Abstract
In multiple myeloma (MM), the growth of primary plasma cells depends not only on interleukin-6 (IL-6), but also on additional unidentified signals delivered by the bone marrow environment. Using Atlas complementary DNA (cDNA) arrays comprising 268 genes coding for intercellular signaling molecules, this study identified genes that are overexpressed in myeloma cells compared to autologous B-lymphoblastoid cell lines. These genes encode the oncogenic Tyro3 tyrosine kinase receptor, the heparin-binding epidermal growth factor–like growth factor (HB-EGF) that is an epithelial autocrine tumor growth factor, the thrombin receptor (TR) that is linked to HB-EGF and syndecan-1 processing and to cell invasion, chemokine receptors CCR1 and CCR2, the Wnt pathway actor Frizzled-related protein (FRZB), and the Notch receptor ligand Jagged 2. These data, obtained with the Atlas cDNA array, were confirmed by reverse transcriptase–polymerase chain reaction or protein analysis or both. Furthermore, Tyro3,HB-EGF, TR, and FRZB gene expression was documented in purified primary malignant plasma cells from patients with plasma cell leukemia or MM. HB-EGF and FRZB were poorly expressed in purified polyclonal plasma cells. Finally, HB-EGF was proved to be an essential autocrine growth factor for the XG-1 myeloma cells. This study shows the potency and the biologic relevance of cDNA arrays used to analyze simultaneously a large panel of intercellular signaling genes and, by identifying several genes overexpressed in malignant plasma cells, opens new fields of investigation in MM biology.
Introduction
Tumor development involves multiple mechanisms that lead to an increase in the survival of tumor cells and a deregulation of the cell cycle. Specifically, cell-to-cell growth signaling is likely to operate between tumor cells and their neighboring stromal cells.1 In multiple myeloma (MM), the tight interactions between malignant plasma cells and their medullar environment exemplify this oncogenic process. As recently reported, myeloma cells produce the angiogenic growth factors, vascular endothelial growth factor (VEGF) and fibroblast growth factor 2 (FGF2), which may promote the neovascularization of the bone marrow observed as the disease activity increases.2-4 Malignant plasma cells express adhesion molecules such as VLA4 and CD44, whose interactions with stromal cells and the extracellular matrix sustain MM cell survival.5-7Partly by producing interleukin-1 (IL-1), MM cells induce bone marrow stromal cells to synthesize IL-6, a major cytokine in MM biology.8-11 However, purified MM cells cannot be rescued, even by IL-6, and require the presence of stromal cells for in vitro survival.12-15 Thus, in the chronic phase of the disease, some molecules produced by stromal cells are critical, combined with IL-6, to promoting MM cell survival and proliferation. Growth factors able to bind heparan sulfate, such as FGFs, could be candidate molecules. Indeed, translocations involving the FGF receptor 3 are found in MM cells from patients with active disease and can confer an increase in the response of MM cells to IL-6 in vitro.16,17 Moreover, MM cells are the only cells in the bone marrow that express syndecan-1, a heparan sulfate proteoglycan that is a coreceptor for FGF.18,19 Insulin growth factors (IGFs) could also be involved because they are potent MM cell survival factors in vitro.20-22 However, the increasing number of growth factors makes it difficult to investigate each one individually. To analyze more comprehensively intercellular communication in MM cell biology, we have studied genes coding for cytokines, their receptors, and closely related genes using Atlas complementary DNA (cDNA) arrays on a panel of 5 human myeloma cell lines (HMCLs) whose growth depends on the addition of exogenous IL-6, one IL-6–sensitive HMCL, and purified primary myeloma cells from one patient. Gene expression in MM cells was compared to that of 4 lymphoblastoid cell lines (LCLs), obtained by Epstein-Barr virus (EBV) infection of polyclonal B cells from the patients with MM from whom these HMCLs were obtained. Differential gene expression analysis between these 2 cell types retrieves the major biologic differences that were previously documented between LCLs and HMCLs. Importantly, this method identifies novel genes that are overexpressed in HMCLs as well as in primary myeloma cells. These genes may be important in MM biology because they comprise an oncogenic tyrosine kinase receptor (Tyro3), a reported autocrine tumor growth factor (heparin-binding epidermal growth factor–like growth factor, HB-EGF), a cell invasion protease (thrombin receptor, TR) and Frizzled-related protein (FRZB), an actor in the Wnt pathway. Data obtained with Atlas cDNA arrays were confirmed by reverse transcriptase–polymerase chain reaction (RT-PCR) or protein analysis or both. Using RT-PCR and flow cytometry, we found expression of these genes in purified primary myeloma cells. In addition, we investigated whether these genes were expressed in purified primary polyclonal plasma cells (PPCs). Finally, it was demonstrated that HB-EGF overexpression had a functional significance because it played a crucial autocrine role in an HMCL that expressed a high level of this cytokine.
Materials and methods
Antibodies
Monoclonal antibodies (MoAbs) to TR (WEDE15), CD19, CD20, CD38, CD40, CD70, and immunoglobulin (Ig) κ and λ light chains, as well as isotype-matched control antibodies were from Immunotech (Marseille, France). Anti-CCR1 MoAb (MAB 145), anti-CCR2 MoAb (MAB 150), and anti–HB-EGF neutralizing antibodies (AF-259-NA) were from R&D Systems (Minneapolis, MN). The MI15 anti–syndecan-1 MoAb was obtained in our laboratory.18
Myeloma cells
The XG-1, XG-6, XG-7, XG-13, XG-14, and XG-16 HMCLs express cytoplasmic Ig and plasma cell antigens (Ag) (CD38 and syndecan-1, except XG-6, which is CD38−syndecan-1+), and lack the usual B-cell Ag (CD19 and CD20) (Table1). They are not infected with EBV. Characterization of XG-1 to XG-7, XG-13, and XG-14 has been reported elsewhere.23-25 XG-16 is a new HMCL obtained from the peripheral blood of a 51-year-old man presenting with relapsing plasma cell leukemia (PCL). Cell lines were grown in RPMI 1640 culture medium supplemented with 10% fetal calf serum (FCS) and 3 ng/mL IL-6 (a generous gift from Novartis, Rueil-Malmaison, France). All cell lines were free of Mycoplasma, as assayed by an enzyme-linked immunosorbent assay (ELISA) kit (Boehringer, Mannheim, Germany). For RNA experiments, primary MM cells from 4 patients with plasma cell leukemia (PCL1-4) and from 2 patients with myeloma (MM1-2) were analyzed. Bone marrow or peripheral blood was obtained after the patients provided informed consent and MM cells were purified (90%-98% purity) using the MI15 antisyndecan-1 MoAb as described.26 For flow cytometry analysis, mononuclear cells from bone marrow from 4 other patients with MM were used (MM3-6).
Comparative phenotypic analysis of myeloma cell lines and lymphoblastoid cell lines
| . | CD19 . | CD20 . | CD38 . | CD138 . | CD40 . | CD70 . | κ . | λ . | HLA . |
|---|---|---|---|---|---|---|---|---|---|
| HMCL | |||||||||
| XG-1 | 1.0 | 0.9 | 80.3 | 75.0 | 1.1 | 1.3 | 3.2 | 0.8 | A2 A29 B40 B44 |
| XG-6 | 1.7 | 1.1 | 1.1 | 357.3 | 7.6 | 1.2 | 0.9 | 43.0 | A2 A2 B15 B37 |
| XG-7 | 1.1 | 0.9 | 65.0 | 30.7 | 1.1 | 89.0 | 11.8 | 1.0 | A2 A24 B7 B40 |
| XG-13 | 0.8 | 0.9 | 38.9 | 110.5 | 9.5 | 6.2 | 0.8 | 36.1 | A32 A68 B8 B40 |
| XG-14 | 1.0 | 0.9 | 134.2 | 107.8 | 9.2 | 21.2 | 50.2 | 0.9 | A2 A26 B38 B51 |
| XG-16 | 1.0 | 1.1 | 21.6 | 178.1 | 5.3 | 15.4 | 27.4 | 1.1 | A2 A29 B40 B49 |
| Mean ± SD | 1.1 ± 0.3 | 0.9 ± 0.1 | 56.8 ± 47.4 | 143.2 ± 115.4 | 5.63 ± 3.8 | 22.3 ± 33.5 | |||
| LCL | |||||||||
| EBV-1 | 45.0 | 91.0 | 6.5 | 9.0 | 46.6 | 150.8 | 80.9 | 2.3 | A2 A29 B40 B44 |
| EBV-13 | 33.3 | 55.7 | 33.8 | 1.2 | 16.3 | 64.5 | 240.5 | 3.1 | A32 A68 B8 B40 |
| EBV-14 | 32.6 | 149.0 | 61.1 | 23.1 | 40.8 | 53.6 | 71.5 | 26.4 | A2 A26 B38 B51 |
| EBV-16 | 18.6 | 53.0 | 2.2 | 17.1 | 23.9 | 71.9 | 132.6 | 89.5 | A2 A29 B40 B49 |
| Mean ± SD | 32.4 ± 10.8 | 87.2 ± 44.7 | 25.9 ± 27.3 | 12.6 ± 9.5 | 31.9 ± 14.2 | 85.2 ± 44.4 |
| . | CD19 . | CD20 . | CD38 . | CD138 . | CD40 . | CD70 . | κ . | λ . | HLA . |
|---|---|---|---|---|---|---|---|---|---|
| HMCL | |||||||||
| XG-1 | 1.0 | 0.9 | 80.3 | 75.0 | 1.1 | 1.3 | 3.2 | 0.8 | A2 A29 B40 B44 |
| XG-6 | 1.7 | 1.1 | 1.1 | 357.3 | 7.6 | 1.2 | 0.9 | 43.0 | A2 A2 B15 B37 |
| XG-7 | 1.1 | 0.9 | 65.0 | 30.7 | 1.1 | 89.0 | 11.8 | 1.0 | A2 A24 B7 B40 |
| XG-13 | 0.8 | 0.9 | 38.9 | 110.5 | 9.5 | 6.2 | 0.8 | 36.1 | A32 A68 B8 B40 |
| XG-14 | 1.0 | 0.9 | 134.2 | 107.8 | 9.2 | 21.2 | 50.2 | 0.9 | A2 A26 B38 B51 |
| XG-16 | 1.0 | 1.1 | 21.6 | 178.1 | 5.3 | 15.4 | 27.4 | 1.1 | A2 A29 B40 B49 |
| Mean ± SD | 1.1 ± 0.3 | 0.9 ± 0.1 | 56.8 ± 47.4 | 143.2 ± 115.4 | 5.63 ± 3.8 | 22.3 ± 33.5 | |||
| LCL | |||||||||
| EBV-1 | 45.0 | 91.0 | 6.5 | 9.0 | 46.6 | 150.8 | 80.9 | 2.3 | A2 A29 B40 B44 |
| EBV-13 | 33.3 | 55.7 | 33.8 | 1.2 | 16.3 | 64.5 | 240.5 | 3.1 | A32 A68 B8 B40 |
| EBV-14 | 32.6 | 149.0 | 61.1 | 23.1 | 40.8 | 53.6 | 71.5 | 26.4 | A2 A26 B38 B51 |
| EBV-16 | 18.6 | 53.0 | 2.2 | 17.1 | 23.9 | 71.9 | 132.6 | 89.5 | A2 A29 B40 B49 |
| Mean ± SD | 32.4 ± 10.8 | 87.2 ± 44.7 | 25.9 ± 27.3 | 12.6 ± 9.5 | 31.9 ± 14.2 | 85.2 ± 44.4 |
Flow cytometry analysis of cell surface antigens and immunoglobulin light chains are expressed as ratios of the MFIs of labeling obtained with an antigen-specific antibody and an isotype-matched control antibody recognizing no human antigen. The specific antibody and control antibody were conjugated with the same fluorochrome or recognized by the same fluorochrome-conjugated second antibody. Because HMCLs did not express surface Ig, cells were permeabilized with saponin before labeling with anti–κ or –λ human Ig light chains to detect cytoplasmic Ig. HLA class I typing demonstrates that the HMCLs and the corresponding LCLs are derived from the same patient.
HMCL indicates human myeloma cell line; LCL, lymphoblastoid cell line; EBV, Epstein-Barr virus; MFI, mean fluorescence intensity; Ig, immunoglobulin.
EBV transformation of peripheral blood B cells
The EBV-1, EBV-13, EBV-14, and EBV-16 LCLs were obtained as described in the report by Commes and colleagues.27Supernatant containing transforming EBV was prepared from the B95-8 cell line cultured at 106 cells/mL in RPMI 1640 medium and 10% FCS for 72 hours. Culture supernatants were harvested, filtered at 0.45 μm and frozen at −70°C until use. Peripheral blood mononuclear cells (4 × 106) from patients with MM were cultured in 2 mL RPMI 1640 medium and 15% FCS to which 0.5 mL EBV supernatant was added. After 18 hours' incubation at 37°C in 5% CO2, cells were washed and resuspended in RPMI 1640 medium and 10% FCS, supplemented with 2 μg/mL cyclosporin A (Sigma, St Quentin Fallavier, France) and 10 ng/mL IL-10 (R&D Systems). Cultures were fed once a week by replacement of half of the culture medium with fresh RPMI 1640, FCS, cyclosporin A, and IL-10. After 7 weeks of culture, cyclosporin A and IL-10 were withdrawn. All cell lines were free of Mycoplasma. Immunophenotypic characteristics of the LCLs are summarized in Table 1.
PPCs
The PPCs were obtained from one patient with reactive plasmacytosis (RP), from 2 tonsil samples, and from the in vitro differentiation of peripheral blood B cells from 3 healthy donors.
The diagnosis of transient RP was established in a patient with drug allergy by cytologic identification and intracellular κ and λ Ig light-chain staining. Peripheral blood mononuclear cells were recovered by centrifugation on a Ficoll-Hypaque cushion and syndecan-1+ PPCs were purified using a MACS separation column (Miltenyi Biotec, Bergisch Gladbach, Germany). Briefly, PPCs were labeled with the MI15 antisyndecan-1 MoAb,18 washed, and incubated with rat antimouse IgG1 MACS microbeads. Cells were passed through a MACS LS separation column and cells retained by the column were flushed out. This selected cell population contained more than 95% syndecan-1+ cells and were purified PPCs as assayed by cytologic examination and Ig light-chain staining.
Tonsils, obtained from patients undergoing routine tonsillectomies at the University Hospital of Montpellier, were minced in buffered Hanks balanced salt solution (HBSS), and the resulting suspension was passed through a 100-μm nylon sieve. This cell population contained on average 1.5% of syndecan-1+ CD38+tonsil plasma cells (TPCs).
We also obtained PPCs by in vitro differentiation of peripheral blood B cells using a methodology we recently developed (manuscript in preparation).28 Peripheral blood B cells from healthy volunteers were purified using CD19 MACS microbeads (Miltenyi Biotec) and a MACS LS column according to the supplier's protocol. Purified B cells were first cultured for 4 days on mitomycin-treated L cells transfected with human CD40 ligand, in the presence of IL-2, IL-4, IL-10, and IL-12. Cells were then cultured for 2 additional days without CD40L transfectant and with IL-2, IL-10, IL-12, IL-6, and soluble IL-6 receptor. At this point, the cell population contained about 50% of cells lacking CD20 and expressing a high density of CD38. These cells were purified by high-speed cell sorting using a FACS Vantage Cell Sorter (Becton Dickinson, San Jose, CA). These purified cells (≥ 99% CD38+ CD20−) were pure plasma cells; they displayed a typical plasma cell morphology on cytologic examination, secreted high levels of polyclonal Ig, and lacked the B-cell markers CD20, CD21, and CD22.
RNA extraction
Cell lines were harvested during the exponential growth phase (5-7 × 105 cells/mL), washed in cold phosphate-buffered saline (PBS), and lysed immediately in 5 M thiocyanate guanidium for RNA extraction. DNA was sheared with a 21-gauge needle and LiCl was added to a final concentration of 3 M. RNA was allowed to precipitate overnight at 4°C and then centrifuged at 10 000g for 10 minutes. RNA was dissolved in TES (10 mM Tris HCl, pH7.5; 5 mM EDTA, 0.2% sodium dodecyl sulfate [SDS]) and then subjected to phenol/chloroform extraction. RNA for primary plasma cells was extracted using the RNeasy Kit (Qiagen, Valencia, CA), except PCL1, which was extracted by the thiocyanate guanidium-LiCl method. The RNA quality was verified by electrophoresis on a formaldehyde 1.2% agarose gel.
Isolation of poly(A) RNA and cDNA hybridization
For each cell line, RNA was prepared on 2 separate occasions and was mixed for each hybridization experiment. Poly(A) messenger RNA (mRNA) was purified through oligo (dT) beads (Dynal, Oslo, Norway) according to the manufacturer's instructions. Radiolabeled cDNA was prepared from each poly(A) RNA sample with the Atlas array kit (Clontech, Palo Alto, CA) by a reverse transcription in the presence of α-32P]dATP (ICN, Orsay, France), except that the Superscript II RT from Life Technologies (Cergy Pontoise, France) was preferred as the reverse transcriptase. The radiolabeled sample was then hybridized to a cytokine/receptor Atlas array nylon membrane (Clontech). After a high-stringency wash, membranes were exposed to a phosphor screen for 1 to 3 days and scanned at an 88 μm resolution using a 445SI PhosphorImager (Amersham Pharmacia Biotech, Saclay, France). A gene list, including array coordinates and GenBank accession numbers, is available at Clontech's Web site (http://www.clontech.com/atlas/genelists/index.html).
Gene array analysis
Quantification was performed with the AtlasImage 1.01 software (Clontech) using the default external background. Data from each different array experiment were normalized by the median value to eliminate the variability due to the sample labeling or to the exposure duration. The normalized median was arbitrarily given the value 100. We checked the validity of this normalization step by showing that gene expression quantification gave identical values when a blot was exposed for various times (data not shown). However, housekeeping genes such asGAPDH (array coordinate: 1D, Figure1) or β-actin (1G) produced very strong signals that saturated rapidly (< 12 hours) with the PhosphorImager detection apparatus, leading to artifactual values for these genes. Housekeeping genes were thus excluded from the analysis. Because the hierarchical clustering requires log transforming the data, we have added 1 to each value to take into account values initially equal to 0. At this step, values ranged from 1 to 78 757. A table displaying this data set is available on the authors' Web site (http://www.u475.montp.inserm.fr). Analysis was done using the Cluster and TreeView hierarchical clustering software developed by Eisen and coworkers29 (http://rana.lbl.gov/). We applied 2 filters. The first filter retained only genes whose expression was above the median value in at least 5 of the 11 samples. The second filter retained genes for which the difference between maximum and minimum values was above twice the median value. Data were log transformed (log base 2), genes were median centered, and clustered by correlation (uncentered) average linkage clustering. The hierarchical clustering was visualized with TreeView.
Expression of 268 genes coding for cytokines, cytokine receptors, and closely related proteins in the XG-14 HMCL and the EBV-14 LCL.
Poly(A)+RNA was extracted from each cell line and used to synthesize radiolabeled cDNA. Radiolabeled cDNAs were hybridized to 2 identical gene array membranes (Atlas Human Cytokine/Receptor Array, ClonTech) and radioactivity was analyzed with a PhosphoImager. Each gene is represented by duplicate spots. Filled arrows highlight several genes that are overexpressed in the myeloma sample, whereas open arrows point out the same genes in the LCL sample.
Expression of 268 genes coding for cytokines, cytokine receptors, and closely related proteins in the XG-14 HMCL and the EBV-14 LCL.
Poly(A)+RNA was extracted from each cell line and used to synthesize radiolabeled cDNA. Radiolabeled cDNAs were hybridized to 2 identical gene array membranes (Atlas Human Cytokine/Receptor Array, ClonTech) and radioactivity was analyzed with a PhosphoImager. Each gene is represented by duplicate spots. Filled arrows highlight several genes that are overexpressed in the myeloma sample, whereas open arrows point out the same genes in the LCL sample.
Western blot
Western blot was done as previously described, using a polyclonal goat anti-Tyro3 antibody (C-20, Santa Cruz Biotechnologies, Santa Cruz, CA) and a monoclonal anti-STAT3 antibody (S21320, Transduction Laboratories, Lexington, KY).30
RT-PCR
We generated cDNA with 2 μg total RNA using the Superscript II reverse transcriptase (Life Technologies) and oligo d(T)12-18 (Amersham Pharmacia Biotech) as primer. Each 25-μL PCR contained 1 μL of the first-strand cDNA, 1 μM of each primer (sense and antisense), 0.2 mM of each dNTP, 1.5 mM MgCl2, 1 × polymerase buffer, 2 U Taqpolymerase (Life Technologies), and 1 μCi α32P-dCTP (Amersham Pharmacia Biotech). The following primers were used: Tyro3 5′-CAC TGA GCT GGC TGA CTA AGC CCC (sense) and 5′-AAT GCA TGC ACT TAA GCA GCA GGG (antisense)31; HB-EGF 5′-TGG TGC TGA AGC TCT TTC TGG (sense) and 5′-GTG GGA ATT AGT CAT GCC CAA (antisense); FRZB 5′-AAG TCT GGC AGG AAC TCG AA (sense) and 5′-ACT TCC TGG TGC TTG ATT GC (antisense); β2-microglobulin (β2-M) 5′-CCA GCA GAG AAT GGA AAG TC (sense) and 5′-GAT GCT GCT TAC ATG TCT CG (antisense). The sizes of the PCR products were Tyro3, 344 bp; HB-EGF, 605 bp; FRZB, 599 bp; β2-M, 269 bp. The amplification profile was 1 minute at 94°C, 45 seconds at 59°C (Tyro3) or 62°C (HB-EGF) or 60°C (FRZB and β2-M) , 1 minute at 72°C, followed by a final extension of 10 minutes at 72°C. The cycle number was 26 for Tyro3, 32 for HB-EGF, and 25 for FRZB and β2-M. Reaction products were electrophoresed on a 4% polyacrylamide gel, dried, and exposed to x-ray films.
Proliferation assay for myeloma cell lines
Cells were washed once with RPMI 1640 culture medium, incubated for 5 hours at 37°C in RPMI 1640 culture medium and 10% FCS, and washed again twice. They were then cultured at 105 cells per well with 3 ng/mL IL-6 with or without graded concentrations of an anti–HB-EGF neutralizing antibody. In some culture groups, the anti–HB-EGF antibody was neutralized by adding an excess of recombinant HB-EGF (1 μg/mL) (259-HE-050, R&D Systems). Cells were cultured in triplicate in 96-well flat-bottomed microplates for 4 days. Tritiated thymidine (0.5 μCi, 25 Ci/mM, NEN, Paris, France) was added for the last 12 hours of culture and tritiated thymidine incorporation was determined as reported previously.30
Flow cytometry analysis
For the detection of TR or CCR1 or CCR2 chemokine receptors on HMCL and LCL cells, 0.5 × 106 cells were labeled with 0.5 μg of the specific murine MoAb for 30 minutes at 4°C in PBS containing 2% FCS. Cells were then washed and a fluorescein isothiocyanate (FITC)-conjugated goat antimurine antibody (dilution 1:50) was added for 30 minutes. After 2 additional washes, cells were suspended in PBS and fluorescence was analyzed with a FACScan flow cytometer (Becton Dickinson). A minimum of 10 000 viable cells, gated according to their forward/side scatter profile, was analyzed. In Tables 1 and 2, flow cytometry results are expressed as ratios of the mean fluorescence intensities (MFI ratio) of labeling obtained with an antigen-specific MoAb and an isotype-matched control MoAb. Expression of TR on normal and malignant primary plasma cells was assessed by 2-color flow cytometry using a phycoerythrin (PE) anti-TR and a FITC-conjugated MI15 antisyndecan-1 MoAb. A minimum of 50 000 viable cells, gated according to their forward/side scatter profile, were analyzed with a FACScan flow cytometer. Plasma cells were identified as the syndecan-1+ cell population within the region of viable mononuclear cells defined by the forward/side scatter profile.
Flow cytometric analysis of TR, CCR1, and CCR2 expression
| . | TR . | CCR1 . | CCR2 . |
|---|---|---|---|
| HMCL | |||
| XG-1 | 7.1 | 0.9 | 1.7 |
| XG-6 | 0.9 | 10.1 | 3.4 |
| XG-7 | 24.4 | 0.8 | 1.1 |
| XG-13 | 12.4 | 12.9 | 1.0 |
| XG-14 | 8.1 | 8.3 | 3.8 |
| XG-16 | 1.2 | 0.9 | 1.4 |
| LCL | |||
| EBV-1 | 0.8 | 0.9 | 1.0 |
| EBV-13 | 3.4 | 1.0 | 1.0 |
| EBV-14 | 0.9 | 0.9 | 1.1 |
| EBV-16 | 1.2 | 0.9 | 0.9 |
| . | TR . | CCR1 . | CCR2 . |
|---|---|---|---|
| HMCL | |||
| XG-1 | 7.1 | 0.9 | 1.7 |
| XG-6 | 0.9 | 10.1 | 3.4 |
| XG-7 | 24.4 | 0.8 | 1.1 |
| XG-13 | 12.4 | 12.9 | 1.0 |
| XG-14 | 8.1 | 8.3 | 3.8 |
| XG-16 | 1.2 | 0.9 | 1.4 |
| LCL | |||
| EBV-1 | 0.8 | 0.9 | 1.0 |
| EBV-13 | 3.4 | 1.0 | 1.0 |
| EBV-14 | 0.9 | 0.9 | 1.1 |
| EBV-16 | 1.2 | 0.9 | 0.9 |
Statisticalanalysis
The MFI ratios for CD40 and for CD70 in HMCLs and LCLs (Table 1) were compared with a Student t test.
Results and discussion
Gene expression and cluster analysis
We studied 6 HMCLs that were obtained as previously described.23,32 The growth of 5 cell lines (XG-1, XG-6, XG-13, XG-14, XG-16) depended strictly on the addition of exogenous IL-6; on removal of IL-6, cells progressively apoptosed within 5 to 10 days. The XG-7 HMCL that displayed a t(6;14) grew autonomously, but its growth was increased by adding IL-6. The 6 HMCLs did not express CD19 and CD20 B-cell antigens but expressed a high density of syndecan-1 (Table 1). They were not infected with EBV and produced either monoclonal κ or λ light chains. Although these HMCLs were obtained from patients with active disease, they were a good model for studying the biology of tumor stem cells that are present in patients with chronic disease in vivo because they still depended on the addition of exogenous cytokine to grow in vitro similarly to primary myeloma cells.8,12 It should be noted that they have been used to identify various genetic abnormalities involving the Ig loci, which were subsequently found at the same frequencies in the chronic or early phases of the disease.33 We also included purified primary myeloma cells (> 98% myeloma cells) from one patient (PCL1) with active disease. Gene expression in MM cells was compared to that of 4 LCLs obtained from MM patients whose tumor cells gave rise to the XG-1, XG-13, XG-14, and XG-16 HMCLs. LCLs were numbered according to their autologous HMCLs, respectively EBV-1, EBV-13, EBV-14, and EBV-16. These LCLs were obtained by EBV infection of polyclonal B cells. We have previously shown that these cell lines are polyclonal B-cell lines and are not related to the MM cell clone, even when they are generated from patients with terminal plasma cell leukemia.27Accordingly, these LCLs had a phenotype of polyclonal B cells and shared the same HLA class I antigens as their autologous HMCLs (Table 1).
The choice of autologous LCL cells as a point of comparison for gene expression in MM cells was supported by the following arguments: (1) plasma cells or normal B lymphocytes from patients or healthy individuals are ethically difficult to harvest in a sufficient amount to perform this RNA-consuming technique (50-100 × 106cells); (2) lymphoblastoid and MM cell lines have a common lymphoid B-lineage origin, thereby restricting differential expression that would only reflect a difference in tissue origin; (3) HMCLs and LCLs are both cell lines that eliminate differential gene expression mainly related to the high in vitro proliferation; (4) the possibility of obtaining tumor samples along with their autologous nonmyeloma B-lymphoid counterpart; (5) previous descriptions of biologic differences between LCLs and HMCLs that make it possible to validate the technique; and (6) an unlimited supply of cells, which is a key requirement for performing further protein or functional analyses.
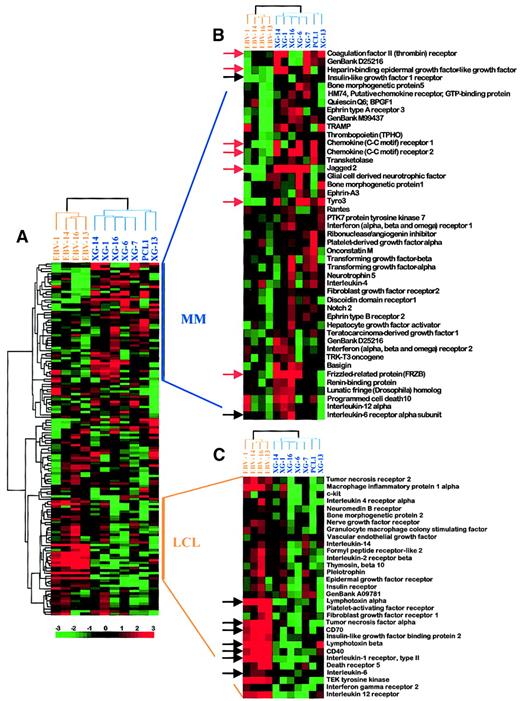
Figure 1 shows the hybridizations of the radiolabeled cDNA from the XG-14 HMCL and the EBV-14 LCL to 2 Atlas human cytokine/receptor array membranes, which comprise cDNAs of 268 genes. Arrows indicate several genes whose overexpression in the MM samples is manifest. Gene expression values from the 6 HMCLs, the primary MM sample, and the 4 LCLs were quantified and processed as detailed in “Materials and methods.” After removal of weakly expressed genes, 138 genes were analyzed using the hierarchical clustering software Cluster developed by Stanford University (Figure2A).29 This software groups genes by similarities in their pattern of expression over all samples and groups samples by similarities in their pattern of expression over all genes. The result is displayed as a matrix where each row represents a gene and each column a sample. The expression level of each gene is color-coded: the median expression across all samples is black, expression below the median is green, and expression greater than the median is red. The magnitude of the expression values is reflected by the degree of color saturation (see color scale). Remarkably, sample clustering according to their gene expression profile separated the HMCLs from the LCLs, assigning each one to a different tree, and genes overexpressed in MM cells or in LCLs were easily identified at first glance (Figure 2A, vertical bars on the right). HMCLs and LCLs originating from a single patient did not group together but segregated according to their cellular ontogeny, that is, malignant plasma cells versus lymphoblastoid cells. Thus MM cells displayed a common gene expression pattern that was different from that of LCLs and this allowed us to focus on genes whose expression was higher in either group.
Hierarchical clustering analysis.
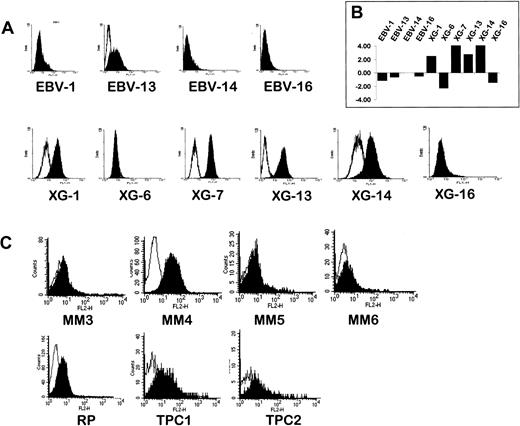
(A) Gene expression values were log transformed (log base 2) and median centered across each sample. Analysis was performed with Cluster and TreeView software. Each row represents a separate gene and each column a separate mRNA sample. The dendrogram at the left shows the relationship of the genes according to the similarity of their expression profile across samples and the dendrogram at the top depicts the relationship of the samples according to the similarity in their gene expression profile. Each value represents the difference from the gene median and is depicted according to the color scale shown at the bottom (−3 to +3 on a log base 2 scale). Vertical bars on the right indicate clusters of coordinately expressed genes in either myeloma samples or LCLs. (B) Enlargement of the MM cluster. Black arrows point out genes overexpressed in MM samples that were previously described in MM. Red arrows point out genes overexpressed in MM samples that were not previously linked to MM. (C) Enlargement of the LCL cluster. Black arrows point out genes overexpressed in LCL samples that were previously described in LCL. See our Web site for full data (http://www.u475.montp.inserm.fr/).
Hierarchical clustering analysis.
(A) Gene expression values were log transformed (log base 2) and median centered across each sample. Analysis was performed with Cluster and TreeView software. Each row represents a separate gene and each column a separate mRNA sample. The dendrogram at the left shows the relationship of the genes according to the similarity of their expression profile across samples and the dendrogram at the top depicts the relationship of the samples according to the similarity in their gene expression profile. Each value represents the difference from the gene median and is depicted according to the color scale shown at the bottom (−3 to +3 on a log base 2 scale). Vertical bars on the right indicate clusters of coordinately expressed genes in either myeloma samples or LCLs. (B) Enlargement of the MM cluster. Black arrows point out genes overexpressed in MM samples that were previously described in MM. Red arrows point out genes overexpressed in MM samples that were not previously linked to MM. (C) Enlargement of the LCL cluster. Black arrows point out genes overexpressed in LCL samples that were previously described in LCL. See our Web site for full data (http://www.u475.montp.inserm.fr/).
Gene expression pattern in LCL and MM samples is in agreement with their known biology
The biologic relevance of the Atlas array analysis was checked by looking at the cluster of genes overexpressed in LCLs (Figure 2C). Lymphotoxin-α, tumor necrosis factor-α, and IL-6 are produced by LCLs and contribute to autocrine growth loops, whereas these cytokines are known to be poorly produced or not produced by MM cells.34-39 Accordingly, these genes were found in the LCL cluster indicating that they were overexpressed in LCLs compared to HMCLs. The presence of IL-1R2 in the LCL cluster is also concordant with its reported lack of expression on MM cells and high expression on lymphoblastoid cells.40CD40 andCD70 genes, coding for 2 major cell membrane proteins involved in B-cell activation, were also overexpressed in LCLs compared to HMCLs (Figure 2C).41 In agreement with the Atlas array analysis, FACS analysis showed that the MFIs for CD40 or CD70 were significantly greater on LCLs than on HMCLs (P < .05), respectively 5.7 and 3.8 times (Table 1).
Among genes located in the MM cluster, we found the receptor of insulinlike growth factor (IGF1-R), again pointing out the relevance of this differential analysis (Figure 2B, black arrow). Indeed, IGF-1 has been reported as a potent survival and proliferation factor for MM cells.20-22 The IL-6 receptor was also located in the MM cluster although the differential expression between HMCLs and LCLs was weak (Figure 2B, black arrow), in agreement with the known expression of this receptor on both HMCLs and LCLs.23 42
The retrieval of some known biologic features of MM or LCLs with the Atlas array analysis provides a good validation of this technique.
Genes overexpressed in myeloma samples
As illustrated in Figure 2B, some genes show a clear overexpression pattern in MM samples (red arrows). These genes encode for proteins that have not been presently shown to be involved in MM biology. Their biologic significance is discussed below. However, by contrast to what is observed in LCLs, none of the 268 genes studied here has a strong overexpression in all MM samples. This may appear as a feature of MM and reflect the heterogeneity of the oncogenic events leading to this cancer. This heterogeneity has already been illustrated by the multiplicity of karyotypic abnormalities observed in MM as well as contrasting growth regulatory effects of cytokines on MM cell samples.33,43,44 Thus, although some biologic processes are common to most malignant plasma cells such as the survival and proliferation roles of IL-6,8 other features, in particular translocations, are only shared by subgroups of MM. One can speculate that the cDNA array will contribute in the near future to defining such subgroups.
Tyrosine kinase receptor Tyro3
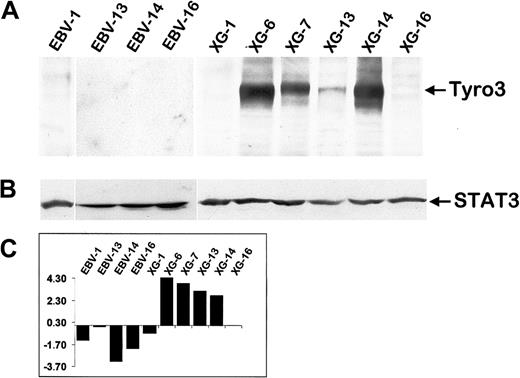
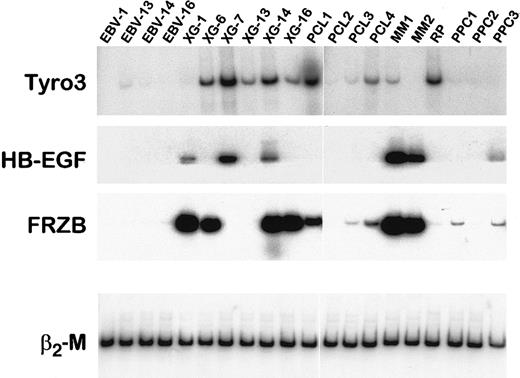
Among genes overexpressed in MM samples, Tyro3 (also termed Rse, Sky, Tif, orDtk) is particularly interesting because it encodes for an oncogenic receptor tyrosine kinase. According to the Atlas array data, the Tyro3 gene was overexpressed in 5 of 7 MM cell samples compared to LCLs (reds of increasing intensity, which represent values above median) (Figure 2B). Using Western blot analysis, we found that Tyro3 protein expression fit the Atlas array gene expression (Figure3). This tyrosine kinase receptor was strongly expressed in 4 of 6 HMCLs and was not detected in 2 HMCLs and 4 LCLs that poorly transcribed the Tyro3 gene. We assessed, using semiquantitative RT-PCR, the expression of Tyro3 in purified malignant plasma cells from 4 patients with PCL and 2 patients with myeloma, in purified plasma cells from one patient with RP, and in PPCs obtained by in vitro differentiation of B cells from 3 healthy individuals. Indeed, we recently developed a double-step culture system that enables the generation of a high number of CD38+ CD20− cells from peripheral blood B cells (manuscript in preparation).28 These CD38+ CD20− cells are genuine plasma cells that display a typical plasma cell morphology by cytologic examination, secrete high levels of polyclonal Ig, and lack the CD20, CD21, and CD22 B-cell markers. RT-PCR results for HMCL and LCL samples match well with those obtained with the Atlas arrays and the Western blot (Figure 4). In primary plasma cells,Tyro3 gene expression was detected in 2 of 4 PCL, 1 of 2 MM, and in 1 of 4 normal plasma cell samples (Figure 4). These observations suggest that Tyro3 might be important in the biology of normal and malignant plasma cells, and prompt us to further study its role in MM. Indeed, Tyro3 is a member of the Axl subfamily of receptor tyrosine kinase, which also comprises Axl (also termed Ufo or Ark) and Mer (also termed Eyk).45 The Axl gene was not expressed in MM samples (see the database on the Web site) and MER cDNA was not included in the Atlas array. These receptor tyrosine kinases are associated with neoplasia. Notably, Axl is overexpressed in leukemia and both Axl and Tyro3 can transform rodent fibroblast when overexpressed.46,47 The transformation process may be mediated by the PI-3 kinase-signaling pathway as demonstrated for Tyro3, and by STAT3 activation, as suggested by v-eyk mutant studies.48,49 It should be noted that the STAT3 pathway is critical in promoting MM cell survival.30,50 Because the ligand for Tyro3, Gas6 (growth arrest-specific), has been reported to be expressed in 4 of 4 HMCLs, an autocrine growth loop involving Tyro3 and Gas6 may be an important feature in MM biology.51Moreover, Gas6 is known to be produced by the bone marrow, the MM tumor cell environment.52 We will further investigate this point, in particular by obtaining recombinant Gas6, not yet commercially available.
Tyro3 protein expression in myeloma and LCL samples.
(A) An equal amount of protein extracts from exponentially growing cell lines were blotted with a specific anti-Tyro3 antibody. (B) As a loading control the same blot was stripped and reprobed with an anti-STAT3 antibody. (C) RNA expression of Tyro3 in the same set of cell lines as assessed by the Atlas array (log base 2 transformed and median centered). Western blots are representative of 2 independent experiments.
Tyro3 protein expression in myeloma and LCL samples.
(A) An equal amount of protein extracts from exponentially growing cell lines were blotted with a specific anti-Tyro3 antibody. (B) As a loading control the same blot was stripped and reprobed with an anti-STAT3 antibody. (C) RNA expression of Tyro3 in the same set of cell lines as assessed by the Atlas array (log base 2 transformed and median centered). Western blots are representative of 2 independent experiments.
RT-PCR analysis of Tyro3, HB-EGF, and FRZB mRNA in primary plasma cells.
RT-PCR analysis of Tyro3, HB-EGF, and FRZB was performed on cDNA from LCLs, HMCLs, purified primary malignant cells from patients with plasma cell leukemia (PCL1-4) or myeloma (MM1-2), and purified PPCs obtained from one patient with RP or by in vitro differentiation of peripheral blood B cells into plasma cells (PPC 1-3). Amplification of β2-M shows the equivalence of the cDNA loading and amplification. Data are representative of at least 2 independent RT-PCRs.
RT-PCR analysis of Tyro3, HB-EGF, and FRZB mRNA in primary plasma cells.
RT-PCR analysis of Tyro3, HB-EGF, and FRZB was performed on cDNA from LCLs, HMCLs, purified primary malignant cells from patients with plasma cell leukemia (PCL1-4) or myeloma (MM1-2), and purified PPCs obtained from one patient with RP or by in vitro differentiation of peripheral blood B cells into plasma cells (PPC 1-3). Amplification of β2-M shows the equivalence of the cDNA loading and amplification. Data are representative of at least 2 independent RT-PCRs.
HB-EGF
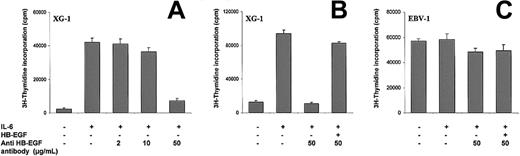
HB-EGF is another gene whose expression may be linked to MM pathobiology. Using the Atlas array we found that theHB-EGF gene was markedly overexpressed in 3 HMCLs (XG-1, XG-7, and XG-14) and in none of the 4 LCLs (Figure 2B). We failed to detect HB-EGF by Western blot, but this assay is poorly sensitive (detection limit 20 ng for 105 cells). We investigatedHB-EGF gene expression by RT-PCR in cell lines and primary cells. HB-EGF mRNA was detected in the XG-1, 7, and 14 HMCLs but in none of the 4 LCLs, confirming the Atlas array data (Figure 4). Interestingly, whereas HB-EGF mRNA could not be amplified by RT-PCR in purified malignant plasma cells from 4 of 4 cases of PCL, a strong expression was found in purified medullary cells from 2 patients with MM. In normal plasma cells, a dim expression was noted in 1 of 4 samples. HB-EGF is a growth factor produced either in a soluble form or as a transmembrane protein.53 This latter splicing variant has been found to be the diphtheria toxin receptor. HB-EGF is a ligand of the epidermal growth factor receptor (ErbB1) and ErbB4.54 It is produced by various tumor cells and acts as an autocrine tumor growth factor.53 The ErbB1gene, unlike the ErbB4 gene, was highly expressed in MM cells as well as in LCLs (see the database on the Web site), suggesting that it could also work as an autocrine growth factor in MM cells. We therefore investigated whether blocking HB-EGF activity could modulate the proliferation of the XG-1 MM cell line that highly expressed theHB-EGF gene. As outlined in Figure5A, adding a neutralizing antibody to HB-EGF blocked the proliferation of XG-1 in a dose-dependent fashion. With 50 μg/mL anti–HB-EGF antibody, inhibition was as high as 80%. This inhibitory effect was reversed by adding an excess of recombinant HB-EGF, demonstrating the specificity of the blocking effects of the antibody (Figure 5B). By contrast, the anti–HB-EGF antibody had no effect on the proliferation of the EBV-1 LCL (Figure 5C). Recombinant HB-EGF was unable to trigger the proliferation of XG-1 cells cultured without IL-6 (data not shown). These observations clearly show that HB-EGF is a novel growth factor that is involved in the IL-6 survival and proliferation of XG-1 myeloma cells. Further studies are required to elucidate the mechanism of action. It is noteworthy that in one prostate cancer cell line, the ErbB2 receptor can be phosphorylated after IL-6 stimulation and coimmunoprecipitated with the gp130 IL-6 transducer.55 Another interesting feature of HB-EGF is its ability to bind heparan sulfate.56 MM cells are the only cells in the bone marrow that express a high level of syndecan-1, a proteoglycan with heparan sulfate chains.18 Syndecan-1 could be an important coreceptor for HB-EGF by facilitating its presentation to membrane ErbB1. Altogether these results document an autocrine HB-EGF loop that controls the growth of XG-1 myeloma cells and a high HB-EGF gene expression in HMCLs and primary myeloma cells.
Effects of blocking HB-EGF in the XG-1 HMCL.
XG-1 (A,B) or EBV-1 (C) cells were cultured at 105cells/well for 4 days in RPMI 1640 medium with 10% FCS, IL-6 (3 ng/mL), and graded concentration of neutralizing anti–HB-EGF antibody (μg/mL). In some culture groups, recombinant HB-EGF (1 μg/mL) was used to reverse the inhibitory effects of the anti–HB-EGF antibody. Results are mean values ± SD of triplicate wells and are representative of 3 independent experiments.
Effects of blocking HB-EGF in the XG-1 HMCL.
XG-1 (A,B) or EBV-1 (C) cells were cultured at 105cells/well for 4 days in RPMI 1640 medium with 10% FCS, IL-6 (3 ng/mL), and graded concentration of neutralizing anti–HB-EGF antibody (μg/mL). In some culture groups, recombinant HB-EGF (1 μg/mL) was used to reverse the inhibitory effects of the anti–HB-EGF antibody. Results are mean values ± SD of triplicate wells and are representative of 3 independent experiments.
TR
The TR (also termed F2R or PAR1) is a member of the G-protein–coupled protease-activated receptors (PAR). Thrombin binds to TR and cleaves its amino-terminal exodomain, unmasking a new receptor amino terminus. This new amino terminus serves as a tethered peptide ligand that binds to and activates the receptor.57The Atlas array results show that TR was overexpressed in 5 of 7 MM samples and in none of the 4 LCLs (Figure 2B). TR protein expression was also investigated using an anti-TR MoAb and FACS analysis. Similarly to Tyro3, we found a close correlation between RNA and protein expression of TR in HMCLs and LCLs (Figure6A and Table 2). Among 4 primary myeloma cells tested for TR expression by 2-color flow cytometry, one sample (MM4) was clearly labeled by the anti-TR MoAb and one (MM6) was weakly labeled, whereas the 2 other samples were negative. We also investigated TR expression in normal plasma cells from one patient with RP and from the tonsils of 2 children by 2-color cytometry. TR was detected in the 3 primary normal plasma cells, though at a lower magnitude than in the HMCL and the MM4 sample (Figure 6C). TR was not expressed on tonsil normal B cells (data not shown). TR expression may have important implications for plasma cell biology. This receptor could be involved in the regulation of syndecan-1 expression on plasma cells. Indeed, syndecan-1 is induced when B cells differentiate into plasma cells and TR activation by thrombin induces syndecan-1 cleavage in vascular endothelial smooth muscle cells.58 Syndecan-1 is shed in a soluble form by myeloma cells and high levels of soluble syndecan-1 are found in patients with MM in association with a poor prognosis.59,60 TR activation could control syndecan-1 shedding in MM cells. Moreover, TR activation by thrombin is involved in the transactivation of ErbB1 by inducing membrane HB-EGF release and could therefore participate in the activation of the autocrine HB-EGF loop in the MM cells we have reported above.61 Finally, TR expression in tumor cells is associated with metastasis potential and could be a marker of disease activity in MM. Taken together, these observations suggest an important role for TR in normal and malignant plasma cells.62
Flow cytometry analysis of TR expression.
(A) Flow cytometry analysis of TR on HMCLs and LCLs. Cells were stained with an FITC-conjugated anti-TR MoAb (filled histograms) or an isotype-matched control antibody (open histograms). (B) RNA expression of TR in the same set of cell lines as assessed by the Atlas array (log base 2 transformed and median centered). (C) Flow cytometry analysis of TR in 4 primary myeloma cell samples (MM3-6), in PPCs from one patient with RP, and in TPC1 and 2. TR expression was assessed by 2-color flow cytometry using a PE anti-TR and an FITC-conjugated MI15 antisyndecan-1 MoAb. Filled histograms show the intensity of the TR staining in the plasma cell population identified as the syndecan-1+ cell population within the region of viable mononuclear cells defined by forward/side scatter profile. Open histograms show the staining by a PE-conjugated isotype-matched control MoAb in the plasma cell population.
Flow cytometry analysis of TR expression.
(A) Flow cytometry analysis of TR on HMCLs and LCLs. Cells were stained with an FITC-conjugated anti-TR MoAb (filled histograms) or an isotype-matched control antibody (open histograms). (B) RNA expression of TR in the same set of cell lines as assessed by the Atlas array (log base 2 transformed and median centered). (C) Flow cytometry analysis of TR in 4 primary myeloma cell samples (MM3-6), in PPCs from one patient with RP, and in TPC1 and 2. TR expression was assessed by 2-color flow cytometry using a PE anti-TR and an FITC-conjugated MI15 antisyndecan-1 MoAb. Filled histograms show the intensity of the TR staining in the plasma cell population identified as the syndecan-1+ cell population within the region of viable mononuclear cells defined by forward/side scatter profile. Open histograms show the staining by a PE-conjugated isotype-matched control MoAb in the plasma cell population.
Chemokine and chemokine receptors
Genes coding for the chemokine receptors CCR1 and CCR2 were overexpressed in MM cells (Figure 2B). FACS analysis confirmed an expression of CCR1 in 3 of 6 HMCLs and CCR2 in 4 of 6 HMCLs (Table 2). None of the 4 LCLs expressed CCR1 or CCR2. CCR1 is a receptor for RANTES, macrophage inflammatory protein (MIP)-1α, and MIP-1β, and CCR2 for the 4 monocyte chemotactic proteins (MCP) 1, 2, 3, and 4.63 MIP-1α and MCP-2 are produced in bone marrow by numerous cells, including monocytes, fibroblasts, and endothelial cells, and may be involved in the attraction of tumor cells close to bone marrow and may thus play a role in myeloma pathogenesis.63 Finally, we also found a strong expression of the CXCR4 gene (see on-line database) in both HMCLs and LCLs, and a high expression of CXCR4 was detected by FACS analysis on the 2 cell types (results not shown). The expression of theCXCR4 gene and protein in MM cells is in good agreement with homing of plasma cells to the bone marrow. Indeed, SDF1 is produced by the bone marrow environment and CXCR4 activation by SDF1 was shown to be critical in attracting hematopoietic stem cells and pre-B cells to the bone marrow environment.64,65 Interestingly, expression of CCR2 and CXCR4 on myeloma cells has been very recently confirmed by other groups.66 67
FRZB
The Wnt signaling pathway is evolutionarily preserved and plays a key role both in embryogenesis and oncogenesis. Involvement of this pathway in malignant cell proliferation regulation is best illustrated in colon cancer.68 Wnt binds to the 7-transmembrane–domain cell surface receptors of the Frizzled family, thereby activating a signaling cascade leading to the accumulation of β-catenin and the activation of the growth-promoting transcription factors of the TCF/LEF family. FRZB possesses a domain with a high degree of sequence identity and structural similarity with the ligand-binding domain of the Drosophila Frizzled. But FRZB is a secreted protein that competes with the membrane-bound Frizzled protein for Wnt binding and consequently acts as a competitive inhibitor of Wnt signaling.69 70 A high expression of FRZB distinguishes MM samples from LCLs because it is present in 4 of 7 cases of MM and no LCLs (Figure 2B). Semiquantitative RT-PCR confirms the Atlas array data and shows that this overexpression is also found in primary myeloma cells; FRZB is highly expressed in 2 of 4 cases of PCL and in 2 of 2 MM patients, whereas it is not present in any LCLs and barely detected in 2 of 4 of the normal plasma cells (Figure 4). It is surprising that an inhibitor of a transduction pathway that has been linked to cancer is overexpressed in neoplastic plasma cells, and therefore a crucial step in understanding the role of FRZB in MM will be to investigate whether the protein is expressed and is functional.
Other genes
Finally, other genes were found overexpressed in MM samples as compared to LCLs, such as Jagged 2, bone morphogenic protein (BMP) 1 and 5, ephrin EFNA3, and ephrin receptorsEPHA3 and EPHB2, as well as miscellaneous molecules such as Basigin, PCD10, TRAMP, and so forth. These genes may be potentially highly interesting in the field of myeloma and deserve further investigation.
Strong evidence has emerged recently to support a role for members of the Notch family in the regulation of hematopoiesis.71Interestingly, the gene coding for the Notch ligand, Jagged 2, was frankly overexpressed in 5 of 7 MM samples when compared to the LCL (Figure 2B). Notch 1 is widely expressed in many normal tissues, most abundantly in lymphoid tissues.72 In agreement, Notch 1 is expressed at the RNA level in both HMCLs and LCLs according to the macroarrays (see database on our Web site). Thus a homotypic signaling between MM cells expressing both the receptor and the ligand from the Notch family may be predicted. Given that Notch signaling usually leads to an inhibition of differentiation processes, and that overexpression of an activated allele of Notch 1 induces T-cell lymphoblastic leukemias and lymphomas, a constitutive activation of the Notch receptor may be involved in the blockage of plasma cell differentiation at the plasmablastic stage in MM as previously discussed.73-75 Moreover, because Notch family members are frequently expressed in hematopoietic lineage, a heterotypic signaling between the malignant plasma cell and its medullar environment cannot be excluded in vivo.71
Expression of BMPs, which have osteogenic potential, is at first glance a surprising finding in an osteolytic disease. But it may make sense given the tight relationship that MM cells weave with their medullary environment. Expression of Ephrin family members is also of interest because these genes encode for receptor tyrosine kinase involved in vascular development and can thus play a role in neoangiogenesis.
In conclusion, 8 years have gone by since our first attempt to study cytokine expression in MM.39 At that time Northern blot was used to analyze the expression of 9 different cytokine genes. The emerging technique of the cDNA array is now rapidly transforming gene expression analysis, allowing analysis of the expression of hundreds or thousands of different genes. Using a cDNA macroarray analysis, we have shown that malignant plasma cells overexpress several genes, which are involved in intercellular communication. These data suggest that several pathways, previously unrelated to MM biology, could be involved in the biology of normal or malignant plasma cells: Tyro3/Axl, ErbB, PAR, CCR, Wnt, and Notch families. When a protein study was possible, a tight correlation between RNA and protein levels was found. This approach presents one limitation: the relationship between gene expression and gene function is correlative, not causal. Therefore, the demonstration of a functional HB-EGF autocrine loop for HB-EGF in the XG-1 HMCL, which overexpresses this cytokine, is a compelling illustration of the pertinence of these cDNA array results.
We thank Bernard Pau and Naomi Taylor for providing access to their laboratory facilities and Eric Legouffe for providing the myeloma samples. We also gratefully acknowledge the technical assistance of Patrick Mensat for the cytometry analysis and Dan Wang for the radioactive RT-PCR.
Supported by grants from the Ligue Nationale Contre le Cancer, Paris, France.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Bernard Klein, INSERM U475, 99 Rue Puech Villa, 34197 Montpellier Cedex 5, France; e-mail: klein@montp.inserm.fr.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal