than block, a hypoxia signal.
Recent work has indicated an unforeseen complexity to mitochondrial signal pathways, for instance involvement in the regulation of apoptosis (for review, see37). In our view, the nature of any mitochondrial involvement in HIF regulation remains unclear. The generation and characterization of further specific genetic defects and the derivation of simpler in vitro systems for the study of HIF regulation should be important in pursuing this question.
The authors thank J. M. Gleadle, C. W. Pugh, and P. H. Maxwell for helpful discussions. They also thank I. E. Scheffler and C. D. Whitfield for generously providing the Chinese hamster fibroblast cell lines, M. P. King for 143B/206ρ0 cell lines, and I. J. Holt for A549/B2ρ0 and B2ρ0 cybrid cell lines.
Supported by research grants from the Wellcome Trust.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Peter J. Ratcliffe, The Henry Wellcome Building of Genomic Medicine, Roosevelt Dr, Oxford, OX3 7BN, United Kingdom; e-mail: peter.ratcliffe@imm.ox.ac.uk.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal