The LW blood group glycoprotein, ICAM-4, is a member of the intercellular adhesion molecule (ICAM) family expressed in erythroid cells. To begin to address the function of this molecule, ligands for ICAM-4 on hemopoietic and nonhemopoietic cell lines were identified. Peptide inhibition studies suggest that adhesion of cell lines to an ICAM-4–Fc construct is mediated by an LDV-inhibitable integrin on hemopoietic cells and an RGD-inhibitable integrin on nonhemopoietic cells. Antibody inhibition studies identified the hemopoietic integrin as α4β1. Antibody inhibition studies on α4β1-negative, nonhemopoietic cell lines suggested that adhesion of these cells is mediated by αVintegrins (notably αVβ1 and αVβ5). The structure of ICAM-4 modeled on the crystal structure of ICAM-2 was used to identify surface-exposed amino acid residues for site-directed mutagenesis. Neither an unusual LETS nor an LDV motif in the first domain of ICAM-4 was critical for integrin binding. ICAM-4 is the first ICAM family member shown to be a ligand for integrins other than those of the β2family, and the data suggest that ICAM-4 has a novel integrin-binding site(s). These findings suggest a role for ICAM-4 in normal erythropoiesis and may also be relevant to the adhesive interactions of sickle cells.
Introduction
Knowledge of cell-cell and cell-extracellular matrix interactions in bone marrow is essential for an understanding of the formation of blood cells and their migration into the peripheral circulation. Such interactions are also relevant to the biology of diseases like sickle cell disease and malaria where adhesive interactions involving red cells and damaged endothelium are crucial features of the pathology.1-3 These adhesive interactions frequently involve molecules belonging to the immunoglobulin superfamily (IgSF) of proteins and the family of proteins known as integrins. Integrins are heterodimers composed of 2 noncovalently associated transmembrane subunits denoted “α” and “β.” Eight different β subunits combine in a restricted manner with 18 α subunits to form some 22 different integrins. Beta subunits complex with several different α subunits, while most α subunits (with the exception of αV, α4, and α6) combine with only one β subunit.4
The LW blood group glycoprotein, or ICAM-4, is a member of the IgSF subfamily known as intercellular adhesion molecules (ICAMs). It has 2 predicted IgSF I-set domains,5-7 and within the subfamily it is most similar to ICAM-2. ICAM-4 has been found expressed only on erythroid cells and weakly in placenta.8,9 ICAM-1, -2, and -3 function in inflammation and immune responses,10 but no function for ICAM-4 has been defined.
The ICAMs are ligands for β2 integrins, and all family members, including ICAM-4, are reported to bind αLβ2.10-13 ICAM-1 and ICAM-4 are also ligands for αMβ2,10,11 while the preferred ligand for ICAM-3 is αDβ2.14Unlike ICAM-1, -2, -3, -5, which have the αLβ2-binding motif (L/I)ET(P/S)L (Figure 1), ICAM-4 has the nonconsensus sequence LR52TPL within the C strand of domain 1 and also has an adjacent potential N-glycosylation site (N48; Figure 1). Another residue (Q) in the G strand is also important for αLβ2binding in ICAM-1, -2, and -3.15-18 ICAM-4 has T91 in this equivalent position (Figure 1), as does ICAM-5. Several other IgSF molecules, including vascular cell adhesion molecule (VCAM)-1, mucosal addressin cell adhesion molecule (MAdCAM)-1, CD31, and L1 are also integrin ligands.10,19-22 VCAM-1 and MAdCAM-1 are related to the ICAM family but are bound by α4 integrins. An aspartic acid residue in the CD loop within the motif QIDSP or GLDTS, respectively, is critical for integrin binding in these molecules.19,23These motifs are all variants of the LDV sequence, which exhibits considerable sequence variation in different ligands.24
The C and G strands of ICAM family N-terminal domains.
Residues critical for αLβ2 binding are boxed; numbering is for ICAM-4.
The C and G strands of ICAM family N-terminal domains.
Residues critical for αLβ2 binding are boxed; numbering is for ICAM-4.
X-ray crystal structures of 2-domain fragments of ICAM-1, ICAM-2, VCAM-1, and MAdCAM-1 provide insights into the structural basis of IgSF-integrin interactions.25 ICAM-1 and -2 are both ligands for the I domain–containing αLβ2integrin. In these ICAMs, a critical glutamic acid residue is located on the relatively flat surface of the CFG face of the molecule.26-28 In contrast, VCAM-1 and MAdCAM-1 are ligands for non–I domain α4 integrins and have a critical aspartic acid residue on a protruding CD loop.29 30
We have modeled ICAM-4 based on the published crystal structure of ICAM-2 to identify the positioning of the ICAM-4 nonconsensus LR52TPL motif and to test whether an LD73V motif located at the end of the predicted E strand is solvent-exposed. Using soluble, recombinant ICAM-4 constructs and by mutating several key residues to restore consensus ICAM-2 sequences, we have explored the integrin-binding properties of the molecule in cell-based adhesion assays. We have also targeted the LD73V motif in domain 1 by site-directed mutagenesis to test for involvement in integrin binding. Our results show that ICAM-4 has an unusual integrin-binding profile in that it binds α4β1 and αV-containing integrins and suggest that the reported binding of αLβ2 is of lesser significance. The results also show that ICAM-4 must have novel ligand-binding motif(s).
Materials and methods
ICAM-4 homology model
A homology model for ICAM-4 was constructed by mutation of the ICAM-2 structure26 followed by energy minimization. Using an alignment of the ICAM-2 and ICAM-4 sequences, the coordinates of ICAM-2 were mutated to match the corresponding sequence for ICAM-4. Where insertion or deletion of residues was necessary, these were constructed using the loop-building facility in InsightII (MSI, San Diego, CA) maintaining maximal homology to ICAM-2 and reasonable peptide geometry. After soaking with an 0.8-nm (8-Å) layer of water, the model structure was energy-minimized while tethering the backbone atoms to their initial positions. The tethering force was reduced during this procedure from 4200 kJ/nm (100 kcal/Å) to a final value of 21 kJ/nm (0.5 kcal/Å). The energy-minimized structures (average derivative < 0.84 kJ/nm [0.02 kcal/Å]) were analyzed using Procheck59 and found to be of similar or better quality compared with the original crystal structure. Structural manipulations were carried out using InsightII97and energy minimizations using the cvff forcefield implemented in Discover (version 2.97, MSI, San Diego, CA).
Mammalian cell lines studied
Cells lines were mostly from European Culture Collection, Wiltshire, United Kingdom. The β1 integrin-negative subclone of JY (Prof M. Humphries, University of Manchester, United Kingdom), fibrosarcoma (FLYRD18 [FLY], Dr C. Porter, Hammersmith Hospital, London, United Kingdom), and endothelial (ECV304 and SK-HEP1, Dr P. Evans, Southampton General Hospital, United Kingdom) lines were gifts. Human umbilical vein endothelial cells (HUVECs) were as described.31 K562 cells transfected with αLβ2 (KL/4) or α4 (KA4C6, which expresses a constitutively active form of α4β1) were as described.32 All cells were maintained in Iscoves modified Eagle medium (IMEM), 10% fetal bovine serum.
Antibodies and peptides
Function-blocking monoclonal antibodies to integrin subunits were anti-β1 (clone13, Becton Dickinson, Oxford, United Kingdom); anti-β2 (MHM23, Dako, Bucks, United Kingdom; YFC118.3, Serotec, Oxford, United Kingdom; 1B4, Alexis, Bingham, United Kingdom; P4H9-A11, Chemicon International, Harrow, United Kingdom); anti-β3 (PM6/13, Harlan Sera-Lab, Loughborough, United Kingdom; RUU-PL 7F12, Becton Dickinson); anti-β4 (ASC-3, Chemicon); anti-αL (38, Dr N Hogg, London, United Kingdom; MHM24, Dako); anti-α1 (FB12, Chemicon); anti-α2 (JA218, Prof M. Humphries, University of Manchester); anti-α3 (C3[VLA3], Immunotech, High Wycombe, United Kingdom); anti-α4 (HP2/1, Serotec; L25.3, Becton Dickinson; Max68P, Dr T. Shock, Celltech, Slough, United Kingdom); anti-α5 (SNAKA55, Prof M. Humphries); anti-α6 (NKI-GoH3, Serotec); anti-αV(69.9.5, Immunotech; CLB-706, Chemicon); anti-αVβ3 (23C6, Serotec; LM609, Harlan Sera-Lab); and anti-αVβ5 (P1F6, Becton Dickinson). Activating antibodies to integrin subunits were anti-β1 (TS2/16, American Type Culture Collection, Manassas, VA); anti-β2 (KIM127, KIM185 as described33 34); and anti-α4 (44H6, Serotec). Antibodies against the first domain of ICAM-4 (BS46, BS56) were from Dr H. Sonneborn, Biotest, Dreieich, Germany. Linear peptides GRGDSPK and EILDVPST were synthesized in-house.
Preparation of fusion proteins
ICAM-4–Fc fusion proteins (ICAM4Fc) used in the study comprised the 2 extracellular domains of ICAM-4 and the hinge region and Fc domains of human IgG1 as described.35 ICAM-4 complementary DNA (cDNA) encoding leader sequence and the 2 extracellular IgSF domains (ICAM-4 amino acid residues −30 to 196) was amplified by polymerase chain reaction (PCR) using sense primer (TTCCCAAGCTTTGCCATGGGGTCTCTGTTCCCT), antisense primer (ACGGATCCACTTACCTGTGGGGCTCCAAGCGAGCATCAGTGT), and full-length ICAM-4 cDNA template. This DNA was subcloned into pBluescript and used for subsequent steps. Mutant ICAM-4 cDNAs encoding consensus ICAM-2 residues at amino acid residues 50, 52, and 91, IC4S50G (removes consensus N48 glycosylation site), IC4R52E, and IC4T91Q were made using inverse PCR.36 Sense primer 5′-ACCCCGCTGCGGCAAGGCAAGACGCTCAGA was used for mutants IC4S50G and IC4R52E. Antisense primer for IC4R52E was 5′-TTCGAGGCTGGAATTCTGCGGCTGGGGACA and for IC4S50G was 5′-GCGGAGGCCGGAATTCTGCGGCTGGGGACA. Sense primer for IC4T91Q was 5′-ACGCTGGGCCACCTCCAGGATCACCGCCTA and antisense primer was 5′-TGTTTTCCTGCGCAGGTCACGAGGCAGTGC. Native and mutant ICAM-4 cDNAs were subcloned into pIg vector as described.35 A mutant encoding IC4D73R was generated by overlap extension PCR36 using native ICAM-4 cDNA in pIg as template with complementary, mutational ICAM-4 primers 5′-GCTGCTCCGCGTGAGGGCCTGG and 5′-CCAGGCCCTCACGCGGAGCAGC together with sense and antisense pIg vector primers 5′-AGAACCCACTGCTTACTGGCT and 5′-TGAGCCTGCTTCCAGCAGACA. All clones were verified by sequence analysis. The cDNA clones encoding the extracellular domains of ICAM-1, -2, and -3 and neural cell adhesion molecule (NCAM) in pIg were gifts from Dr D. Simmons, SmithKline Beecham, Harlow, United Kingdom. CAMFc proteins were expressed in COS-7 cells as described35 and extracted from culture supernatant on protein A–Sepharose. Fc fusion proteins containing the first 2 domains of VCAM-1 (VCAMFc) or MAdCAM-1 (MAdCAMFc) were as described.37 ICAM4Fc and mutant ICAM4Fc proteins migrated with Mr about 100 000 kd on sodium dodecyl sulfate–polyacrylamide gel electrophoresis under reducing conditions. IC4S50G migrated with somewhat lowerMr, suggesting an absence of N-glycan at residue N48. Western blotting (nonreducing conditions) using antihuman IgG and anti–ICAM-4 antibodies BS46 and BS56 was also performed (not shown). Protein concentrations were determined by the “Nano-orange” technique (Molecular Probes, Leiden, The Netherlands).
Flow cytometry
Cells were analyzed for antigen expression as described.38 Mean fluorescence intensity was used as a measure of antibody binding.
Cell adhesion assay
Immulon-4 96-well plates (Dynex Technologies, Billingshurst, United Kingdom) were coated with 1 μg/well goat-antihuman–Fc (Sigma, Poole, United Kingdom) for 18 hours at 4°C, blocked with phosphate-buffered saline (pH 7.4), 0.4% bovine serum albumin (Fraction V, Sigma) for 2 hours at 22°C, and coated with chimeric proteins in phosphate-buffered saline (1 μg/well unless stated otherwise) for 2 hours at 37°C. Hemopoietic cells were washed once in assay buffer (IMEM, 2 mM EGTA, 5% human group AB serum [National Blood Service, Bristol, United Kingdom]). Nonhemopoietic cells were lifted in phosphate-buffered saline containing 2 mM ethylenediaminetetraacetic acid (EDTA), 0.1% bovine serum albumin and washed once in IMEM containing 0.1% (wt/vol) bovine serum albumin. Cells (107/mL in assay buffer) were labeled with 10 μg/mL 2′,7′-bis(2-carboxyethyl)-5(6)-carboxyfluorescein acetoxymethyl ester (Sigma) for 15 minutes at 37°C and washed thrice in assay buffer. Cells were activated with 80 μM phorbol myristate acetate (PMA, Sigma) in assay buffer for 15 minutes at 37°C and washed twice in assay buffer containing cations. Cells were incubated for 15 minutes at 0°C with 10 μg/mL antibodies (KIM and anti–ICAM-4 antibodies at 25 μg/mL), 500 μM peptides, or 25 μg/mL ICAM4Fc in assay buffer containing 2 mM Mn2+ or 10 mM Mg2+. Cells were added to CAMFc-coated plates (5 × 104/well in 100 μL) for 30 minutes at 37°C. Plates were read on a fluorescence microplate reader (excitation 485 nm, emission 530 nm, Bio-Tek Instruments, VT), given standardized washes in assay buffer, and read after each wash. Washing was performed by flooding the plates with assay buffer at 37°C and then vigorously decanting the buffer to waste by rapid inversion of the plate. The percentage of input cells bound was calculated. Each data point is the mean of 3 or more replicates, and assays were performed on at least 3 independent occasions.
Results
ICAM-4 homology model
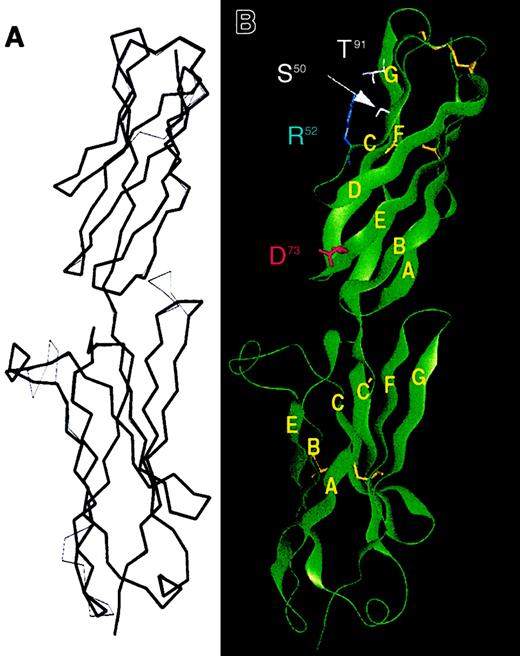
A homology model of ICAM-4 was constructed (Figure2A) based on the crystal structure of ICAM-2,26 which, in the region modeled, has 29% amino acid sequence identity with ICAM-4. Because this tentative model is derived from sequence homology with ICAM-2, it closely follows the reported fold for ICAM-2 with domain 1 adopting an I-1 fold and domain 2 belonging to the I-2 subset.25 Numbering of residues within the ICAM-4 model follows the numbering reported for the N-terminal amino acid sequence derived from ICAM-4 isolated from red cells.6,7 The N-terminal region of ICAM-4 has an additional 15 residues not present in the ICAM-2 crystal structure, but these have not been included in the model, although it is possible that they form an additional strand adjacent to the existing A/A′ edge strand. The model of ICAM-4 therefore starts at residue 16 (VPF), which is equivalent to residue 1 (KVF) in the structure of ICAM-2.26 After energy minimization, the root mean square deviation of 185 equivalent Cα positions between the ICAM-4 model and the ICAM-2 crystal structure is 0.065 nm (0.65 Å). Like ICAM-2, ICAM-4 also has an additional disulphide bond in domain 1 between the B/C loop and the end of the F strand, a feature commonly associated with IgSF domains that act as integrin ligands.25 There are 2 regions in domain 1 of the model for ICAM-4 where the conformation differs significantly from that of ICAM-2. These are the D/E loop, which forms a prominent protrusion from the top of the molecule, and the E/F loop, which is in close proximity to domain 2, where ICAM-4 has an insertion of one additional residue. In the model, residue R52, which is equivalent to the proposed integrin-binding E37 in the LETSL motif of ICAM-2, adopts a similar location and conformation to the glutamic acid side chain in ICAM-2. Significant differences are also observed in domain 2 at the C-terminal end of the A strand and in the C′/E and F/G loops both at the top of domain 2, where contacts are made with domain 1. These latter changes suggest the contacts between the domains—and hence relative orientations of the domains—are likely to differ between ICAM-2 and ICAM-4. Our model makes no attempt to predict these movements between whole domains, which are beyond the limitations of current modeling techniques. A similar model for ICAM-4 has also recently been described.39 Although it is not possible to directly compare these models because the coordinates are not available, the energy minimization of our model appears to have generated marginally more loop movements in regions of poor sequence homology. For 122 equivalent Cα positions, the model of Hermand et al differs from the ICAM-2 crystal structure coordinates by 0.05 nm (0.85 Å), whereas the difference for our model is 0.10 nm (1.0 Å). Nonetheless, the placement of key residues is essentially the same in both model structures.
Model of ICAM-4.
Domain 1 is at the top of the figure. (A) Model of ICAM-4 (Cα trace in bold) is overlaid on the equivalent trace for the crystal structure of ICAM-2 (thin lines). (B) Ribbon diagram of the modeled ICAM-4 structure, with β strands labeled A-G and the residues mutated in this study highlighted.
Model of ICAM-4.
Domain 1 is at the top of the figure. (A) Model of ICAM-4 (Cα trace in bold) is overlaid on the equivalent trace for the crystal structure of ICAM-2 (thin lines). (B) Ribbon diagram of the modeled ICAM-4 structure, with β strands labeled A-G and the residues mutated in this study highlighted.
The exposure and conformation of residues forming the adhesion surface for domain 1 of ICAM-4 are largely similar to those reported for ICAM-2, although ICAM-4 has a paucity of acidic residues in the expected binding surfaces. Indeed, a distinctive feature of domain 1 in the ICAM-4 model is the concentration of basic residues in these regions, in particular on the CFG domain face. The acidic residue within the LD73V motif at the end of the E strand is exposed at the domain surface in the modeled structure (Figure 2B). The location and interactions of other residues mutated in this study were also examined to ensure, as far as possible, that the substituted side chains would not be deleterious for the protein conformation. S50, R52, and T91 are all surface residues in the model; none appear to be involved in critical hydrogen or other bonds at the domain surface (Figure 2B); and, from the model, no significant rearrangements of the protein surface would be expected to accompany their mutation to the ICAM-2 concensus residues G, E, and Q, respectively.
Hemopoietic and nonhemopoietic cells show integrin-mediated adhesion to ICAM-4
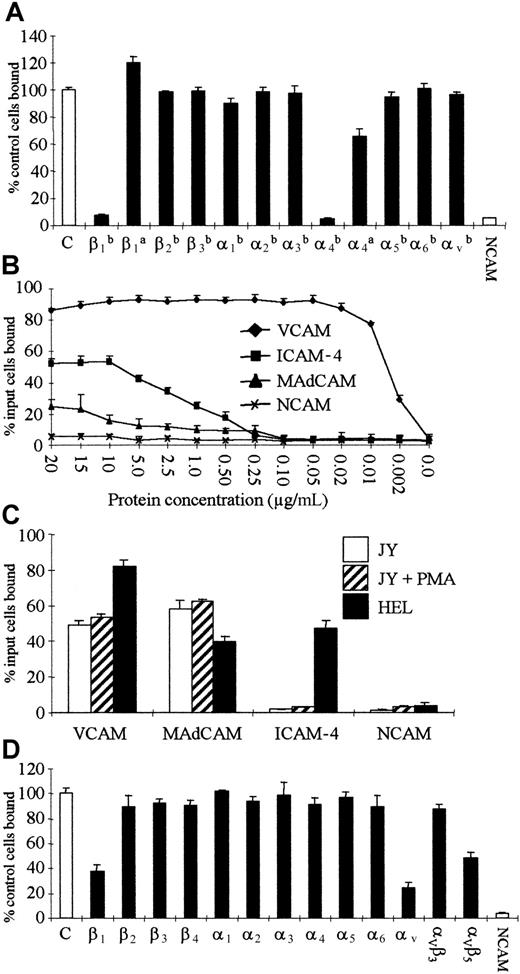
A panel of 12 hemopoietic and 10 nonhemopoietic cell lines was tested for adhesion to soluble recombinant ICAM4Fc. To define optimal activating conditions, cells were tested in the presence of cations and after stimulation with phorbol ester. Six hemopoietic (HEL, THP1, KG1a, Raji, Jurkat, and Molt-4) and all nonhemopoietic cell lines bound to ICAM4Fc at levels that were characteristic of each cell line (Figure 3A, Tables 1 and2). Adhesion of these cells was abrogated in the presence of EDTA, suggesting that binding was integrin-mediated (Figure 3B). Binding of hemopoietic HEL cells was markedly inhibited by LDV-containing peptide, whereas binding of nonhemopoietic FLY cells was partially inhibited by LDV peptide but was abrogated in the presence of RGD-containing peptide (Figure 3B). Two anti–ICAM-4 antibodies, BS46 and BS56, did not inhibit HEL or FLY cell adhesion to ICAM-4 (data not shown). A measure of the relative avidity of adhesion to ICAM-4 was obtained by examining binding of several hemopoietic and nonhemopoietic cell lines to titrated ICAM4Fc. For most hemopoietic lines that bound to ICAM-4, a plateau of binding was reached at 10 μg/mL (Figure 3C), while the plateau of binding for the nonhemopoietic cell lines ranged between 2.5 μg/mL (DX3) and 15 μg/mL (HT29) (Figure 3D).
Hemopoietic and nonhemopoietic cell lines adhere to ICAM4Fc.
Adhesion assays were as described in “Materials and methods.” Results are input cells bound ± SD (n = 6). (A) Binding of hemopoietic cell lines (HEL, KG1a, THP1, IM9) and nonhemopoietic cell lines (FLY, HUVEC, 293, COS) to ICAM4Fc (▪) or to NCAMFc (control, ■). Activating conditions were cations (HEL, HUVEC) and PMA+ cations (IM9, KG1a, THP1, FLY, 293, COS). (B) Binding of cation-activated HEL cells (■) or PMA+ cation–activated FLY cells (▪) to ICAM4Fc in the presence of 2 mM EDTA, LDV peptide, or RGD peptide (500 μM). (C) Relative avidity of adhesion of hemopoietic cell lines to ICAM4Fc. Results are adjusted by subtraction of the baseline binding to NCAMFc (for maximal levels, see Table 1). Activation conditions were cations (HEL,♦; Raji) and PMA+ cations (Jurkat, ▪; THP1, ▴; Molt-4, ⋄; EBV-LCL, ▵; U937, ■). (D) Relative avidity of adhesion of nonhemopoietic cell lines to ICAM4Fc. Results are adjusted by subtraction of the baseline binding to NCAMFc (for maximal levels, see Table 2) Activation conditions were cations (HT29, ⋄) and PMA+ cations (DX3, ♦; FLY, ▪; SK-HEP1, ▴; HFFF, ■).
Hemopoietic and nonhemopoietic cell lines adhere to ICAM4Fc.
Adhesion assays were as described in “Materials and methods.” Results are input cells bound ± SD (n = 6). (A) Binding of hemopoietic cell lines (HEL, KG1a, THP1, IM9) and nonhemopoietic cell lines (FLY, HUVEC, 293, COS) to ICAM4Fc (▪) or to NCAMFc (control, ■). Activating conditions were cations (HEL, HUVEC) and PMA+ cations (IM9, KG1a, THP1, FLY, 293, COS). (B) Binding of cation-activated HEL cells (■) or PMA+ cation–activated FLY cells (▪) to ICAM4Fc in the presence of 2 mM EDTA, LDV peptide, or RGD peptide (500 μM). (C) Relative avidity of adhesion of hemopoietic cell lines to ICAM4Fc. Results are adjusted by subtraction of the baseline binding to NCAMFc (for maximal levels, see Table 1). Activation conditions were cations (HEL,♦; Raji) and PMA+ cations (Jurkat, ▪; THP1, ▴; Molt-4, ⋄; EBV-LCL, ▵; U937, ■). (D) Relative avidity of adhesion of nonhemopoietic cell lines to ICAM4Fc. Results are adjusted by subtraction of the baseline binding to NCAMFc (for maximal levels, see Table 2) Activation conditions were cations (HT29, ⋄) and PMA+ cations (DX3, ♦; FLY, ▪; SK-HEP1, ▴; HFFF, ■).
Adhesion of hemopoietic cells to Fc fusion proteins
| Cell line (activation) . | Adhesion to Fc fusion proteins, percentage of input cells . | Expression of integrin subunits, MFI . | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ICAM1Fc . | ICAM2Fc . | ICAM3Fc . | ICAM4Fc . | IC4R52EFc . | IC4S50GFc . | IC4T91QFc . | NCAMFc . | αL . | β2 . | α4 . | β1 . | αV . | C . | |
| HEL (−PMA) | 20 | 16 | 10 | 62 | 64 | 59 | 60 | 11 | 14 | 8 | 283 | 308 | 43 | 4 |
| K562 (+PMA) | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 8 | 4 | 3 | 112 | 9 | 3 |
| KG1a (+PMA) | 89 | 78 | 73 | 34 | 27 | 27 | 30 | 9 | 285 | 252 | 379 | 280 | 4 | 2 |
| HL-60 (+PMA) | 21 | 8 | 5 | 5 | 5 | 5 | 4 | 3 | 31 | 26 | 158 | 248 | 4 | 4 |
| THP1 (+PMA) | 96 | 64 | 38 | 53 | 50 | 40 | 43 | 23 | 151 | 141 | 95 | 155 | 5 | 3 |
| U937 (+PMA) | 51 | 12 | 4 | 5 | 3 | 5 | 4 | 4 | 28 | 22 | 365 | 385 | 3 | 2 |
| Jurkat (+PMA) | 67 | 34 | 24 | 26 | 21 | 23 | 22 | 4 | 60 | 51 | 363 | 558 | 15 | 3 |
| Molt-4 (+PMA) | 5 | 5 | NT | 29 | 32 | 18 | 19 | 4 | 34 | 27 | 580 | 452 | 5 | 2 |
| EBV-LCL (+PMA) | 42 | 27 | 19 | 10 | 10 | 9 | 9 | 10 | 94 | 75 | 47 | 172 | 13 | 3 |
| IM9 (+PMA) | 61 | 19 | 9 | 9 | 9 | 8 | 9 | 7 | 77 | 56 | 288 | 144 | 15 | 3 |
| Daudi (−PMA) | 24 | 14 | NT | 9 | 9 | 10 | 10 | 6 | 31 | 21 | 107 | 129 | 2 | 2 |
| Raji (−PMA) | 28 | 7 | NT | 10 | 14 | 9 | 13 | 4 | 15 | 11 | 172 | 120 | 3 | 3 |
| Cell line (activation) . | Adhesion to Fc fusion proteins, percentage of input cells . | Expression of integrin subunits, MFI . | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ICAM1Fc . | ICAM2Fc . | ICAM3Fc . | ICAM4Fc . | IC4R52EFc . | IC4S50GFc . | IC4T91QFc . | NCAMFc . | αL . | β2 . | α4 . | β1 . | αV . | C . | |
| HEL (−PMA) | 20 | 16 | 10 | 62 | 64 | 59 | 60 | 11 | 14 | 8 | 283 | 308 | 43 | 4 |
| K562 (+PMA) | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 8 | 4 | 3 | 112 | 9 | 3 |
| KG1a (+PMA) | 89 | 78 | 73 | 34 | 27 | 27 | 30 | 9 | 285 | 252 | 379 | 280 | 4 | 2 |
| HL-60 (+PMA) | 21 | 8 | 5 | 5 | 5 | 5 | 4 | 3 | 31 | 26 | 158 | 248 | 4 | 4 |
| THP1 (+PMA) | 96 | 64 | 38 | 53 | 50 | 40 | 43 | 23 | 151 | 141 | 95 | 155 | 5 | 3 |
| U937 (+PMA) | 51 | 12 | 4 | 5 | 3 | 5 | 4 | 4 | 28 | 22 | 365 | 385 | 3 | 2 |
| Jurkat (+PMA) | 67 | 34 | 24 | 26 | 21 | 23 | 22 | 4 | 60 | 51 | 363 | 558 | 15 | 3 |
| Molt-4 (+PMA) | 5 | 5 | NT | 29 | 32 | 18 | 19 | 4 | 34 | 27 | 580 | 452 | 5 | 2 |
| EBV-LCL (+PMA) | 42 | 27 | 19 | 10 | 10 | 9 | 9 | 10 | 94 | 75 | 47 | 172 | 13 | 3 |
| IM9 (+PMA) | 61 | 19 | 9 | 9 | 9 | 8 | 9 | 7 | 77 | 56 | 288 | 144 | 15 | 3 |
| Daudi (−PMA) | 24 | 14 | NT | 9 | 9 | 10 | 10 | 6 | 31 | 21 | 107 | 129 | 2 | 2 |
| Raji (−PMA) | 28 | 7 | NT | 10 | 14 | 9 | 13 | 4 | 15 | 11 | 172 | 120 | 3 | 3 |
Expression of integrin subunits αL, β2, α4, β1, and αV determined by flow cytometry.
ICAM indicates intercellular adhesion molecule; NCAMFc, neural cell adhesion molecule control protein; PMA, phorbol myristate acetate; MFI, mean fluorescence intensity measured by flow cytometry; NT, not tested; C, control antibody.
A comparison of the adhesion of nonhemopoietic cell lines to intercellular adhesion molecule-4Fc with their integrin profiles determined by flow cytometry
| Cell line (activation) . | Adhesion to Fc fusion proteins, percentage of input cells . | Integrin expression, MFI . | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ICAM4Fc . | NCAMFc . | β1 . | β2 . | β3 . | β4 . | α1 . | α2 . | α3 . | α4 . | α5 . | α6 . | αV . | αVβ3 . | αVβ5 . | C . | |
| FLY (+PMA) | 71 | 5 | 743 | 7 | 21 | 7 | 15 | 434 | 274 | 6 | 296 | 73 | 116 | 49 | 93 | 2 |
| HFFF (+PMA) | 51 | 7 | 307 | 3 | 18 | 4 | 94 | 74 | 65 | 76 | 142 | 9 | 90 | 27 | 33 | 4 |
| ECV304 (−PMA) | 28 | 3 | 754 | 5 | 47 | 170 | 5 | 147 | 163 | 9 | 43 | 30 | 155 | 55 | 136 | 4 |
| SK-HEP1 (+PMA) | 48 | 3 | 1098 | 3 | 4 | 75 | 9 | 243 | 353 | 126 | 90 | 70 | 50 | 5 | 71 | 4 |
| HUVEC (−PMA) | 18 | 6 | 389 | NT | NT | NT | 13 | 163 | 69 | 5 | 170 | 5 | NT | 165 | 33 | 4 |
| DX3 (+PMA) | 65 | 7 | 347 | 3 | 17 | 5 | 33 | 107 | 105 | 68 | 37 | 10 | 36 | 25 | 26 | 3 |
| HT29 (−PMA) | 39 | 3 | 332 | 3 | 3 | 311 | 117 | 158 | 150 | 3 | 3 | 76 | 109 | 3 | 134 | 3 |
| 293 (+PMA) | 51 | 20 | 413 | 3 | 5 | 11 | 146 | 98 | 74 | 28 | 82 | 6 | 60 | 7 | 12 | 3 |
| COS (+PMA) | 80 | 9 | NT | NT | NT | NT | NT | NT | NT | NT | NT | NT | NT | NT | NT | NT |
| CHO (−PMA) | 33 | 3 | NT | NT | NT | NT | NT | NT | NT | NT | NT | NT | NT | NT | NT | NT |
| Cell line (activation) . | Adhesion to Fc fusion proteins, percentage of input cells . | Integrin expression, MFI . | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ICAM4Fc . | NCAMFc . | β1 . | β2 . | β3 . | β4 . | α1 . | α2 . | α3 . | α4 . | α5 . | α6 . | αV . | αVβ3 . | αVβ5 . | C . | |
| FLY (+PMA) | 71 | 5 | 743 | 7 | 21 | 7 | 15 | 434 | 274 | 6 | 296 | 73 | 116 | 49 | 93 | 2 |
| HFFF (+PMA) | 51 | 7 | 307 | 3 | 18 | 4 | 94 | 74 | 65 | 76 | 142 | 9 | 90 | 27 | 33 | 4 |
| ECV304 (−PMA) | 28 | 3 | 754 | 5 | 47 | 170 | 5 | 147 | 163 | 9 | 43 | 30 | 155 | 55 | 136 | 4 |
| SK-HEP1 (+PMA) | 48 | 3 | 1098 | 3 | 4 | 75 | 9 | 243 | 353 | 126 | 90 | 70 | 50 | 5 | 71 | 4 |
| HUVEC (−PMA) | 18 | 6 | 389 | NT | NT | NT | 13 | 163 | 69 | 5 | 170 | 5 | NT | 165 | 33 | 4 |
| DX3 (+PMA) | 65 | 7 | 347 | 3 | 17 | 5 | 33 | 107 | 105 | 68 | 37 | 10 | 36 | 25 | 26 | 3 |
| HT29 (−PMA) | 39 | 3 | 332 | 3 | 3 | 311 | 117 | 158 | 150 | 3 | 3 | 76 | 109 | 3 | 134 | 3 |
| 293 (+PMA) | 51 | 20 | 413 | 3 | 5 | 11 | 146 | 98 | 74 | 28 | 82 | 6 | 60 | 7 | 12 | 3 |
| COS (+PMA) | 80 | 9 | NT | NT | NT | NT | NT | NT | NT | NT | NT | NT | NT | NT | NT | NT |
| CHO (−PMA) | 33 | 3 | NT | NT | NT | NT | NT | NT | NT | NT | NT | NT | NT | NT | NT | NT |
The monkey (COS) and hamster (CHO) cell lines were not tested (NT) with antibodies against human integrins.
MFI indicates mean fluorescence intensity.
The main ICAM-4–binding integrin of hemopoietic cell lines is not αLβ2
We tested whether the LDV-inhibitable hemopoietic integrin was αLβ2 and whether adhesion to any of the mutant ICAM4Fc fusion proteins containing consensus ICAM-2 residues (IC4S50G, IC4R52E, and IC4T91Q) was different compared with native ICAM4Fc. Each mutant was titrated and tested, in comparison with native ICAM4Fc, for adhesion of HEL cells. Similar results were obtained with wild-type and all mutant ICAM4Fc proteins, demonstrating that each mutant was functionally active (Figure 4A). Hemopoietic cell lines were tested for adhesion to ICAM4Fc or mutant ICAM-4–Fc proteins in comparison with ICAM-1–Fc, -2–Fc, and -3–Fc or NCAMFc under optimal activating conditions for each cell line (Table 1). Every cell line that adhered to ICAM4Fc also bound to all mutant ICAM4Fc proteins, and no cell line that failed to adhere to native ICAM4Fc bound mutant ICA4Fc proteins. The level of αLβ2 integrin expression did not correlate with adhesion to ICAM4Fc. HEL and Molt-4 cells expressed low levels of αLβ2; adhered poorly to ICAM-1–Fc, -2–Fc, or -3–Fc; but adhered well to ICAM4Fc. Conversely, IM9 and EBV-LCL expressed high levels of αLβ2; adhered to ICAM-1–Fc, -2–Fc, or -3–Fc; but did not adhere to ICAM4Fc or to mutant ICAM4Fc proteins (Table 1).
Adhesion of hemopoietic cells to ICAM4Fc is not αLβ2-mediated.
Adhesion assays were as described in “Materials and methods.” Results are input cells bound ± SD (n = 6). (A) Adhesion of cation-activated HEL cells to ICAM-4 and mutant ICAM-4–Fc proteins (coated at 0.5 μg/well). (B) Binding of PMA+ cation–activated THP1 cells to ICAM-1–Fc, -2–Fc, and -3–Fc, ICAM4Fc, and mutant ICAM-4–Fc proteins (coated at 0.5 μg/well) in the presence of inhibiting anti-αL (mab 38, hatched), anti-β2 (1B4, black) antibodies or a control antibody (white). (C) Adhesion of PMA+ cation–activated IM9 cells to ICAMs–Fc, -2–Fc, and -3–Fc, ICAM4Fc, and mutant ICAM4Fc proteins (coated at 1 μg/well). Cells were treated with either control antibody (white) or activating β2antibodies, KIM127 (hatched) or KIM185 (black).
Adhesion of hemopoietic cells to ICAM4Fc is not αLβ2-mediated.
Adhesion assays were as described in “Materials and methods.” Results are input cells bound ± SD (n = 6). (A) Adhesion of cation-activated HEL cells to ICAM-4 and mutant ICAM-4–Fc proteins (coated at 0.5 μg/well). (B) Binding of PMA+ cation–activated THP1 cells to ICAM-1–Fc, -2–Fc, and -3–Fc, ICAM4Fc, and mutant ICAM-4–Fc proteins (coated at 0.5 μg/well) in the presence of inhibiting anti-αL (mab 38, hatched), anti-β2 (1B4, black) antibodies or a control antibody (white). (C) Adhesion of PMA+ cation–activated IM9 cells to ICAMs–Fc, -2–Fc, and -3–Fc, ICAM4Fc, and mutant ICAM4Fc proteins (coated at 1 μg/well). Cells were treated with either control antibody (white) or activating β2antibodies, KIM127 (hatched) or KIM185 (black).
Under conditions where adhesion to ICAM-1–Fc, -2–Fc, or -3–Fc was markedly inhibited, function-blocking antibodies to integrin subunits αL and β2 did not abrogate binding of hemopoietic cell lines to ICAM4Fc or to mutant ICAM4Fc proteins (Figure4B; several αL and β2 antibodies were tested and representative results are shown). A small reduction of THP1 cell adhesion to ICAM4Fc in the presence of αL or β2 antibodies was observed, suggesting there may be some αLβ2-mediated adhesion of THP1 cells. An alternative interpretation is that this apparent blocking was nonspecific because a similar reduction in background THP1 cell adhesion to NCAMFc control protein was also observed (Figure 4B). Treatment with β2-activating antibodies (KIM127 and KIM185) increased the percentage of IM9 and EBV-LCL cells adhering to ICAM-1–Fc, -2–Fc, or -3–Fc proteins but not to ICAM4Fc or to mutant ICAM4Fc proteins (Figure 4C and not shown). An αLβ2-transfected cell line (KL/4) also adhered to ICAM-1–Fc, -2–Fc, or -3–Fc but failed to adhere to ICAM4Fc or mutant ICAM4Fc proteins under maximally activating conditions (not shown).
ICAM-4 is a ligand for α4β1 of hemopoietic cells
All hemopoietic cell lines that adhered to ICAM4Fc were found to express integrins of the β1 family (Table 1). In adhesion assays, a β1 blocking antibody (clone 13) abrogated binding while an activating antibody (TS2/16) stimulated binding (β1b, β1a, Figure5A). Other β subunit antibodies had no effect. Blocking α4 antibodies (HP2/1, Max68P, and L25.3) totally inhibited binding while an activating antibody (44H6) partially inhibited binding (α4a, α4b, Figure 5A). Other antibodies against α subunits of β1-family integrins had no effect.
Adhesion of hemopoietic cells to ICAM4Fc is α4β1-mediated, and adhesion to nonhemopoietic cells is αV integrin–mediated.
Adhesion assays were as described in “Materials and methods.” (A) Adhesion of cation-activated, hemopoietic, HEL cells to ICAM4Fc in the presence of blocking (b) or activating (a) antibodies to integrin α or β subunits as described in “Materials and methods.” Results are cells bound ± SD (n = 6), in comparison with control antibody (C, 100%). Antibodies used were β1b, clone 13; β1a, TS2/16; β2b, MHM23; β3b, RUU-PL; α1, FB12; α2, JA218; α3, C3(VLA3); α4b, HP2/1; α4a, 44H6; α5b, SNAKA55; α6b, NKI-GoH3; and αV b, 69.9.5. ICAM4Fc was coated at 0.5 μg/well. (B) Comparison of the relative avidity of adhesion of cation-activated HEL cells to VCAMFc (diamonds), MAdCAMFc (triangles), ICAM4Fc (squares), and NCAMFc (crosses) by titration of CAMFc protein as described in “Materials and methods.” Results are input cells bound ± SD (n = 6). (C) Adhesion of cation-activated β1-negative JY cells and HEL cells to VCAMFc, MAdCAMFc, ICAM4FcFc, and NCAMFc. Fc fusion proteins were coated at 0.05 μg/well (VCAMFc, MAdCAMFc) and 1 μg/well (ICAM4Fc, NCAMFc). Results are the percentage of input cells bound ± SD (n = 6). (D) Adhesion of PMA+ cation–activated, nonhemopoietic, FLY cells to ICAM4Fc in the presence of blocking antibodies to integrin α or β subunits or integrin complexes as described in “Materials and methods.” Results are cells bound ± SD (n = 6), in comparison with control antibody (C, 100%). Blocking antibodies were as above except β4, ASC-3; αVβ3, LM609; and αVβ5, P1F6. ICAM4Fc was coated at 0.25 μg/well.
Adhesion of hemopoietic cells to ICAM4Fc is α4β1-mediated, and adhesion to nonhemopoietic cells is αV integrin–mediated.
Adhesion assays were as described in “Materials and methods.” (A) Adhesion of cation-activated, hemopoietic, HEL cells to ICAM4Fc in the presence of blocking (b) or activating (a) antibodies to integrin α or β subunits as described in “Materials and methods.” Results are cells bound ± SD (n = 6), in comparison with control antibody (C, 100%). Antibodies used were β1b, clone 13; β1a, TS2/16; β2b, MHM23; β3b, RUU-PL; α1, FB12; α2, JA218; α3, C3(VLA3); α4b, HP2/1; α4a, 44H6; α5b, SNAKA55; α6b, NKI-GoH3; and αV b, 69.9.5. ICAM4Fc was coated at 0.5 μg/well. (B) Comparison of the relative avidity of adhesion of cation-activated HEL cells to VCAMFc (diamonds), MAdCAMFc (triangles), ICAM4Fc (squares), and NCAMFc (crosses) by titration of CAMFc protein as described in “Materials and methods.” Results are input cells bound ± SD (n = 6). (C) Adhesion of cation-activated β1-negative JY cells and HEL cells to VCAMFc, MAdCAMFc, ICAM4FcFc, and NCAMFc. Fc fusion proteins were coated at 0.05 μg/well (VCAMFc, MAdCAMFc) and 1 μg/well (ICAM4Fc, NCAMFc). Results are the percentage of input cells bound ± SD (n = 6). (D) Adhesion of PMA+ cation–activated, nonhemopoietic, FLY cells to ICAM4Fc in the presence of blocking antibodies to integrin α or β subunits or integrin complexes as described in “Materials and methods.” Results are cells bound ± SD (n = 6), in comparison with control antibody (C, 100%). Blocking antibodies were as above except β4, ASC-3; αVβ3, LM609; and αVβ5, P1F6. ICAM4Fc was coated at 0.25 μg/well.
The integrin α4β1 has 2 other well-characterized IgSF ligands, VCAM-1 and MAdCAM-1, both of which are related to the ICAM family. A measure of the relative avidity of α4β1-expressing cells for the 3 ligands ICAM-4, VCAM-1, and MAdCAM-1 was obtained by defining the minimum concentration of ligand that promoted adhesion of HEL cells in titrations. Values of 10 μg/mL for ICAM4Fc, 0.05 μg/mL for VCAMFc, and 15 μg/mL for MAdCAMFc were obtained (Figure 5B). These findings are consistent with those of Newham et al,40 who also reported that the relative avidity of α4β1-mediated adhesion to VCAM-1 was more than 10 times the avidity for MAdCAM-1. The avidity of α4β1/ICAM-4–mediated adhesion was therefore either greater or of a similar order to α4β1/MAdCAM-1–mediated adhesion. A β1-negative, β7-positive subclone of the JY cell line that expresses α4β7 adhered to VCAMFc or MAdCAMFc but did not adhere to ICAM4Fc (Figure 5C). The data suggest that ICAM-4 may not be a ligand for α4β7 in this B-cell line.
ICAM-4 is a ligand for αV integrins of nonhemopoietic cells
Every nonhemopoietic cell line tested, whether of human or animal origin, bound to ICAM4Fc (Figure 3A,D; Table 2). Several cell lines did not express the α4 integrin subunit (FLY, ECV304, HUVEC, and HT29), whereas the integrins αVβ1, α2β1, α3β1, and αVβ5were consistently expressed (Table 2). Function-blocking integrin antibodies were tested for inhibition of adhesion of these α4-negative cell lines to ICAM4Fc (Figure 5D and not shown). Of the β subunit antibodies tested, only β1antibody had an effect, causing partial inhibition of binding. Antibodies to α subunits of the β1 family were tested, and only αV antibodies had an effect, causing partial inhibition of binding (irrespective of the antibody or concentration used). The αV complex antibodies were tested, and an αVβ5 antibody partially inhibited adhesion. Complete inhibition of binding was not observed with any combination of inhibiting antibodies tested. The integrin profiles of the α4-negative cell lines, together with antibody inhibition data, suggest that ICAM-4 is a ligand for both αVβ1 and αVβ5integrins (the profiles of FLY, HT29, and 293 cells are informative; Table 2). Expression levels of αV integrins did not correlate with percentage of cells adhering or with the ICAM-4 coating concentration at which this was maximal (Figure 3D, Table 2). The restriction of the α4β1/ICAM-4 interaction to a subset of hemopoietic cell lines contrasts with the observation that all nonhemopoietic lines expressing α4β1 adhered to ICAM4Fc (DX3, HFFF, SK-HEP-1, 293; Table 2). This adhesion was partially inhibited by β1, αV, αVβ5, and α4 antibodies, but no combination of antibodies completely inhibited binding (data not shown). Expression levels of α4 and αV integrins did not correlate with percentage of cells adhering or the ICAM-4 coating concentration at which this was maximal (Figure 3D, Table 2). Untransfected monkey (COS-7) or hamster (CHO) cells also showed integrin-mediated adhesion to ICAM-4 (Figure 3A, Table 2). Titration studies with 3 α4β1-negative lines (FLY, HT29, ECV304; Figure 3D) indicated that the avidity of ICAM-4/αVintegrin–mediated adhesion was of a similar order to ICAM-4/α4β1–mediated adhesion and was equal to or greater than MAdCAM-1/α4β1–mediated adhesion, which suggests biological relevance.
ICAM-4 site-directed mutation studies
Mutation of residues on the CFG face of ICAM-4 (IC4R52E, IC4T91Q, and IC4S50G), homologous to those found to be important for αLβ2 binding in ICAM-1, -2, and -3, did not reduce or increase α4β1-mediated adhesion of hemopoietic cell lines (Figure 4A, Table 1) or αV-mediated binding of nonhemopoietic, FLY cells (not shown). The ICAM-4 model predicts a solvent-accessible, consensus LDV site on domain 1, located on the E strand, toward the bottom of the domain (Figure 2B). α4β1-mediated adhesion of HEL cells and αV integrin–mediated adhesion of FLY cells were unaffected by mutation of this motif (IC4D73R) (Figure 6).
Comparison of the relative avidity of adhesion of HEL cells and FLY cells to native ICAM4Fc and IC4D73RFc.
Binding of cation-activated HEL cells or PMA+ cation–activated FLY cells was compared by titration of ICAM4Fc, IC4D73RFc, and NCAMFc as described in “Materials and methods.” Adhesion to ICAM4Fc, IC4D73RFc, or NCAMFc of HEL cells is indicated by triangles, crosses, or lines, respectively. Adhesion to ICAM4Fc, IC4D73RFc, or NCAMFc of FLY cells is indicated by diamonds, squares, or stars, respectively.
Comparison of the relative avidity of adhesion of HEL cells and FLY cells to native ICAM4Fc and IC4D73RFc.
Binding of cation-activated HEL cells or PMA+ cation–activated FLY cells was compared by titration of ICAM4Fc, IC4D73RFc, and NCAMFc as described in “Materials and methods.” Adhesion to ICAM4Fc, IC4D73RFc, or NCAMFc of HEL cells is indicated by triangles, crosses, or lines, respectively. Adhesion to ICAM4Fc, IC4D73RFc, or NCAMFc of FLY cells is indicated by diamonds, squares, or stars, respectively.
Discussion
Our results indicate that ICAM-4 is unique in the ICAM family because it binds through novel motifs to α4β1 and αV integrins. We have demonstrated adhesion of hemopoietic (LDV peptide–inhibitable) and nonhemopoietic (RGD peptide–inhibitable) cells to soluble, recombinant ICAM-4. LDV- and RGD-containing peptides have been used extensively as inhibitors of integrin-mediated interactions because these motifs, or their variants, are found in many integrin ligands.24 41 Given the short incubation times in our experiments and our observation that EDTA also inhibited adhesion, we concluded that adhesion was integrin-mediated and hypothesized that there are 2 different integrin ligands for ICAM-4: an LDV-inhibitable hemopoietic integrin and a nonhemopoietic cell integrin that has a preference for RGD- over LDV-containing sequences.
Adhesion of hemopoietic cells to ICAM4Fc was markedly affected in the presence of activating or inhibiting antibodies against α4 or β1, which strongly suggests that ICAM-4 is bound by the hemopoietic integrin α4β1. The observation that an activating anti-α4 caused partial inhibition of HEL cell adhesion might indicate that the binding site for ICAM-4 on α4β1 is qualitatively different from that occupied by VCAM-1 or MAdCAM-1. There is evidence for overlapping but distinct binding sites for ICAM-1 and ICAM-3 on αLβ2, which might represent an analogous situation.42 The ICAM-4/α4β1interaction was seen only with certain α4β1-expressing hemopoietic lines (but was seen with all α4β1-positive nonhemopoietic lines; see above) and did not correlate with α4β1 expression levels. An activating β1 antibody did not induce ICAM-4 binding in the nonbinding cell lines, suggesting that in hemopoietic cells interaction with ICAM-4 is dependent on additional factors. Masumoto and Hemler43 suggest that the activation state of α4β1 is regulated in each cell type by unknown cell-specific constraints and that not all cell types can achieve the highest states of activation and bind the full repertoire of ligands. Our data are consistent with the notion that not all α4β1-positive hemopoietic cell lines can achieve the activation state required for binding to ICAM-4. The restriction of the α4β1/ICAM-4 interaction to a subset of hemopoietic cell lines contrasts with the observation that all nonhemopoietic lines expressing α4β1 adhered to ICAM4Fc. An α4β1-expressing, K562, α4-transfectant (KA4C6, PMA-activated–positive cations) adhered to VCAMFc but not to MAdCAMFc or ICAM4Fc (not shown). This result might reflect the cell-specific activation state of α4β1 in this transfectant. We were unable to test transfectants of COS or CHO origin because untransfected cells adhered to ICAM4Fc (Figure 3A, Table 2).
Notably, HEL cells that bound with the highest avidity to ICAM-4 under minimal activating conditions are of erythroid origin. Given that ICAM-4 expression appears to be restricted to erythroid cells and placenta,8 it is tempting to speculate that the molecule has a particular function in erythropoiesis. ICAM-4 is first expressed early in erythropoiesis, at the colony-forming unit–erythroid stage.9,44 In mammalian bone marrow, erythropoiesis occurs in discrete anatomic units known as erythroblastic islands.45,46 The integrity of these structures is maintained at least in part by the interaction of α4β1 expressed on erythroblasts, with VCAM-1 expressed on a central macrophage.47 In vivo treatment of mice with VLA-4 antibodies specifically induces anemia.48 Our results show that the apparent avidity of α4β1-mediated adhesion of an erythroblast cell line to ICAM-4 is 200-fold lower than that estimated for α4β1-mediated adhesion to VCAM-1. We have postulated elsewhere that the interaction of ICAM-4 with its ligand α4β1 on adjacent erythroblasts may help to stabilize erythroblastic islands.49 It is likely that there are multiple interactions between adjacent cells in these structures, and further studies will be necessary to assess the physiologic relevance of those involving ICAM-4.
Our finding that the adhesion of various cell lines to ICAM4Fc does not correlate with the level of αLβ2expression; that adhesion is not abrogated by function-blocking, αLβ2 antibodies; that adhesion is not enhanced by activating αLβ2 antibodies; and that an αLβ2 transfectant does not adhere all suggests that adhesion to ICAM-4 or ICAM-4 mutants is not mediated by αLβ2 in the hemopoietic cell lines examined. In contrast to these findings, others have reported that ICAM-4 is a ligand for both αLβ2 and αMβ2 on peripheral blood lymphocytes and transfected cell lines.11,39 In these other studies, αLβ2-mediated adhesion was observed only in the presence of 2 mM CaCl2, and the maximum adhesion observed was only about 12% bound cells. In adhesion assays using ICAM4Fc and α4β1-expressing hemopoietic cell lines, we found that adhesion was inhibited in the presence of 2 mM CaCl2 and was maximal in the presence of Mg2+ or Mn2+ ions and EGTA (see “Materials and methods”). It is also reported that Ca2+ ions are negative regulators of αLβ2-mediated adhesion to ICAM-1.50 We suggest that the adhesive interactions of ICAM-4 with β2 integrins are of lower avidity than those between ICAM-4 and α4β1- or αV-family integrins described here and reflect the rather promiscuous nature of ICAM-4–ligand interactions. Clearly, further studies are needed to establish whether any of these interactions observed in vitro are physiologically significant.
Antibody inhibition studies suggested that the adhesion of nonhemopoietic cells is mediated by αV integrin(s) (probably αVβ1 and αVβ5), though complete inhibition of binding was not observed with any combination of inhibiting antibodies tested. A possible explanation for this observation is that the ICAM-4 binding site on αV integrins may be qualitatively different from that occupied by its other ligands, as suggested for α4β1, above. It is also possible that ICAM-4 is bound by other αV integrins, αVβ6 and αVβ8, that might be expressed by these cell lines but may not be inhibited by these αV antibodies. We did not find αVβ3-mediated adhesion of nonhemopoietic cells to ICAM-4. Adhesion to ICAM-4 of the αV-positive, hemopoietic cell line HEL (which expresses only αVβ3, data not shown) was not blocked by inhibiting αV antibodies. These findings were not unexpected because the assays were performed in the presence of EGTA, which is reported to inhibit αVβ3-mediated binding to CD31.21 Monkey and hamster cells also showed integrin-mediated adhesion to ICAM-4. CHO cells express αV integrins, and CHO transfectants expressing hamster αV/human β5 hybrid integrins are active in functional assays with human-derived ligands.51 52 It is therefore likely that adhesion of both COS-7 and CHO cells is also αV integrin–mediated. Indeed, our data are consistent with the hypothesis that ICAM-4 is a truly promiscuous αVintegrin ligand and is bound by all αV-family integrins.
These results raise the possibility that interactions between ICAM-4 on erythroblasts and αV integrins on the central macrophage of erythroblastic island structures in bone marrow might also contribute to the integrity of the island structures. It is also possible that ICAM-4 has a wider role in erythrocyte recognition by αV integrin-expressing cells of the reticuloendothelial system and may thereby serve as an “erythrocyte addressin.” In sickle cell disease, episodes of acute pain are linked to vascular occlusion caused by the abnormal adhesion of sickle red cells to vascular endothelial cells.1,2,53,54 There are some data suggesting that ICAM-4 expression is abnormally elevated on the erythrocytes of patients with sickle cell disease and that antibodies to ICAM-4 partially inhibit adhesion of sickle red blood cells to tumor necrosis factor-α–activated endothelial cells.49 These observations raise the possibility that ICAM-4/αVintegrin interactions may contribute to the abnormal adhesion of sickle red cells to activated endothelium during sickle cell disease crisis.
Our results strongly suggest that ICAM-4 is a ligand for α4β1 and αV integrins, although we cannot rule out the possibility of crosstalk between these integrins and other integrins or unidentified molecules not targeted by antibody inhibition studies and that these molecules, rather than α4β1 or αV integrins, are ligands for ICAM-4. In hemopoietic cell lines, α4β1 antibodies, an LDV peptide, or EDTA completely blocked adhesion, strongly suggesting that ICAM-4 was bound by α4β1 and the absence of crosstalk. In nonhemopoietic cell lines, both the RGD peptide and EDTA completely blocked adhesion, but no combination of inhibiting antibodies completely blocked binding. In these cells it is possible that crosstalk between αV integrins and other molecules underlies this observation. In this context αVβ3 is reported to regulate both α4β1-mediated lymphocyte migration and α5β1-mediated adhesion and endocytosis of phagocytic cells.55 56
ICAM-4 is clearly an unusual molecule because it can be a ligand for α4β1, which has a preference for LDV-based sequences, and also for αV integrins that usually bind ligands containing the RGD sequence.24,41 The αV integrins generally bind extracellular matrix molecules, and αVβ3 has the largest repertoire of ligands.24 αVβ3is the only αV integrin previously shown to have IgSF ligands and binds CD31 and L1.21,22 The integrin-binding site for L1 has been identified as an RGD sequence in domain 6,22 while that of CD31 has been shown to reside largely on domain 1.21 This domain has no RGD motif, suggesting that αVβ3 can use other sequences.57 This is clearly also the case for the αV integrin–binding site of ICAM-4 which, like CD31, has no RGD sequences. Most integrin-binding sites on IgSF molecules contain an acidic residue, and they are commonly located on the N-terminal domain.21,25,58 Unlike other ICAMs, ICAM-4 domain 1 does not have the consensus αLβ2-binding motif in the C strand. Unlike VCAM-1 and MAdCAM-1, ICAM-4 also lacks a consensus α4β1-binding motif in the C-D loop of this domain. IgSF molecules are highly versatile and can use almost any surface for ligand recognition.25 It is therefore possible that integrin-binding motif(s) may be located in domain 2 of ICAM-4, where there are 7 acidic residues. With the exception of D173 the model shows that all of these residues are exposed at the surface and are potentially available for interaction. Further mutational analysis of domain 2 is clearly necessary to define the integrin-binding site of this novel ICAM.
We thank Dr D. Simmons and Dr M. K. Robinson for helpful discussions, P. Martin for DNA sequence analysis, and Dr W. Mawby for peptide synthesis.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Frances A. Spring, Bristol Institute for Transfusion Sciences, Southmead Rd, Bristol, BS10 5ND, United Kingdom; e-mail: fran.spring@nbs.nhs.uk.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal