Abstract
This study identified a new substitution in the Bβ chain of an abnormal fibrinogen, denoted Longmont, where the residue Arg166 was changed to Cys. The variant was discovered in a young woman with an episode of severe hemorrhage at childbirth and a subsequent mild bleeding disorder. The neo-Cys residues were always found to be disulfide-bridged to either an isolated Cys amino acid or to the corresponding Cys residue of another abnormal fibrinogen molecule, forming dimers. Removing the dimeric molecules using gel filtration did not correct the fibrin polymerization defect. Fibrinogen Longmont had normal fibrinopeptide A and B release and a functional polymerization site “a.” Thus, the sites “A” and “a” can interact to form protofibrils, as evidenced by dynamic light-scattering measurements. These protofibrils, however, were unable to associate in the normal manner of lateral aggregation, leading to abnormal clot formation, as shown by an impaired increase in turbidity. Therefore, it is concluded that the substitution of Arg166→Cys-Cys alters fibrinogen Longmont polymerization by disrupting interactions that are critical for normal lateral association of protofibrils.
Introduction
The fibrinogen molecule is composed of 2 copies of 3 polypeptide chains called Aα, Bβ, and γ. The molecule consists of a central domain (E) linked to 2 distal domains (D) by a coiled-coil connector. The N-terminal ends of the 6 chains form the E domain, whereas the C-terminal parts of the γ and Bβ chains and a short fragment of the C-terminal domain of the Aα chain constitute the D domain.
The fibrinogen-to-fibrin conversion is an ordered reaction. It is initiated by the cleavage of fibrinopeptides A and B (FpA and FpB) by thrombin, which unmasks polymerization sites “A” and “B” in the neo–N-terminal ends of the α and β chains, respectively.1 Thereafter, the fibrin monomers interact spontaneously and form a fibrin clot. Concomitantly, thrombin-activated factor XIII stabilizes the clot by introducing interchain cross-links between γ chains and between α chains.2-4
Fibrin polymerization is a 2-step process. First, half-staggered, end-to-end interactions form double-stranded protofibrils, which result from interactions between site “A” and its complementary site “a,”5 always present in the γ chain part of the D domain.6 In fact, the peptide Gly-Pro-Arg-Pro (GPRP), which resembles the natural Gly-Pro-Arg-Val sequence of the N-terminus extremity of the α chain depleted in FpA, prevents clot formation.7 In the second step, termed “lateral association,” the protofibrils are assembled into thick fibers of fibrin that branch to form a fibrin network.8,9 The interactions involved in the lateral association are less well characterized. It has been proposed that the release of FpB enhances lateral association.10,11 Alternatively, the release of FpB may be a consequence of fibrin formation.12 Moreover, the C-terminal domains of the Aα chain may also participate in lateral association.13 14
In this report, we describe fibrinogen Longmont, a new Bβ chain defect (Arg166→Cys) in a dysfibrinogen discovered in a heterozygous female.15 No free sulfhydryl group was found; rather the neo-Cys residue forms a disulfide bridge with either another abnormal molecule or with a Cys residue. These post-translational changes alter the polymerization process, particularly the lateral association of protofibrils.
Materials and methods
Materials
All reagents were obtained from Sigma (St Louis, MO) unless otherwise noted. Human α-thrombin was obtained from Enzyme Research Labs (South Bend, IN). Coagulation factor XIII was a generous gift from Dr Kevin Siebenlist (Sinai Samaritan Medical Center, Milwaukee, WI). Plasminogen was purified from human plasma as described by Deutsch et al.16 Streptokinase was purchased from American Diagnostica (Greenwich, CT). Rabbit polyclonal antiserum to human fibrinogen was from Dako (Carpinteria, CA); monoclonal antihuman serum albumin was from Sigma; and the goat antirabbit conjugated to horseradish peroxidase and the goat antimouse conjugated to horseradish peroxidase were from Calbiochem (Darmstadt, Germany). UV-transparent 96-well plates were purchased from Corning Costar (Cambridge, MA).
Fibrinogen purification
Fibrinogen was purified from the citrated plasma of healthy donors and the proband essentially according to the method of Holm et al.17 Briefly, the plasma was supplemented with a cocktail of protease inhibitors (pepstatin, leupeptin, and phenylmethylsulfonyl fluoride at 2 μM final concentration each) and then precipitated by adding an equal volume of 6 M β-alanine containing 0.1 M ε-amino-n-caproic acid. The pellet was harvested by centrifugation at 10 000g at 4°C for 30 minutes and resuspended in 0.3 M NaCl. After repeating the precipitation once, the fibrinogen solutions were dialyzed extensively against 0.3 M NaCl. The purity of fibrinogen was verified by 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). The solutions were aliquoted and stored at −20°C until use.
Fibrinogen structural studies
Electrophoresis.
SDS-PAGE was performed according to Laemmli.18 SDS-agarose gel was prepared using Seakem Gold ultra pure agarose (FMC, Rockland, ME) in Laemmli running buffer and run under constant voltage in the same buffer. Mouse laminin (Gibco, Gaithersburg, MD) was used as 850-kd molecular weight (Mol wt) marker.
Size-exclusion chromatography.
Normal and the higher Mol wt fibrinogen molecules were separated by gel filtration with a column of 1.6 × 51 cm packed with Spectra/gel TSK HW-55S (Spectrum Medical, Los Angeles, CA) according to the manufacturer's instructions. MES (2-morpholinoethanesulfonic acid) buffer (50 mM MES [pH 6.0] and 200 mM NaCl) was used to equilibrate the column, and the different fibrinogen species were eluted at a flow rate of 0.4 mL/min. Samples of purified normal or Longmont fibrinogens were loaded and the fractions collected, concentrated using a centrifugal filter device (Millipore, Bedford, MA), dialyzed extensively against HEPES buffer (20 mM HEPES [pH 7.4] and 150 mM NaCl), and analyzed on a 2% SDS-agarose gel.
Plasmin protection assay.
Plasmin was generated by activation of 100 μg/mL plasminogen by 100 U/mL streptokinase. Samples of 0.2 mg/mL normal fibrinogen or fibrinogen Longmont in HEPES buffer containing either 5 mM CaCl2, 5 mM ethylenediaminetetraacetic acid (EDTA), or 2 mM GPRP peptide were incubated with plasmin at 10 μg/mL final concentration for 24 hours at 37°C. Following electrophoresis in a 7.5% gel, the proteins were either stained with Coomassie brilliant blue or electroblotted onto a nitrocellulose membrane. The membrane was blocked overnight at 4°C in 20 mM Tris (pH 7.4), 150 mM NaCl (TBS) containing 5% nonfat milk and then incubated with a 1:7000 dilution of rabbit polyclonal antiserum to fibrinogen for 2 hours in TBST (TBS supplemented with 0.05% Tween 20) containing 1% bovine serum albumin. After washing 3 times with TBST, the membrane was incubated with a 1:5000 dilution of goat antirabbit immunoglobulin G conjugated to horseradish peroxidase for 1 hour in TBST with 1% bovine serum albumin, washed 3 times, and developed with ECL Western Blotting Detection Reagent (Amersham Pharmacia Biotech, Piscataway, NJ).
Albumin binding to fibrinogen.
Purified normal or Longmont fibrinogen and the corresponding whole plasma were run on 5% SDS-PAGE and then transferred to a nitrocellulose sheet. After saturating the membrane with TBS containing 5% nonfat milk, it was incubated with a 1:2000 dilution of a monoclonal antibody to serum human albumin in TBST. After washing 4 times with TBST, the membrane was incubated with a 1:5000 dilution of goat antimouse immunoglobulin G conjugated to horseradish peroxidase in TBST and developed as described above.
DNA sequence analysis.
The DNA fragment coding exon IV of the fibrinogen Bβ chain gene was amplified by the polymerase chain reaction (PCR) using the following primers: Bβ-IVF, corresponding to nucleotides 4921 to 4942 in intron 3 (5′-CTGCTTGGTG ATAGCTCAGTG-3′), and Bβ-IVR primer (5′-GGTGTGTGAGTTCTTCTGGA-3′), corresponding to the complementary nucleotides 5322 to 5302.19 The amplification was performed in a 50-μL reaction volume containing 45 μL PCR SuperMix buffer (Gibco, Grand Island, NY) supplemented by 0.2 μM each primer and 0.1 μg genomic DNA for 37 cycles. Each cycle consisted of 1 minute at 95°C, 0.5 minutes at 57°C, and 1 minute at 72°C. The PCR product was run on 1% agarose gel, the band with the expected size purified with the Gel Extraction Kit (Qiagen, Santa Clarita, CA), and the purified products directly sequenced using both Bβ-IVF and Bβ-IVR primers (GlaxoWellcome DNA sequence facility, University of North Carolina, Chapel Hill).
Mass spectrometric analysis.
Unfractionated fibrinogen Longmont (1.3 mg) was dissolved in 300 μL 6 M guanidine hydrochloride, pH 8.5, containing 10 μL 4-vinylpyridine to derivatize free sulfhydryl groups.20 After 30 minutes, the protein was precipitated with 10 vol methanol. The dried precipitate was dissolved in 200 μL concentrated formic acid containing 20 mg cyanogen bromide (CNBr), incubated for 2 hours at room temperature,21 and the digest fractionated on a 1 × 110–cm column of Sephadex G-50sf in 0.2% trifluoroacetic acid and monitored at 215 nm.22 Quantitative N-terminal amino acid sequence analysis was determined with a Hewlett-Packard G1005A protein sequencer. Trypsin digestion of the pooled fractions was followed by mass spectrometric analysis in a Perseptive Biosystems Voyager DE Pro matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) instrument (Foster City, CA). The pools were concentrated to a small volume for the trypsin digestion and then adjusted to pH 8 with ammonia. Trypsin was added to an estimated 1:50 (wt/wt) enzyme:substrate ratio and the digests left for 20 hours at room temperature. Part of each digested sample was subsequently incubated overnight with mercaptoethanol (5% final concentration) at 37°C to cleave any disulfide bonds that might be present. For all mass spectrometric analyses, α-cyano-4-hydroxycinnamic acid was used as matrix.
Fibrinopeptide release.
Release of FpA and FpB was performed essentially as described by Mullin et al.23
Turbidity measurements.
Polymerization at ambient temperature was monitored continuously at 350 nm in a SpectraMax-340PC 96-well microplate reader (Molecular Devices, Sunnyvale, CA). The reaction was initiated by adding 10 μL thrombin at a concentration of 1 NIH U/mL to 200 μL fibrinogen solution (0.4 mg/mL) in HEPES buffer. Each reaction was carried out in triplicate.
Factor XIIIa–catalyzed cross-linking of fibrin.
Fibrinogen (0.2 mg/mL in HEPES containing 2 mM CaCl2) was incubated at ambient temperature with 0.1 NIH U/mL thrombin and 2 μg/mL factor XIII. The reaction was stopped at timed intervals by adding an equal volume of Laemmli sample buffer containing 10% β-mercaptoethanol and heating at 100°C for 5 minutes. The samples, 0.8 μg protein, were analyzed by immunoblot following 7.5% SDS-PAGE using a rabbit polyclonal antiserum to fibrinogen as described above.
Dynamic light scattering.
Samples for light scattering were prepared by gel filtration chromatography with a 10 × 50 HR column packed with Superose 6B resin (Amersham Pharmacia Biotech) using HEPES buffer and a constant flow rate of 1 mL/min. Fractions corresponding to single fibrinogen molecules were concentrated using a centrifugal filter device. Polymerization of normal fibrinogen and fibrinogen Longmont at ambient temperature was performed in quadruplicate using 0.2 mg/mL fibrinogen and thrombin at 0.001 NIH U/mL in HEPES buffer. Static and dynamic light-scattering data were collected simultaneously from each sample, as previously described by Hogan et al.24 The static light-scattering signal reports changes in intensity due to increases in Mol wt distribution during fibrin polymerization. Time-dependent changes in intensity were fit with the equation y = y0 + aek(t − t0), where k is the rate constant, t0 the lag time, y0 the intensity at time zero, and a the increase of intensity between time zero and t0. Fitting was done by nonlinear regression (Sigmaplot, SPSS, Chicago, IL). Dynamic light scattering monitors decreases in translational diffusion coefficient (D) as fibrin protofibrils assemble; these data were expressed as D/D0and the kinetic traces compared to changes for normal protofibril growth reported by Knoll.25 This analysis yielded the protofibrils' time. Each set of kinetic parameters was averaged and the means compared by an unpaired t test using Statview software (Abacus Concepts, Berkeley, CA).
Results
Case report and coagulation data
The proband is a 37-year-old female; the family hemostatic disorders and coagulation data were described in a previous report.15 Routine coagulation studies of fibrinogen Longmont were remarkable for a dysfibrinogenemia. Briefly, both the functional and the antigenic plasma fibrinogen levels of the proband were within normal range. The thrombin clotting time was normal when measured by an electromechanical detection method, but there was not an end point when it was measured by an optical detection method. The findings indicated that fibrinogen Longmont was totally clottable, but the clot was translucent.
Fibrinogen Longmont structure
Electrophoresis and size-exclusion chromatography analysis.
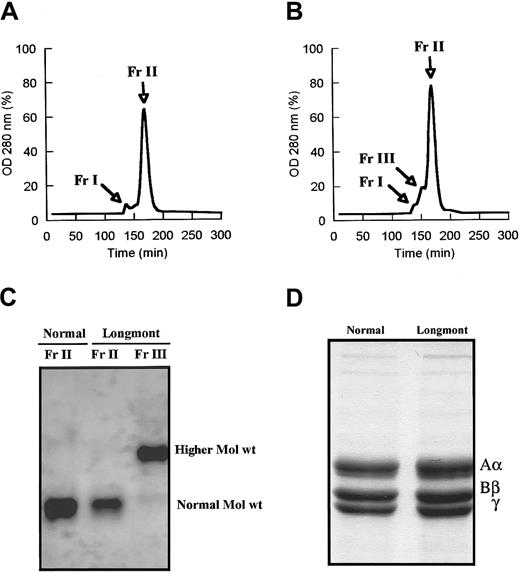
As shown by SDS-agarose gel analysis, fibrinogen Longmont contained a fraction of higher Mol wt molecules amounting to 23% ± 6% (densitometry) of the purified preparation (Figure1). The presence of larger molecules was confirmed by size-exclusion chromatography. Under conditions where normal fibrinogen showed 2 peaks, a minor peak (Fr I) corresponding to aggregates and a major peak (Fr II) corresponding to fibrinogen monomeric molecules (Figure 2A), fibrinogen Longmont displayed 3 peaks, the third novel peak (Fr III) having a shorter retention time (Figure 2B). SDS-agarose gel analysis of these fractions showed that fibrinogen Longmont contained 2 stable proteins: one was similar to normal fibrinogen and the second was with higher Mol wt (Figure 2C). When examined under reduced conditions, fibrinogen Longmont showed 3 polypeptide chains with similar electrophoretic migration to normal fibrinogen chains (Figure 2D). We concluded that the higher Mol wt fraction of fibrinogen Longmont consisted of normal Mol wt polypeptide chains.
Identification of a higher Mol wt species in fibrinogen Longmont.
Normal fibrinogen and fibrinogen Longmont along with mouse laminin were analyzed on 2% SDS-agarose gel. The gel was stained with Coomassie brilliant blue. Normal Mol wt and the higher Mol wt species are indicated.
Identification of a higher Mol wt species in fibrinogen Longmont.
Normal fibrinogen and fibrinogen Longmont along with mouse laminin were analyzed on 2% SDS-agarose gel. The gel was stained with Coomassie brilliant blue. Normal Mol wt and the higher Mol wt species are indicated.
Isolation of the higher Mol wt species of fibrinogen Longmont.
Normal fibrinogen (A) and fibrinogen Longmont (B) were run over a 1.6 × 51–cm column packed by TSK HW-55S gel, and the products eluted were monitored at 280 nm. The fraction Fr II from normal fibrinogen and the fractions Fr II and Fr III from fibrinogen Longmont were collected, concentrated, and analyzed on 2% SDS-agarose gel (C). (D) SDS-PAGE analysis of normal fibrinogen and fibrinogen Longmont polypeptide chains.
Isolation of the higher Mol wt species of fibrinogen Longmont.
Normal fibrinogen (A) and fibrinogen Longmont (B) were run over a 1.6 × 51–cm column packed by TSK HW-55S gel, and the products eluted were monitored at 280 nm. The fraction Fr II from normal fibrinogen and the fractions Fr II and Fr III from fibrinogen Longmont were collected, concentrated, and analyzed on 2% SDS-agarose gel (C). (D) SDS-PAGE analysis of normal fibrinogen and fibrinogen Longmont polypeptide chains.
Albumin binding to fibrinogen.
Immunoblotting of purified fibrinogen or the whole plasma did not show any albumin bound to fibrinogen Longmont (data not shown).
Plasmin protection assay.
In the presence of calcium ions or GPRP peptide, plasmin digestion of normal fibrinogen generates the fragments D1 and E, whereas in the presence of EDTA the C-terminal part of the γ chain is further degraded with fragment D1 transformed to D2 and D3.26 27In the presence of Ca++ ions or GPRP, plasmin digests of fibrinogen Longmont were similar to normal, except for an additional fragment with a Mol wt of approximately 200 kd (Figure3A). Immunoblotting analysis with an antihuman fibrinogen confirmed that this novel fragment was derived from fibrinogen (Figure 3B). In the presence of EDTA, the immunoblot showed 3 fragments larger than D1, presumably arising from further digestion of the 200-kd fragment (Figure 3B). We concluded from these data that (1) the 200-kd fragment may be a dimer of D1 and that (2) calcium ions bind to the high-affinity calcium-binding site while the GPRP peptide binds to the polymerization site “a.” Thus, we expected no change in the C-terminal part of the γ chain domain.
Fibrinogen Longmont is protected by both GPRP peptide and calcium ions against further digestion by plasmin.
Normal fibrinogen and fibrinogen Longmont (0.2 mg/m) were digested with 10 μg/mL plasmin in the presence of 5 mM EDTA, 5mM CaCl2, or 2 mM GPRP peptide. The plasmin digests were analyzed on 7.5% SDS-PAGE in nonreduced conditions. (A) The gel was stained with Coomassie brilliant blue. (B) A Western blot of the plasmin digests in the presence of 5 mM EDTA or 5mM CaCl2 using a polyclonal antibody to human fibrinogen.
Fibrinogen Longmont is protected by both GPRP peptide and calcium ions against further digestion by plasmin.
Normal fibrinogen and fibrinogen Longmont (0.2 mg/m) were digested with 10 μg/mL plasmin in the presence of 5 mM EDTA, 5mM CaCl2, or 2 mM GPRP peptide. The plasmin digests were analyzed on 7.5% SDS-PAGE in nonreduced conditions. (A) The gel was stained with Coomassie brilliant blue. (B) A Western blot of the plasmin digests in the presence of 5 mM EDTA or 5mM CaCl2 using a polyclonal antibody to human fibrinogen.
DNA sequence analysis.
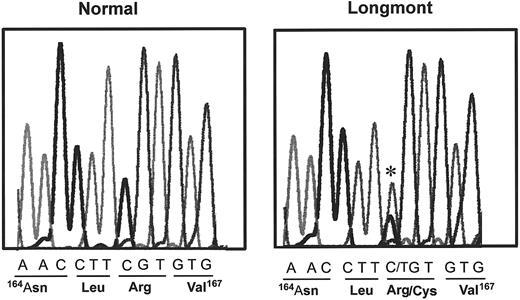
The presence of dimeric molecules of D1 suggested that the structural defect of fibrinogen Longmont is located in the D domain. Therefore, we determined the DNA sequence of PCR-amplified genomic DNA encoding the Bβ and γ chain segments within the D domain. The sequence of the γ chain was normal, but a single substitution (C→T) was identified in the exon IV of the Bβ chain (Figure4), changing Bβ 166Arg (CGT) to a Cys (TGT). The same mutation was identified in the mother of the proband, whereas no mutation was found in the father. Both the mother and the proband were heterozygous for the substitution.
Identification of a substitution in the Bβ chain gene of the proband.
DNA sequence of a part of the exon IV of the Bβ fibrinogen gene of the proband and a control. The mutation is indicated by a star, and the corresponding amino acid sequence of normal and the proband are indicated underneath the DNA sequences.
Identification of a substitution in the Bβ chain gene of the proband.
DNA sequence of a part of the exon IV of the Bβ fibrinogen gene of the proband and a control. The mutation is indicated by a star, and the corresponding amino acid sequence of normal and the proband are indicated underneath the DNA sequences.
Mass spectrometric analysis.
To identify the modifications associated with the neo-Cys residue, tryptic peptides isolated from fibrinogen Longmont were analyzed by mass spectrometry. After protecting the free sulfhydryl by vinylpyridine treatment, purified fibrinogen Longmont was cleaved with CNBr and the products separated on Sephadex G-50sf. N-terminal sequence analysis identified the relevant CNBr peptide Bβ119-190 sequence, Tyr-Leu-Leu-Lys-, in 2 peaks corresponding to the normal expected-sized peptide and a larger peptide. Approximately 22% of this sequence was found in the larger peptide. Assuming that half the Bβ chains contained Cys, we concluded that 22% of the abnormal chains were found in dimers and 28% in monomers.
Peptides isolated from these 2 peaks were digested with trypsin and examined by mass spectroscopy; the data are presented in Table1. The normal tryptic fragment Bβ His149-Arg166 was not present in the larger peptide, as expected if the disulfide bonds between fibrinogen molecules occurred between 2 neo-Cys residues. Following reduction of this peptide with mercaptoethanol, a larger tryptic fragment with mass 2443.1, corresponding to Bβ His149-Arg169, was found. The extension of the 3 residues (166-169) results from the replacement of Arg by Cys at position 166 and its subsequent resistance to trypsin digestion. The normal tryptic fragment Bβ His149-Arg166 was present in the trypsin digestion of the normal peak, consistent with the presence of normal Bβ chains. In addition, an abnormal fragment with mass 2561.8, corresponding to Bβ His149-Arg169 with a disulfide-linked to Cys residue, was found. Following reduction with mercaptoethanol, a new fragment corresponding to B149-169 with a sulfhydryl group was present. No component corresponding to B149-169 with an attached glutathione moiety or a fragment equivalent to the pyridylethylated form of B149-169 was found. We concluded that the normal CNBr peak contained only normal Bβ 149-166 and Bβ 149-169 with a disulfide-linked Cys residue. Thus, the neo-Cys residues in fibrinogen Longmont existed as disulfide-linked Cys and disulfide-linked oligomers between neo-Cys residues belonging to 2 abnormal Bβ chains. The oligomers are likely to be fibrinogen dimers, because the Mol wt of the higher Mol wt fraction of fibrinogen Longmont was less than 850 kd (Figure 1), which is consistent with the expected Mol wt of fibrinogen dimers (680 kd).
Mass spectrometric analysis of tryptic fragments from the Bβ-chain of fibrinogen Longmont
| Fragment corresponding to positions . | Calculated mass . | Observed mass . |
|---|---|---|
| 149-166, normal | 2127.3 | 2127.0 |
| 149-169, abnormal, with pyridylethylated Cys | 2547.9 | Not found |
| 149-169, abnormal, with additional, disulfide-linked Cys | 2561.9 | 2561.8 |
| 149-169, abnormal, with disulfide-linked glutathione | 2748.1 | Not found |
| 149-169, abnormal, after disulfide cleavage | 2442.8 | 2443.1 |
| Fragment corresponding to positions . | Calculated mass . | Observed mass . |
|---|---|---|
| 149-166, normal | 2127.3 | 2127.0 |
| 149-169, abnormal, with pyridylethylated Cys | 2547.9 | Not found |
| 149-169, abnormal, with additional, disulfide-linked Cys | 2561.9 | 2561.8 |
| 149-169, abnormal, with disulfide-linked glutathione | 2748.1 | Not found |
| 149-169, abnormal, after disulfide cleavage | 2442.8 | 2443.1 |
Fibrin polymerization studies
Turbidity measurements.
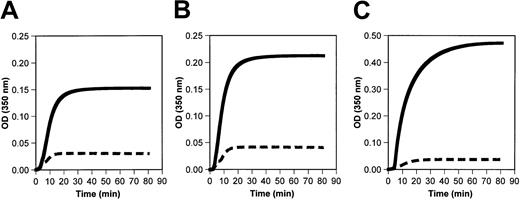
Polymerization of fibrinogen Longmont was markedly impaired in the presence or absence of calcium (Figure5A-B). Moreover, the polymerization of the normal Mol wt fraction (Fr II) of fibrinogen Longmont was impaired to a similar extent as the whole purified fibrinogen Longmont (Figure5C). These data indicated that the abnormal polymerization of fibrinogen Longmont is due to a structural defect present in the normal-sized fibrinogen molecules.
Fibrinogen Longmont polymerization is impaired.
Fibrin polymerization reaction was initiated by adding 0.05 NIH U/mL thrombin to 0.4 mg/mL normal fibrinogen (solid lines) or fibrinogen Longmont (dashed lines) in the absence of calcium (A) or in the presence of 10 mM calcium (B). (C) The fibrin polymerization curve of the Fr II (monomeric fraction) of normal fibrinogen and fibrinogen Longmont. Each curve is an average of 3 experiments.
Fibrinogen Longmont polymerization is impaired.
Fibrin polymerization reaction was initiated by adding 0.05 NIH U/mL thrombin to 0.4 mg/mL normal fibrinogen (solid lines) or fibrinogen Longmont (dashed lines) in the absence of calcium (A) or in the presence of 10 mM calcium (B). (C) The fibrin polymerization curve of the Fr II (monomeric fraction) of normal fibrinogen and fibrinogen Longmont. Each curve is an average of 3 experiments.
Fibrinopeptide release.
The HPLC elution profiles of FpA and FpB released on thrombin treatment of fibrinogen Longmont were indistinguishable from those of normal fibrinogen (profiles not shown). Moreover, the time course of thrombin-catalyzed FpA and FpB release was virtually identical to that found in the normal (data not shown).
Factor XIIIa–catalyzed cross-linking of fibrin.
Despite severely impaired fibrin polymerization, factor XIIIa–catalyzed cross-linking of fibrinogen Longmont proceeded normally. The formation of the γ dimers of fibrinogen Longmont was similar to normal and, as judged from the disappearance of the α chains, the α polymers were formed normally (data not shown). We concluded that fibrinogen Longmont formed protofibrils.
Static and dynamic light scattering.
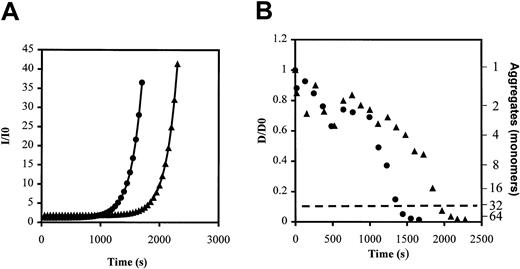
To study in more detail the kinetics of the protofibril formation, we used static and dynamic light scattering to monitor protofibril growth during the early step of the fibrin polymerization process. Kinetic analysis of time-dependent intensity changes (Figure6A) showed that Longmont protofibrils grew with a similar rate constant to normal fibrinogen (5.07 × 103 ± 0.46 × 103seconds−1 vs 6.02 × 103 ± 0.45 × 103seconds−1; P = .45). However, the lag time was significantly longer (12.32 × 103 ± 0.99 × 103 seconds vs 8.06 × 103 ± 1.101 × 103 seconds; P = .028). This delayed protofibril growth with fibrinogen Longmont was further documented by dynamic light-scattering measurements such as those shown in Figure 6B. Note that the protofibril time, the time required for a 10-fold decrease in translational diffusion coefficient (corresponding to the formation of elongated, rodlike structures indicated by dashed line in Figure 6B), was significantly longer for Longmont fibrinogen compared with normal. The mean protofibril time was 2.00 × 103 ± 0.28 × 103 seconds for Longmont fibrinogen versus 1.43 × 103 ± 0.31 × 103seconds for normal fibrinogen (P = .029). From these complementary indices of protofibril growth provided by static and dynamic light scattering, we concluded that thrombin treatment of fibrinogen Longmont leads to protofibril formation. The reaction takes a longer time to start, but the final product was similar in size to normal protofibrils.
Static and dynamic light scattering: thrombin treatment of fibrinogen Longmont leads to protofibril formation.
Thrombin (0.001 NIH U/mL) was added to purified Fr II of normal fibrinogen (closed circles) or of fibrinogen Longmont (closed triangles), and the intensity of the light scattered was monitored for 30 minutes. Representative curve of the intensity of light scattered (I/I0) over time (A); representative curve of the change in translational diffusion coefficient (D/D0) and the equivalent of the growing protofibrils size (second y-axis) over time (B). The dashed line indicates the formation of protofibrils.
Static and dynamic light scattering: thrombin treatment of fibrinogen Longmont leads to protofibril formation.
Thrombin (0.001 NIH U/mL) was added to purified Fr II of normal fibrinogen (closed circles) or of fibrinogen Longmont (closed triangles), and the intensity of the light scattered was monitored for 30 minutes. Representative curve of the intensity of light scattered (I/I0) over time (A); representative curve of the change in translational diffusion coefficient (D/D0) and the equivalent of the growing protofibrils size (second y-axis) over time (B). The dashed line indicates the formation of protofibrils.
Discussion
Sequence analysis of the proband's DNA identified a point mutation altering the codon CGT for BβArg166 to TGT coding for Cys. Both she and her mother were heterozygous for the variant gene. Because the post-translational nature of neo-Cys residues in abnormal fibrinogens is variable, we examined the nature of this neo-Cys. Mass spectrometric analysis of tryptic peptides of CNBr-cleaved fibrinogen Longmont demonstrated that this neo-Cys residue was entirely disulfide-bridged either to a free Cys amino acid or to another abnormal Bβ chain. The fraction of dimeric molecules, 22% to 23%, was the same whether measured by densitometry of SDS-agarose gels or quantitative amino acid sequence analysis. This indicates that the disulfide-bridged Bβ chains were intermolecular, forming dimers with 2 abnormal fibrinogen molecules, and we assume that the 78% of the remaining abnormal Bβ chain have to be in the monomeric fraction.
Previously, neo-Cys residues in abnormal fibrinogens have been found as free sulfhydryls, or disulfide-bridged to other molecules such as albumin or, as shown here, to Cys or a second abnormal fibrinogen molecule. Analysis of the neo-Cys in these dysfibrinogens is not predictive of the post-translational modification of any specific new Cys residue. Remarkably, although neo-Cys residues have been identified in all 3 chains, all abnormal fibrinogens to date with intermolecular, disulfide-bridged fibrinogen molecules have neo-Cys located in the Bβ chain. The neo-Cys residues in fibrinogens Fukuoka28 and IJmuiden29 are located in the N-terminal end of the Bβ chain, whereas in Osaka VI30 the neo-Cys is in the C-terminal end. Perhaps there is a feature of chain assembly that permits formation of intermolecular disulfides through the Bβ chains but not through the Aα or γ chains. Because the impact of the abnormal fibrinogen dimers was not examined in these cases, whether the dimeric molecules contributed significantly to the defective polymerization of these dysfibrinogens remains unknown.
Analysis of polymerization of fibrinogen Longmont, or only the monomeric fraction of fibrinogen Longmont, showed that protofibril formation was normal in that the rate of protofibril growth and the final protofibril size were similar to normal fibrinogen. We did find that the lag time was significantly longer with this variant, suggesting that short protofibrils of fibrinogen Longmont are less stable than normal. We interpret this result as suggesting that the substitution impairs a part of protofibril formation that is independent of the “A-a” interactions. Perhaps the recruitment and alignment of the abnormal molecules in a dimer or trimer is less favorable, such that the initiation of protofibrils is delayed. Nevertheless, the delay in protofibril formation alone cannot explain the impaired polymerization. Indeed, delayed protofibril formation, which was achieved by decreasing the thrombin concentration, leads to formation of a clot with thicker fibers, as evidenced by higher than normal turbidity.9 In contrast, the final turbidity with fibrinogen Longmont is lower than normal. We conclude that Longmont fibrin monomers polymerize via normal “A-a” interactions to form protofibrils, but the subsequent lateral aggregation of the Longmont protofibrils is impaired by the BβArg166 Cys-Cys substitution.
As shown in the crystal structure of the D domain, the residue BβArg166 is located in the coiled-coil region, between the globular E and D domains and far from the polymerization site “a.” Two other abnormal fibrinogens with changes in this region, fibrinogen Lima31 and Niigata,32 have been reported. In both, an extra carbohydrate is linked to a residue near the mutation, so the impaired polymerization in these cases is likely due to steric hindrance. Because removal of the abnormal disulfide-bridged molecules from fibrinogen Longmont did not correct the polymerization defect, the presence of Cys-Cys in the abnormal molecules must alter polymerization. Therefore, we conclude that the β chain within the distal part of the coiled-coil contains sites to mediate the formation of fibers from protofibrils.
Together, these data show clearly that the structural integrity of the Bβ chain within the distal part of the coiled-coil region is critical for fibrin polymerization, particularly for the association of protofibrils. We suggest that this critical region participates in lateral aggregation as proposed in the fibrin polymerization model of Fowler et al.5 These authors proposed a new type of interaction, DD-lateral (DD-lat), necessary for lateral association of the protofibrils. DD-lat contacts are thrombin independent and involve 2 D domains belonging to 2 distinct protofibrils. The DD-lat bonds must be relatively weak because there is no significant association between fibrinogen molecules. Nevertheless, in the association of protofibrils, several of these contacts will be present simultaneously so that bonds form in a cooperative fashion. The stability of the resulting fiber depends on the length of the protofibril, consistent with the increased number of DD-lat bonds. We propose that the neo-Cys-Cys at position Bβ166 residue interferes with the DD-lat contacts.
In summary, our data show that a single substitution of BβArg166 to Cys induced a dramatic effect on fibrin polymerization. This result points to a new region of the molecule that may play an important role in polymerization, specifically in the lateral association of protofibrils.
We gratefully acknowledge Dr D. Lavrinets for providing us blood samples, Dr J. Mullin for purifying plasminogen, and Dr J. Graham for helping in the preparation of the paper.
Supported in part by National Institutes of Health grant HL-31048, National Science Foundation grant MCB 9728122, and a fellowship from the Mid-Atlantic Affiliate of the American Heart Association, no. 0020166U.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Susan T. Lord, Dept of Pathology and Laboratory Medicine, University of North Carolina at Chapel Hill, Rm 603 Brinkhous-Bullitt Bldg, CB#7525, Chapel Hill, NC, 27599-7525; e-mail:stl@med.unc.edu.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal