Little is known about the molecular characteristics of alloantigens recognized by alloreactive T cells mediating hematologic stem cell graft rejection. In particular, it has never been shown that such alloantigens can be encoded by HLA-DPβ alleles. Indeed, matching for HLA-DP antigens is generally not considered to be of functional importance for the outcome of allogeneic bone marrow or peripheral blood stem cell transplantation. In this study, a case of peripheral blood stem cell allograft rejection was investigated in which the patient and donor differed for a single mismatch at HLA-DP in the rejection direction. Patient-derived T lymphocytes circulating at the time of rejection showed direct ex vivo cytotoxic activity against donor-derived B-lymphoblastoid cells as well as other HLA-DPβ1*0901–expressing targets. The presence of HLA-DPβ1*0901–specific effectors in vivo was further confirmed by in vitro stimulation experiments. CD4+ T-cell lines and clones with specific cytotoxic activity against HLA-DPβ1*0901–expressing targets including donor B-lymphoblastoid cells were generated both by nonspecific and by donor-specific in vitro stimulation. Taken together, these data demonstrate that HLA-DP can be the target antigen of cytotoxic CD4+ T lymphocytes involved in peripheral blood stem cell allograft rejection.
Introduction
Allogeneic transplantation of bone marrow or peripheral blood stem cells (PBSCs) is increasingly being used as a therapeutic approach for a variety of diseases, including hematologic malignancies, solid tumors, and inborn genetic defects. One of the main caveats of hematologic stem cell transplantation is the histocompatibility barrier between patient and donor. In fact, 2 severe clinical complications, graft-versus-host (GVH) disease and graft rejection, are frequently caused by alloreactive T lymphocytes specific for nonself major or minor histocompatibility antigens. GVH disease is one of the major causes of death after hematologic stem cell transplantation, with an incidence of up to 50%. The risk of graft failure or rejection is about 1% to 6% in transplants from HLA-matched unrelated donors.1 Previous studies have shown that alloreactive T cells causing GVH disease or rejection can recognize as little as a single amino acid difference between patient and donor major histocompatibility antigens.2,3 In line with these findings, it is now generally accepted that molecular matching for HLA class I A and B and class II DRβ1 and DQβ1 alleles is strongly correlated with the outcome of transplantation.4-10 For the HLA class II locus, the role of HLA-DP is still a matter of debate. Initial studies did not detect a correlation between HLA-DP matching and patient survival or risk of GVH disease.11 Maybe due to improved HLA typing techniques, this view has recently been challenged by the observation that HLA-DPβ but not HLA-DPα matching was correlated with the risk of severe GVH disease and patient survival in a different patient cohort.12 In concordance with this, HLA-DPβ–specific CD4+ T cells have been isolated from the skin of patients presenting with acute GVH disease in 2 studies.13-15 The role of HLA-DP for graft rejection is currently unknown.
In the present study, we have investigated a case of PBSC allograft rejection in which the patient and donor differed for a single mismatch at HLA-DP in rejection direction. Donor-specific cytotoxic T lymphocytes recognizing the mismatched HLA-DPβ antigen were found to be circulating in the patient during the time of rejection. These data demonstrate that HLA-DPβ can be the target of alloreactive T lymphocytes involved in rejection of hematologic stem cell transplantation in vivo.
Patient, materials, and methods
Case report
Patient.
A 28-year-old female affected by Philadelphia chromosome–positive chronic myeloid leukemia (CML) diagnosed in February 1997 was treated first with hydroxyurea (Teofarma, Pavia, Italy) and cytarabine (Pharmacia and Upjohn, Milan, Italy). Her blood counts stabilized to normal values, and at the time of PBSC transplantation she was still in the first chronic phase.
PBSC transplantation.
An extended family search detected the patient's 56-year-old father as a possible donor. High-resolution HLA class I and II genomic typing (indicated for the patient and donor in Table1) was performed by standard polymerase chain reaction (PCR) sequence–specific oligonucleotide probing according to protocols of the XIth and XIIth International Histocompatibility Workshop.16,17 A cytotoxic T-lymphocyte precursor frequency (CTLpf) assay was performed in the host-versus-graft (HVG) direction and mixed lymphocyte culture in both HVG and GVH directions according to standard protocols described elsewhere.18 19
HLA typing of patient, donor, and allogeneic B-lymphoblastoid cell lines used in this study
| BLCL . | A* . | B* . | Cw* . | DRβ1* . | DQβ1* . | DPα1* . | DPβ1* . |
|---|---|---|---|---|---|---|---|
| Patient | 0201/3001 | 3901/4901 | 0701/1203 | 0701/1601 | 0202/0502 | 0103 | 0201/0402 |
| Donor | 3001/3001 | 3901/4901 | 0701/1203 | 0701/1601 | 0202/0502 | 0103/0201 | 0201/0901 |
| OMW | 0201 | 1401/4501 | 1601 | 1301 | 0603 | 0201 | 0101 |
| AKIBA | 2402 | 5201 | 1202 | 1502 | 0601 | 0201 | 0901 |
| LG-2 | 2402/3201 | 3503/4403 | 0401 | 0701/1401 | 0202/0503 | 0103 | 0301/0401 |
| BLCL . | A* . | B* . | Cw* . | DRβ1* . | DQβ1* . | DPα1* . | DPβ1* . |
|---|---|---|---|---|---|---|---|
| Patient | 0201/3001 | 3901/4901 | 0701/1203 | 0701/1601 | 0202/0502 | 0103 | 0201/0402 |
| Donor | 3001/3001 | 3901/4901 | 0701/1203 | 0701/1601 | 0202/0502 | 0103/0201 | 0201/0901 |
| OMW | 0201 | 1401/4501 | 1601 | 1301 | 0603 | 0201 | 0101 |
| AKIBA | 2402 | 5201 | 1202 | 1502 | 0601 | 0201 | 0901 |
| LG-2 | 2402/3201 | 3503/4403 | 0401 | 0701/1401 | 0202/0503 | 0103 | 0301/0401 |
OMW is on HLA-typed BLCL from the XIth and XIIth International Histocompatibility Workshop. LG-2 as well as patient and donor BLCLs were HLA-typed by standard PCR sequence–specific oligonucleotide probing as described in “Patient, materials, and methods.”
BLCL indicates B-lymphoblastoid cell line.
Indicates genomic denomination of HLA alleles determined by molecular typing.
Conditioning regimen.
The patient was treated with busulfan (GlaxoWellcome Segix, Pomezia, Italy), 16 mg/kg administered over 4 days; cyclophosphamide (Asta Medica, Milan, Italy), total dose 120 mg/kg administered over 2 days; and antithymocyte globulin (Imtix, Milan, Italy), 5 mg/kg/d administered for 4 days before transplantation.
Characteristics of the graft.
Donor CD34+ cells were mobilized by 5 days of administration of granulocyte colony-stimulating factor (Rhone Poulenc, Lyon, France), 15 μg/kg/d, and harvested by leukapheresis. CD34+ cells were selected by immune affinity column and further depleted of CD4+ and CD8+ T cells. The patient received 5 × 106/kg CD34+ cells with 2.5 × 105/kg CD3+ T cells (<1%).
Evolution.
White blood cell (WBC) count was more than 1 × 109/L (1000/μL) (neutrophils 95%) on day +17. On the same day, platelets were at 34 × 109/L (34 000/μL) with no need for transfusion support. No further platelet reconstitution was observed until death. On day +40, the WBC count was still 2 × 109/L (2000/μL) but with 30% CD3+ T lymphocytes, which rose to 98% by day +45. A bone marrow aspirate performed on day +45 showed general hypoplasia with lymphocytic and plasmacellular infiltration. The patient's peripheral blood had converted to a host-phenotype by immunofluorescence staining of peripheral blood mononuclear cells (PBMCs) with an HLA-A2–specific monoclonal antibody (mAb) (see below). An attempt to reverse acute rejection by administration of antithymocyte globulin, steroids, and cyclophosphamide was unsuccessful. On day +51, a second transplant using autologous PBSCs (3.8 × 106/kg CD34+) mobilized by granulocyte colony-stimulating factor during the chronic phase of the disease was administered. WBC and platelet counts at the time of the second transplantation were 0.6 × 109/L (600/μL) (with no neutrophils) and 7 × 109/L (7000/μL), respectively. The second transplant did not result in evidence of WBC or platelet engraftment until the patient's death. The patient expired with multiple organ failure on day +70 after the first transplantation, with the WBC count at 0.4 × 109/L (400/μL) (no neutrophils) and platelet count at 31 × 109/L (31 000/μL).
In vitro culture of PBMCs circulating at the time of graft rejection and T-cell cloning.
PBMCs were isolated by Ficoll (Nycomed Pharma, Milan, Italy) gradient centrifugation from a heparinized peripheral blood sample obtained from the patient at day +45 after PBSC transplantation and frozen in 10% dimethyl sulfoxide (CH3SOCH3) in liquid nitrogen until further use. For in vitro analysis, the PBMCs were thawed and plated in culture medium consisting of Iscoves modified Dulbecco medium (BioWhittaker, Milan, Italy), antibiotics, and 10% pooled human serum supplemented with 100 U/mL interleukin-2 (IL-2) (Chiron Italia, Milan, Italy). Cells were kept for 24 hours in a 5% CO2 atmosphere in 24-well plates (Corning, Corning, NY) at 2 × 106/mL. Subsequently, they were either used directly for in vitro functional assays (see below) or subjected to in vitro restimulation. To obtain a T-cell line, 5 × 105 PBMCs were plated in a 48-well plate (Corning) in culture medium in the presence of 100 U/mL IL-2 and 106 irradiated (30 Gy [3000 rad]) donor PBMCs. To generate a panel of clones, PBMCs were plated at 0.3 or 1 cell per well in 96-microwell round-bottomed culture plates (Corning) in culture medium supplemented with 100 U/mL IL-2 and a pool of irradiated (30 Gy [3000 rad]) allogeneic feeder PBMCs (105/well); irradiated (100 Gy [10 000 rad]) allogeneic feeder cell line LG-2 (2 × 104/well); an Epstein-Barr virus–transformed B-lymphoblastoid cell line (BLCL) kindly provided by Dr Thierry Boon, Ludwig Institute for Cancer Research, Brussels, Belgium (Table 1 shows HLA typing); and 1 μg/mL phytohemagglutinin (PHA; Roche, Milan, Italy). In addition, antigen-specific limiting dilution cloning was performed using irradiated (100 Gy [10 000 rad]) donor BLCLs (2 × 104/well) and the allogeneic feeder BLCL LG-2 (3 × 104/well).
Flow cytometry and mAb.
T cells were stained in 2-color immunofluorescence using the following phycoerythrin (PE)– or fluorescein isothiocyanate (FITC)-conjugated mAbs (Becton Dickinson, Erembodegem, Belgium): CD4-PE, CD8-FITC, CD3-PE, CD56-FITC, CD19-PE. The HLA-A2–, HLA-A69–specific mAb BB7.2 20 was used in a 2-step staining with goat-antimouse FITC mAb (Becton Dickinson). Labeled cells were analyzed on a FACScan (Becton Dickinson).
In vitro functional assays
Cytotoxicity assays.
The cytotoxic activity of effector T lymphocytes was analyzed in a standard chromium51 (51Cr) release assay as described in detail elsewhere.21 Briefly, 100051Cr-labeled (NEN Dupont, Milan, Italy) target BLCLs were incubated for 4 hours with effector cells at various effector:target (E/T) ratios at 37°C in a 5% CO2 atmosphere. Subsequently, the supernatant was removed and counted on a γ-scintillation counter. Percentage of specific release was calculated according to the formula: 100 × (51Cr release into supernatant − spontaneous release)/(total release into detergent − spontaneous release). Samples were tested in duplicate, and the mean value of both duplicates is indicated.
Cloning of the full-length HLA-DPβ1*0901 cDNA.
The full-length complementary DNA (cDNA) coding region of HLA-DPβ1*0901 and HLA-DPβ1*0201 was PCR-amplified from reverse-transcribed cDNA obtained from the donor BLCL. The HLA-DPβ–specific sense primer 5DPβ-COD was derived from base pair (bp) −18 in the 5′ untranslated region to bp +8 in the coding region of HLA-DPβ alleles22 and contained an EcoRI site in the 10 bp 5′ tail. The reverse primer 3DPβ-COD was derived from bp +755 to the TAA stop codon at bp 783 of HLA-DPβ alleles and contained a XhoI site in the 10 bp 5′ tail. The sequences were as follows: 5DPβ-COD 5′-TACGGAATTCGCCATCCTTTTCCAGCTCCATGATGGT-3′ and 3DPβ-COD 5′-CATGCTCGAGTTATGCAGATCCTCGTTGAACTTTCTTG-3′. The PCR product was digested by EcoRI/XhoI (Roche) and cloned into the corresponding restriction sites of the retroviral vector LXSΔN.23 The 2 HLA-DPβ alleles were discriminated by digestion with ScaI, which cuts at bp +281 of the DPβ1*0201 but not of the DPβ1*0901 cDNA. An HLA-DPβ1*0901 clone designated LDPβ9SΔN was sequenced on both strands by automated sequencing using the dye primer method (Primm, Milan, Italy).
Retroviral vector-mediated transfer of HLA-DPβ1*0901 into a BLCL.
An amphotropic packaging cell line for LDPβ9SΔN was generated as described elsewhere.23 BLCLs were transduced by 48 to 72 hours of coculture with 104 irradiated (100 Gy [10 000 rad]) AM12 packaging cells in the presence of 0.8 mg/mL polybrene (Abbott Laboratories, Irving, TX). Transduced cells were purified by magnetic immunobead selection using the human nerve growth factor receptor–specific mAb 20.4 as described.23
Results
Patient and donor matching
PBSC transplantation for CML in the chronic phase was performed on a patient using a haploidentical family donor who was matched at the molecular level for HLA-B, -C, -DRβ, -DQα, and -DQβ alleles also on the unrelated chromosome. At the HLA-A locus, there was a single mismatch in GVH direction, with the patient expressing HLA-A*0201/A*3001 while the donor was homozygous for HLA-A*3001 (Table 1). This allowed tracing of patient-derived cells by immunofluorescence staining with an HLA-A2–specific mAb. At the HLA-DP locus, the patient and donor were mismatched for both HLA-DPα and HLA-DPβ, with DPα1*0201 and DPβ1*0901 being the donor-specific HLA-DP alleles not shared by the patient (Table 1). Results of the mixed lymphocyte culture were 4.7% in the GVH direction and 11.3% in the HVG direction. The CTLpf assay in the HVG direction revealed a donor-specific precursor frequency of about 1:105(data not shown). This value is considered to be borderline positive and has previously been reported to be at the lower limit of CTLpf values associated with poor transplantation outcome when present in GVH direction.4
Characterization of patient PBMCs circulating during the time of graft rejection
Around day +40 after PBSC transplantation, a rise in patient-derived CD3+ T lymphocytes was observed concomitantly with a loss of donor-derived neutrophils. PBMCs were obtained by Ficoll density separation from a blood sample drawn from the patient on day +45 after transplantation and are referred to hereafter as PBMCs–ex vivo. As demonstrated by immunofluorescence staining, most of these PBMCs were CD3+ T lymphocytes with a CD8/CD4 ratio of 9:1 (Table 2). Most of these cells were of patient origin, because they stained positive for the HLA-A2 antigen (Table 2). The T-cell receptor–Vβ repertoire of these T lymphocytes was analyzed by reverse transcription PCR and found to be highly polyclonal, with representation of almost all Vβ specificities (data not shown).
Immunophenotype, as determined by FACS analysis, of patient peripheral blood mononuclear cells (PBMCs) circulating at day +45 after PBSC transplantation (PBMC–ex vivo) or after a single round of in vitro stimulation with donor PBMC (TCL + 1)
| Responder . | No. st . | Positive cells, % . | |||||
|---|---|---|---|---|---|---|---|
| CD3+ . | CD4+ . | CD8+ . | CD56+ . | CD19+ . | HLA-A2+ . | ||
| PBMC–ex vivo | 0 | 88.7 | 9.4 | 80.2 | 4.7 | 0 | 86.7 |
| TCL + 1 | 1 | ND | 98.6 | 0.5 | ND | ND | 97.9 |
| Responder . | No. st . | Positive cells, % . | |||||
|---|---|---|---|---|---|---|---|
| CD3+ . | CD4+ . | CD8+ . | CD56+ . | CD19+ . | HLA-A2+ . | ||
| PBMC–ex vivo | 0 | 88.7 | 9.4 | 80.2 | 4.7 | 0 | 86.7 |
| TCL + 1 | 1 | ND | 98.6 | 0.5 | ND | ND | 97.9 |
No. st indicates the number of in vitro stimulations; ND, not done.
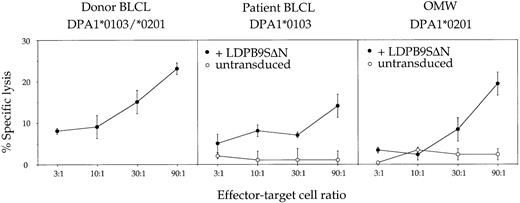
PBMCs–ex vivo were used without further in vitro restimulation in a cytotoxicity assay against donor- or patient-derived target BLCLs. Weak but specific cytotoxic activity against donor but not against patient BLCLs was observed (Figure 1). Transduction of patient BLCLs with the retroviral vector LDPβ9SΔN encoding the HLA-DPβ1*0901 cDNA was sufficient to allow recognition, demonstrating that cytotoxic activity was at least in part directed against the donor-specific HLA-DPβ allele. The donor-specific DPα allele was not recognized on an allogeneic BLCL sharing only DPα1*0201 with the donor cells (Figure 1). Finally, the epitope recognized on HLA-DPβ1*0901 was not dependent on the nature of the associated DPα chain, because BLCLs homozygous for either DPα1*0103 or DPα1*0201 were recognized after transduction with LDPβ9SΔN (Figure 1).
Functional characterization of patient PBMCs circulating at day +45 after PBSC transplantation (PBMCs–ex vivo).
Effector cells were used ex vivo without further in vitro resimulation and tested at various effector-target ratios in a standard 4-hour chromium release assay using donor BLCLs, patient BLCLs, or the allogeneic BLCL OMW as targets. The latter 2 BLCLs were used untransduced (○) or transduced (●) with retroviral vector LDPβ9SΔN encoding HLA-DPβ1*0901. For complete HLA typing of BLCLs, see Table 1. Values are means of duplicates, with error bars indicating SDs.
Functional characterization of patient PBMCs circulating at day +45 after PBSC transplantation (PBMCs–ex vivo).
Effector cells were used ex vivo without further in vitro resimulation and tested at various effector-target ratios in a standard 4-hour chromium release assay using donor BLCLs, patient BLCLs, or the allogeneic BLCL OMW as targets. The latter 2 BLCLs were used untransduced (○) or transduced (●) with retroviral vector LDPβ9SΔN encoding HLA-DPβ1*0901. For complete HLA typing of BLCLs, see Table 1. Values are means of duplicates, with error bars indicating SDs.
Isolation of a donor-specific T-cell line
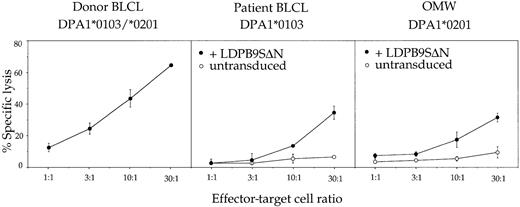
PBMCs–ex vivo were stimulated once in vitro with irradiated donor PBMCs in the presence of exogenous IL-2. After 30 days in culture, an oligoclonal T-cell line referred to hereafter as TCL+1 was obtained that contained more than 95% CD4+ T cells (Table 2). These CD4+ T cells showed the same target specificity as PBMCs–ex vivo, ie, they recognized HLA-DPβ1*0901 in association with either HLA-DPα1 or HLA-DPα2 on donor BLCLs as well as on LDPβ9SΔN-transduced patient-derived or allogeneic BLCLs (Figure2). However, the absence of 1 of the 2 HLA-DPα chains considerably reduced the efficiency of recognition, suggesting the presence in the T-cell line of clones with differential affinity for heterodimers of HLA-DPβ1*0901 with either of the 2 HLA-DPα chains.
Functional characterization of patient PBMCs after a single round of in vitro stimulation with irradiated donor PBMCs (TCL+1).
T cells were tested at various effector-target ratios in a standard 4-hour chromium release assay using donor BLCLs, patient BLCLs, or the allogeneic BLCL OMW as targets. The latter 2 BLCLs were used untransduced (○) or transduced (●) with retroviral vector LDPβ9SΔN encoding HLA-DPβ1*0901. For complete HLA typing of BLCLs, see Table 1. Values are means of duplicates, with error bars indicating SDs.
Functional characterization of patient PBMCs after a single round of in vitro stimulation with irradiated donor PBMCs (TCL+1).
T cells were tested at various effector-target ratios in a standard 4-hour chromium release assay using donor BLCLs, patient BLCLs, or the allogeneic BLCL OMW as targets. The latter 2 BLCLs were used untransduced (○) or transduced (●) with retroviral vector LDPβ9SΔN encoding HLA-DPβ1*0901. For complete HLA typing of BLCLs, see Table 1. Values are means of duplicates, with error bars indicating SDs.
To analyze the antigen specificity of donor-reactive T cells at the clonal level, PBMCs–ex vivo were cloned by limiting dilution without prior polyclonal restimulation in vitro. Two of 117 and 8 of 144 T-cell clones obtained by nonspecific stimulation via PHA and by antigen-specific stimulation with irradiated donor BLCLs, respectively, showed specific recognition of donor BLCLs in cytotoxicity and interferon-γ release assays (data not shown). The antigenic determinant for all clones was HLA-DPβ1*0901, as demonstrated by adoptive transfer of HLA-DPβ1*0901 into targets used for functional characterization of these clones. Of note, all donor-specific clones had the CD4+ phenotype, whereas most of the randomly chosen clones without donor specificity had the CD8+ phenotype (19 of 21 and 24 of 25 clones obtained by nonspecific or donor-specific limiting dilution cloning, respectively, were CD8+; data not shown).
Discussion
The data from the present study suggest that the case of allogeneic PBSC rejection characterized here was at least in part mediated by alloreactive CD4+ T cells recognizing a donor-specific mismatched HLA-DP molecule. This is indicated by the recovery of donor-specific cytolytic activity against HLA-DPβ1*0901 from the patient's blood at the time of rejection, without prior restimulation in vitro. Furthermore, HLA-DPβ1*0901–specific CD4+ T-cell clones could be obtained by nonspecific restimulation in vitro, albeit with low frequency. Finally, a host-derived CD4+ T-cell line recognizing HLA-DPβ1*0901 was obtained by a single round of in vitro restimulation with donor cells. Others have shown that alloreactive T-cell lines could be obtained by donor-specific stimulation of recipient PBMCs in the first 60 days after transplantation only in those cases where significant numbers of host cells were circulating in association with graft failure.24 It is possible that, in addition to HLA-DPβ1*0901, other alloantigens were involved in the rejection process described here. Indeed, the HLA-DPβ1*0901–specific CD4+ T cells might have mediated not only direct cytotoxicity but also helper functions to CD8+ T cells with an as yet undefined specificity. In this context, it is of interest that most of the CD3+ T cells circulating at the time of rejection were of the CD8+ phenotype. In line with these observations, it has previously been shown that graft rejection is frequently associated with the emergence of host-derived CD8+ T cells without apparent donor-specific cytolytic activity as assessed by functional in vitro analysis.25 26However, the lack of evidence for a functional importance of the CD8+ T cells for donor-specific alloreactivity does not rule out a potential role of these cells in the rejection process. Indeed, the culture conditions and/or read-out targets could have been inappropriate for their function to be expressed. For instance, such effectors might recognize a minor histocompatibility antigen differentially expressed on donor-derived hematopoietic stem cells but not on mature PBMC or Epstein-Barr virus–transformed BLCLs.
In addition to CD8+ T cells, natural killer (NK) cells might also have contributed to the process of graft rejection reported here. Indeed, the direct cytotoxicity against donor cells observed might be at least in part mediated by NK cells whose activity might have been favored by missing self on donor cells lacking the patient-specific HLA-A2 molecule. In this context, it should be noted that the donor-recipient pair analyzed here was mismatched in the GVH direction both for the “classical” HLA-A2 antigen and for HLA-DPβ1*0402. Nevertheless, there was no appreciable immune response in the GVH direction in vivo. This could be due to T-cell depletion of the graft or to the absence of total body irradiation in the conditioning regimen, which might have facilitated patient-derived lymphoid reconstitution. Moreover, transplantation for CML in the chronic phase is known to be associated with a higher risk of rejection as compared to other hematopoietic disorders.1
In line with previous observations that matching for HLA-DPα is not important for the outcome of hematologic stem cell transplantation,12 no evidence for direct immunogenicity of the donor-specific HLA-DPα1*0201 was observed in this study. It could be speculated that the antigenic effect of HLA-DPβ1*0901 might have been increased by the simultaneous presence of an allogeneic HLA-DPα chain. However, the data from this study do not support this hypothesis. In fact, killing by the donor-specific T-cell line of targets coexpressing the mismatched HLA-DPα and HLA-DPβ chains was not generally more efficient than of targets mismatched for only HLA-DPβ.
The finding that HLA-DPβ can be an alloantigen involved in PBSC graft rejection is in contrast with the classical assumption that graft failure is predominantly mediated by HLA class I–restricted alloreactive CD8+ T cells. This assumption is based on early clinical observations describing CD8+ T cells in association with failure of HLA-identical sibling marrow grafts.25,26 One possible reason for the emergence of CD8+ and not CD4+ T cells in these patients could be that the predominant source of alloantigen target for rejection of genotypically identical PBSCs are minor histocompatibility antigens, ie, antigenic peptides presented mostly by HLA class I to cytotoxic CD8+ T cells. Indeed, more recent studies of unrelated donor-recipient pairs have shown that both CD8+ T cells specific for mismatched HLA class I subtypes3 and CD4+ T lypmphocytes recognizing mismatched HLA-DR or HLA-DQ antigens24 can be associated with graft failure. Alloreactive CD4+ T cells recognizing a single mismatch for HLA-DPβ have also been identified in association with acute GVH disease.13-15 Taken together, these findings suggest that alloreactive CD4+ T cells may be important players in the rejection of HLA class II–mismatched PBSC grafts, either mediating direct cytotoxicity and/or providing helper functions to other cytotoxic effectors.
The authors thank Dr Catia Traversari and Dr Maria Grazia Roncarolo for useful discussions and Nadia Nobili for excellent assistance in tissue culture. Dr Francesca Poli is gratefully acknowledged for help in HLA-DP typing and Dr Thierry Boon for providing the LG-2 BLCL.
Supported by grants from the Italian Association for Cancer Research and from the Association “Antonio Castelnuova,” Cermenate, Italy.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Katharina Fleischhauer, HLA Tissue Typing Laboratory, Istituto Scientifico H.S. Raffaele, via Olgettina 58, I-20132 Milan, Italy; e-mail: katharina.fleischhauer@hsr.it.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal