Abstract
Primary mediastinal B-cell lymphoma (PMBL) is a subentity of diffuse large B-cell lymphoma with characteristic clinical, histomorphologic, immunophenotypical, and genetic features. Unlike other B-cell lymphomas, PMBL has not yet been the subject of comprehensive molecular studies on the rearranged immunoglobulin (Ig) gene. Such investigations have proved essential to obtaining information about the differentiation stage of the lymphomagenic B cell. In the present study, the clonally rearranged immunoglobulin heavy-chain gene of 13 PMBL cases is analyzed by polymerase chain reaction (PCR) in conjunction with cloning and DNA sequencing. Twelve of 13 rearrangements were potentially functional. All clonally rearranged immunoglobulin genes bore a high load of somatic mutations (average, 13.0%), which appeared to be selected for a functional antibody in the majority of cases. The comparison of cloned PCR products revealed no evidence of ongoing mutation of the immunoglobulin variable gene. By means of reverse-transcriptase PCR, lymphoma-specific immunoglobulin transcripts were detected in 8 of 13 cases, all of which were of the postswitched type, whereas immunoglobulin protein expression was undetectable except for 1 case. A PMBL cell line, MedB-1, generated from an IgG− parental tumor, constitutively expressed IgG protein in a subset of cells, which was moderately suppressed by interleukin-4 and up-regulated in the presence of dexamethasone. PMBL is thus characterized by a heavily mutated, class-switched immunoglobulin gene without evidence of ongoing mutational activity. Moreover, our data indirectly suggest that regulation by extrinsic signals contributes to the immunoglobulin-negative phenotype of PMBL.
Introduction
Primary mediastinal B-cell lymphoma (PMBL) is categorized as a subentity of diffuse large B-cell lymphoma (DLBCL) in the Revised European American Lymphoma classification of malignant lymphoma1 and in the pending World Health Organization classification.2 Characteristic clinical features of PMBL are a comparatively young age at the time of presentation with a median age in the early thirties; a male-to-female ratio of 1:2; and local aggressiveness of the tumor with frequent invasion of adjacent structures, such as lung, vena cava, and pericardium.3 The discovery of B cells in the thymic medulla promoted the now widely accepted hypothesis of thymic medullary B cells as precursors of PMBL.4,5 This assumption has been corroborated by a high degree of immunophenotypical similarities between PBML and thymic B cells.6 Recent reports on recurrent chromosomal imbalances with amplifications on chromosome 9p,7,8 the low prevalence of bcl-6 rearrangements and lack of bcl-2 rearrangements,9 and the frequent expression of the MAL gene10 further supported the notion of PMBL as a distinct entity separate from other DLBCLs.
Analysis of the rearranged immunoglobulin heavy-chain variable (IgVH) gene is a commonly used method to gain insight into the differentiation stage of the B lymphocyte giving rise to the neoplastic clone of a lymphoma.11,12 Such analyses have been extensively applied to various types of B-cell non-Hodgkin lymphoma, including DLBCL. However, until now there has been only one study explicitly dealing with different subclasses of DLBCL.13 In this study, which also included 5 cases of PMBL, direct sequencing of amplified variable-diversity-joining (VDJ) rearrangements prevented the assessment of ongoing mutation. Ongoing mutational activity within a B-cell lymphoma leads to clonal diversification and is informative with respect to a germinal center origin or post–germinal center origin of the precursor B cell.14 In addition, no consistent data exist relating to whether PMBL cells underwent immunoglobulin isotype recombination, which is another crucial step on the pathway of B-cell ontogeny.
In the present study, we analyzed clonal VDJ rearrangements of 13 cases of PMBL for evidence of somatic hypermutation and ongoing mutation of the IgVH gene. To obtain information on isotype switching in this constitutively Ig−tumor,5,6 15-17 we determined the presence of lymphoma-specific IgH messenger RNA (mRNA). Finally, since DNA sequence analysis was inconclusive in explaining the lack of immunoglobulin protein in PMBL, we measured antibody protein expression of a mediastinal B-cell lymphoma cell line, MedB-1, and assessed its amenability to physiologic regulators of immunoglobulin expression.
Patients, materials, and methods
Patients
Twenty patients with a diagnosis of PMBL were randomly selected on the basis of the availability of frozen material. Of these patients, those cases with detectable clonal IgVH gene rearrangements were further investigated in the present study. Clinical data of these patients are summarized in Table 1. The age at diagnosis ranged from 21 to 59 years (mean, 38 years). Six patients were female, and 7 were male. The stage of disease was between 2 and 4.
Clinical data
| Case no. . | Sex . | Age at diagnosis . | Stage . | Primary site . |
|---|---|---|---|---|
| 1 | M | 21 | 4 | Mediastinum |
| 2 | M | 43 | 2 | Mediastinum |
| 3 | F | 43 | 2 | Mediastinum |
| 4 | F | 46 | 2 | Mediastinum |
| 5 | M | 38 | 2 | Mediastinum |
| 6 | M | 34 | 2 | Mediastinum |
| 7 | F | 23 | 2 | Mediastinum |
| 8a | M | 27 | 2 | Lung (recurrence) |
| 8b* | — | — | — | — |
| 9 | F | 59 | 4 | Mediastinum |
| 10 | M | 52 | 3 | Mediastinum |
| 11 | F | 30 | ND | Mediastinum |
| 12 | F | 36 | ND | Mediastinum |
| 13a | M | 42 | 2 | Mediastinum |
| 13b† | M | 42 | 4 | Mediastinum |
| Case no. . | Sex . | Age at diagnosis . | Stage . | Primary site . |
|---|---|---|---|---|
| 1 | M | 21 | 4 | Mediastinum |
| 2 | M | 43 | 2 | Mediastinum |
| 3 | F | 43 | 2 | Mediastinum |
| 4 | F | 46 | 2 | Mediastinum |
| 5 | M | 38 | 2 | Mediastinum |
| 6 | M | 34 | 2 | Mediastinum |
| 7 | F | 23 | 2 | Mediastinum |
| 8a | M | 27 | 2 | Lung (recurrence) |
| 8b* | — | — | — | — |
| 9 | F | 59 | 4 | Mediastinum |
| 10 | M | 52 | 3 | Mediastinum |
| 11 | F | 30 | ND | Mediastinum |
| 12 | F | 36 | ND | Mediastinum |
| 13a | M | 42 | 2 | Mediastinum |
| 13b† | M | 42 | 4 | Mediastinum |
ND indicates not determined.
8b is a cell line generated from case no. 8a.
13b is a second sample taken from the patient in case 13a.
Tissue and cell line
One tissue sample was taken from lymphoma invading the lung of a patient who had developed recurrent disease after receiving antitumor chemotherapy (case 8a). A stable cell line previously generated from this tumor (MedB-1,18 designated case 8b) was also included in this study. From one patient, 2 sequential samples were available. The first was taken at lymphoma diagnosis from a tumor mass located in the anterior mediastinum (case 13a), and the second was taken 1 year later at autopsy following chemotherapy and lymphoma progression to extensive disease (case 13b). All other specimens derived from primary tumors localized in the mediastinum without preceding chemotherapy. The diagnosis of PMBL was made according to established clinical, histomorphologic, and immunophenotypical criteria.19 Histologically, all cases exhibited a diffuse growth pattern and a variable degree of sclerosis. Tumor cells were of moderate to large size with abundant and often pale-staining cytoplasm and irregularly shaped nuclei lacking prominent nucleoli. The immunophenotype was consistently CD10−CD19+CD20+CD21−CD22+.
DNA extraction
Frozen tissue was cut into 25-μm slices on a cryostat microtome; a clean blade was used for every new tissue sample. Five tissue sections per case or pellets of 107 MedB-1 cells or peripheral blood leukocytes from healthy donors as polyclonal control samples were digested with 100 μg/mL Proteinase K (Sigma-Aldrich, Deisenhofen, Germany) at 50°C for 16 hours in a lysis buffer (10 mM Tris-HCl, pH 7.6; 10 mM EDTA; 50 mM NaCl; 0.5% sodium dodecyl sulfate [SDS]). After the addition of 4 volumes of a saturated NaCl solution and centrifugation at 10 000g, DNA was ethanol-precipitated from the supernatant and washed twice in 70% ethanol.
Polymerase chain reaction
We submitted 500 ng DNA to a polymerase chain reaction (PCR). Amplification of the VDJ rearrangement of the IgVHgene was performed with 2 separate primer sets. The first primer set (designated frame region 1 [FR1]) encompassed a consensus primer (FR1c) of FR1,20 in conjunction with a JHprimer mix21 containing primers 3′JH1245, 3′JH3, and 3′JH6 at a molar ratio of 4:1:1. PCR was carried out in a 50-μL standard reaction buffer with 12.5 pmol FR1c and JH primer mix, 0.2 pmol each deoxynucleoside 5′-triphosphate (dNTP), and 0.5 U Taq polymerase (catalog No. 27-0799-01; Amersham Pharmacia Biotech, Freiburg, Germany). To avoid elongation of nonspecifically annealed primers, a “hot start” was performed by preheating the reaction mix to 94°C before adding the enzyme. Amplification was carried out for 35 cycles at an annealing temperature of 65°C. Cases that did not yield a clonal band were reanalyzed with a second primer set (designated FR2) as previously published,22 with the use of an FR2 region consensus primer (FR2A) combined with a JH consensus primer (LJH) for a first amplification round of 35 cycles at an annealing temperature of 55°C. Primer concentrations were 1 pmol/μL; all other reaction conditions were as described above. We submitted 1 μL PCR product to a second amplification round in a seminested fashion with primer FR2A and a JH-consensus primer (VLJH) located internally to LJH. The second amplification round was carried out for 25 cycles at 60°C. Then 10 μL PCR product was size-fractionated on a 2% agarose gel and stained with ethidium bromide. All PCR analyses were performed at least in duplicate.
Cloning and sequencing of PCR products
PCR products were size-separated on a 2% agarose gel and specific bands excised from the gel. DNA was isolated and purified by gel chromatography (QIAquick gel extraction kit) (catalog No. 28104; Qiagen, Hilden, Germany); ligated into a PCR-cloning vector (pGEM-T-Vector-System) (catalog No. A3610; Promega, Mannheim, Germany) at a molar insert-to-vector ratio of 3:1; and subsequently transformed into competent JM 109 bacteria. Bacteria were plated on Luria Bertani (LB)–agar plates containing ampicillin, X-Gal (Sigma-Aldrich), and isopropylthiogalactoside (Sigma-Aldrich) at concentrations recommended by the manufacturer, and were grown overnight at 37°C. White colonies were picked, and the presence of an insert was confirmed by restriction enzyme analysis with SacI and ApaI. Plasmid DNA was prepared according to an established alkaline lysis protocol. Sequencing was always performed on both strands with an Alf Express Sequencer (Amersham Pharmacia Biotech) by means of the Auto Cycle Kit (catalog No. 27-2693-02; Amersham Pharmacia Biotech) as recommended by the manufacturer.
Analysis of sequencing data
Sequences of clonal VDJ rearrangements were compared with published germline sequences of the human IgVH gene by means of DNAPLOT Version 2.0.1 and V BASE Version 1.0 (available at:http://www.mrc-cpe.cam.ac.uk/imt-doc/restricted/ok.html. Accessed August 6, 2001). Case 3 did not yield an alignment by DNAPLOT owing to sequence deletions and was therefore analyzed by means of the IgBLAST-software (available at: http://www.ncbi.nlm.nih.gov/igblast/. Accessed August 6, 2001). To determine whether mutations of the IgVH gene were random or selected for, the ratio between replacement and silent mutations (R/S ratio) of the framework regions was determined and compared with the R/S ratio to be expected if the mutations merely happened by chance.23 These calculations were carried out with the computer software Inh Sus Calc V 1.0 and R S Probability, kindly provided by Dr Paolo Casali and Dr Hong Zan (Division of Molecular Immunology, Department of Pathology, Cornell University Medical College, New York, NY). In codons with 2 or 3 substitutions, each mutation was classified separately as R or S in comparison with the germline sequence of the codon.11
Reverse-transcriptase PCR
Ten slices of 25 μm each were cut off frozen tissue blocks, collected in a FastRNA Tube (catalog No. 6040-601; Bio 101, Carlsbad, CA), and mechanically homogenized in Trizol reagent (catalog No. 15596-026; Life Technologies, Karlsruhe, Germany) by a FastPrep 120 device (Bio 101). RNA was extracted according to the instructions by the manufacturer of the Trizol reagent. Then, 5 μg RNA was annealed to 1 pmol oligo(dt)15 primer, denatured at 80°C, and reverse transcribed with 200 U Super Script reverse transcriptase (catalog No. 18053-017; Life Technologies) in 25 μL reaction mix containing buffer supplied by the manufacturer, 10 mM dithiothreitol, 40 U ribonuclease inhibitor (RNAsin) (catalog No. N2111; Promega), and 1 mM each dNTP. The reverse transcription was carried out at incremental temperature steps ranging from 25°C to 95°C. We submitted 1 μL reverse-transcriptase (RT) reaction product to a PCR analogous to the above description. Clone-specific 5′ primers were designed complementary to complementarity-determining region (CDR)–3 or intensely hypermutated areas of the rearranged VH gene. In 5 separate reactions, the clone-specific oligonucleotides were combined with 3′ primers, each specific for the first exon of the constant region of 1 of the 5 immunoglobulin isotypes (μ, TTC TCA CAG GAG ACG AGG GG; δ, GAT GGG GAA CAC ATC CGG AG; γ, TAG TCC TTG ACC AGG CAG CC; α, AGG CTC AGC GGG AAG ACC TT; ε, GTC ACG GAG GTG GCA TTG GA). Amplification was carried out at precalculated annealing temperatures ranging from 60°C to 65°C. PCR products were size-separated on a 2% agarose gel and stained with ethidium bromide.
Southern blot
Southern blotting of the RT-PCR products was carried out according to established protocols. Briefly, agarose gels were incubated in 0.5 M NaOH and 1.5 M NaCl to denature the DNA, followed by neutralization in 0.5 M Tris buffer (pH 7.5) and 3 M NaCl. PCR products were transferred to a Hybond N+ membrane (catalog No. RPN1520B; Amersham Pharmacia Biotech) by a conventional capillary blot with 10 × SSC as a transfer buffer and were subsequently cross-linked to the membrane by baking for 2 hours at 80°C. Specific PCR products were detected via hybridization to 5′-digoxigenin–labeled isotype-specific oligonucleotide probes (μ, GGA GTG CAT CCG CCC CAA CC; δ, CAC CCA CCA AGG CTC CGG AT; γ, AAG GGC CCA TCG GTC TTC CC; α, CAT CCC CGA CCA GCC CCA AG; ε, CCT CCA CAC AGA GCC CAT CC) (MWG Biotech, Ebersberg, Germany) located internally to the 3′ primers. Hybridization was carried out for 16 hours at 68°C in 5 × SSC containing 0.1% N-lauroyl-sarcosine, 0.02% SDS, 2% blocking reagent (catalog No. 1175041; Boehringer Mannheim), and 5 pmol/mL labeled probe. After 2 posthybridization washes in 2 × SSC/0.1% SDS at room temperature for 15 minutes, bound probe was detected by antidigoxigenin Fab fragments and chemiluminescent detection with CSPD by means of the DIG Luminescent Detection Kit (catalog No. 1363514; Boehringer Mannheim) according to the manufacturer's instructions. Chemiluminescence was visualized by exposing the membranes to x-ray film for 15 to 120 minutes at room temperature.
Immunomorphology
Paraffin sections were dewaxed in xylene, rehydrated in decreasing concentrations of ethanol, and subsequently incubated with polyclonal rabbit antibodies against IgM (catalog No. A 0426; Dako Diagnostika, Hamburg, Germany), IgD (catalog No. A 0093; Dako), IgG (catalog No. A 0423; Dako), IgA (catalog No. A 0262; Dako), Igκ (catalog No. A 191; Dako), Igλ (catalog No. A 0193; Dako), and mouse monoclonal antibodies specific for IgE (clone CIA-E-7.12) (catalog No. M7071; Dako) and J-chain (clone M19-9J) (catalog No. SM1013P; DPC Biermann, Bad Nauheim, Germany), diluted in phosphate-buffered saline (PBS) buffer at pretested concentrations. Acetone-fixed cryosections of cases 6, 9, and 12 and cytospin preparations of the MedB-1 cell line were treated with 3% H2O2 to quench endogenous peroxidase and incubated with monoclonal mouse antibodies against IgM (clone R1/69) (catalog No. M 0702; Immunotech, Hamburg, Germany) IgD (clone EgD26) (catalog No. M 703; Dako), IgG (clone 8a4) (catalog No. 0279; Immunotech), and IgA (clone NIF2) (catalog No. 0278; Immunotech) instead of the respective polyclonal antibodies. Sections were then washed in PBS and incubated with a polyclonal swine anti–rabbit immunoglobulin antibody (catalog No. Z 0196; Dako) followed by rabbit peroxidase-antiperoxidase complex (catalog no. Z 0113; Dako) to label bound polyclonal rabbit antibody. Monoclonal primary antibodies were detected with biotinylated sheep anti–mouse immunoglobulin (catalog No. RPN 1001; Amersham Pharmacia Biotech) followed by streptavidin–horseradish-peroxidase complex (catalog No. RPN 1051; Amersham Pharmacia Biotech). Peroxidase activity was visualized by the substrate 3-amino-9-ethylcarbazole (0.1 mg/mL in 0.17 M sodium acetate, pH 5.2, plus 0.01% H2O2). Sections were counterstained in hematoxylin.
Stimulation of MedB-1 cells
We incubated 5 × 105/mL MedB-1 cells with 100 U/mL interleukin-4 (IL-4) (catalog No. 50444; Biomol, Hamburg, Germany) or 1 to 100 μg/mL dexamethasone (catalog No. D2915; Sigma-Aldrich) in RPMI 1640 medium (catalog No. 21875, Life Technologies) to which 100 IU/mL penicillin (catalog No. 15140-114; Life Technologies), 100 μg/mL streptomycin (catalog No. 15140-114; Life Technologies), and 2 mM L-glutamine (catalog No. 21051-016; Life Technologies) had been added. After stimulation for 48 hours, cells were washed twice in PBS buffer. Then, 2 × 106 cells were resuspended in a lysis buffer (50 mM Tris-HCl, pH 7.5; 150 mM NaCl; 1% Nonidet P 40; 0.5% sodium deoxycholate) containing 1 tablet of Complete protease inhibitor cocktail (catalog No. 1697498; Boehringer Mannheim) per 50 mL buffer and mechanically disrupted on ice in a dounce homogenizer with the use of a 2-mL polished tube with an appropriate Teflon pestle. Cellular debris was removed by centrifugation at 13 000g for 5 minutes at 4°C. The supernatant was transferred to a fresh tube and the protein concentration measured according to the method described by Lowry et al.24
Enzyme-linked immunosorbent assay
To quantify IgG in stimulated MedB-1 cells, an IgG enzyme-linked immunosorbent assay (ELISA) kit (catalog No. K 6510; Cell Concepts, Umkirch, Germany) was used according to the manufacturer's instructions. Cell lysates containing 100 μg protein were added to the wells of a microtiter plate precoated with a polyclonal anti–human IgG rabbit antibody. After washing, a peroxidase-conjugated polyclonal anti–human IgG rabbit antibody was added. Bound antibody was detected by the addition of tetramethylbenzidine substrate and measurement of the extinction in an MRX ELISA-reader (Dynatech Laboratories, Denkendorf, Germany) at 450 nm.
Results
Detection of clonal VDJ rearrangements
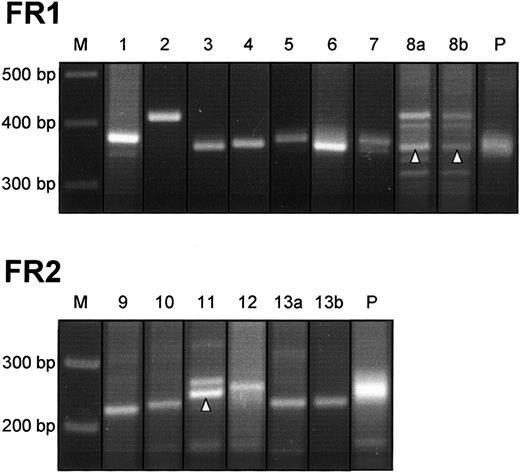
Clonal IgVH gene rearrangements were amplified from 13 of 20 cases (65%) (Figure 1). Eight cases yielded clonal signals with primer combination FR1. Five cases were negative with this primer set but showed clonal PCR bands after analysis with the alternative primer combination FR2. All clonal rearrangements were monoallelic; additional bands amplified from cases 8a, 8b, and 11 proved to be nonspecific upon sequence analysis. Seven specimens (35%) generated polyclonal amplification products from which dominant clonal bands were absent. Undetected monoclonality in PCR assays of the VDJ rearrangement can be attributed to alterations of the target DNA by somatic hypermutation, impairing the hybridization of the primers. In fact, a rate of 30% to 50% of false-negative results is commonly found in analysis of VDJ rearrangement by PCR with the use of consensus primers.25 In a recent study,26 no clonal VDJ rearrangements could be amplified from 40% of DLBCL when VH gene family–specific primers were applied.
Detection of clonal VDJ rearrangements in PMBL.
DNA from PMBL tissue was submitted to a PCR with primer combination FR1. Cases that did not reveal clonal signals with FR1 were reanalyzed with an alternative primer set FR2. Monoclonal rearrangements are identified by a sharp, distinct band in comparison with diffuse signals generated from polyclonal control samples (P). In cases 8a, 8b, and 11, specific PCR products are indicated by an arrowhead. Additional bands detectable in these cases proved to be nonspecific upon sequence analysis. M indicates molecular size marker.
Detection of clonal VDJ rearrangements in PMBL.
DNA from PMBL tissue was submitted to a PCR with primer combination FR1. Cases that did not reveal clonal signals with FR1 were reanalyzed with an alternative primer set FR2. Monoclonal rearrangements are identified by a sharp, distinct band in comparison with diffuse signals generated from polyclonal control samples (P). In cases 8a, 8b, and 11, specific PCR products are indicated by an arrowhead. Additional bands detectable in these cases proved to be nonspecific upon sequence analysis. M indicates molecular size marker.
Rate of somatic mutation
The results of the DNA sequence analysis are summarized in Table2. All cases bore a high mutational load, ranging from 5.6% to 30.9% (mean, 13.0%). Case 3, which was heavily mutated (18.8%), showed a 19–base pair (bp) deletion at the border between FR1 and CDR1 involving codons 26 through 31a of the corresponding V4-30.4 germline sequence and an 8-bp deletion within the FR3 region (codons 72 through 75) (Figure2). These deletions are likely to have been introduced during the process of somatic hypermutation,27 especially as the VDJ rearrangement of case 3 was out of frame and displayed 2 stop codons.
Analysis of VH gene sequences
| Case no. . | Primer combination . | Corresponding VH germline . | In frame . | Percentage mutation . | Observed mutations in FR . | R/S . | P . | No. clones sequenced . | Ongoing mutation . | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| R . | S . | Observed . | Expected . | ||||||||
| 1 | FR1 | V3-21 | + | 7.4 | 7 | 5 | 1.4 | 2.8 | .0327* | 7 | − |
| 2 | FR1 | V3-48 | + | 5.6 | 4 | 3 | 1.3 | 2.9 | .0156* | 8 | − |
| 3 | FR1 | V4-30.4 | − | 18.8 | NA‡ | NA‡ | NA‡ | NA‡ | NA‡ | 8 | − |
| 4 | FR1 | V3-09 | + | 6.3 | 8 | 5 | 1.6 | 3.0 | .1434 | 7 | − |
| 5 | FR1 | V5-a | + | 14.4 | 17 | 5 | 3.4 | 3.3 | .0252* | 8 | − |
| 6 | FR1 | V3-33 | + | 6.7 | 7 | 3 | 2.3 | 2.9 | .0651 | 9 | − |
| 7 | FR1 | V1-69 | + | 9.3 | 7 | 7 | 1.0 | 3.0 | .0026* | 6 | − |
| 8a | FR1 | V4-39 | + | 16.9 | 14 | 14 | 1.0 | 2.6 | .0007* | 8 | − |
| 8b | FR1 | V4-39 | + | 16.9 | 14 | 14 | 1.0 | 2.6 | .0007* | 7 | + |
| 9† | FR2 | V4-34 | + | 14.9 | 8 | 12 | 0.7 | 2.8 | .0069* | 3 | − |
| 10 | FR2 | V2-05 | + | 10.5 | 4 | 7 | 0.6 | 3.2 | .0088* | 6 | − |
| 11 | FR2 | V3-30 | + | 30.9 | 21 | 12 | 1.8 | 3.2 | .0396* | 5 | − |
| 12† | FR2 | V1-02 | + | 10.5 | 9 | 10 | 0.9 | 3.4 | .0339* | 3 | − |
| 13a | FR2 | V3-20 | + | 17.3 | 8 | 7 | 1.1 | 3.2 | .0033* | 5 | − |
| 13b | FR2 | V3-20 | + | 17.3 | 8 | 7 | 1.1 | 3.2 | .0033 | 3 | − |
| Case no. . | Primer combination . | Corresponding VH germline . | In frame . | Percentage mutation . | Observed mutations in FR . | R/S . | P . | No. clones sequenced . | Ongoing mutation . | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| R . | S . | Observed . | Expected . | ||||||||
| 1 | FR1 | V3-21 | + | 7.4 | 7 | 5 | 1.4 | 2.8 | .0327* | 7 | − |
| 2 | FR1 | V3-48 | + | 5.6 | 4 | 3 | 1.3 | 2.9 | .0156* | 8 | − |
| 3 | FR1 | V4-30.4 | − | 18.8 | NA‡ | NA‡ | NA‡ | NA‡ | NA‡ | 8 | − |
| 4 | FR1 | V3-09 | + | 6.3 | 8 | 5 | 1.6 | 3.0 | .1434 | 7 | − |
| 5 | FR1 | V5-a | + | 14.4 | 17 | 5 | 3.4 | 3.3 | .0252* | 8 | − |
| 6 | FR1 | V3-33 | + | 6.7 | 7 | 3 | 2.3 | 2.9 | .0651 | 9 | − |
| 7 | FR1 | V1-69 | + | 9.3 | 7 | 7 | 1.0 | 3.0 | .0026* | 6 | − |
| 8a | FR1 | V4-39 | + | 16.9 | 14 | 14 | 1.0 | 2.6 | .0007* | 8 | − |
| 8b | FR1 | V4-39 | + | 16.9 | 14 | 14 | 1.0 | 2.6 | .0007* | 7 | + |
| 9† | FR2 | V4-34 | + | 14.9 | 8 | 12 | 0.7 | 2.8 | .0069* | 3 | − |
| 10 | FR2 | V2-05 | + | 10.5 | 4 | 7 | 0.6 | 3.2 | .0088* | 6 | − |
| 11 | FR2 | V3-30 | + | 30.9 | 21 | 12 | 1.8 | 3.2 | .0396* | 5 | − |
| 12† | FR2 | V1-02 | + | 10.5 | 9 | 10 | 0.9 | 3.4 | .0339* | 3 | − |
| 13a | FR2 | V3-20 | + | 17.3 | 8 | 7 | 1.1 | 3.2 | .0033* | 5 | − |
| 13b | FR2 | V3-20 | + | 17.3 | 8 | 7 | 1.1 | 3.2 | .0033 | 3 | − |
Sequences have been submitted to the EMBL/GenBank/DDBJ database under accession numbers AJ292059 through AJ292070. See Table 1 footnote for descriptions of cases 8b and 13b.
FR indicates frame region; R, replacement mutation; S, silent mutation; NA, not analyzed.
P < .05.
No intraclonal diversity was detectable, but this was of limited significance owing to low number of sequenced clones.
Not analyzed because of nonfunctional rearrangement.
Deletions within the rearranged IgVH gene of case 3.
Two deletions (indicated by dots and gray background) are detectable: a 19-bp deletion is found at the border between FR1 and CDR1 involving codons 26 through 31a of the corresponding V4-30.4 germline sequence, and an 8-bp deletion is detectable within FR3 (codons 72 through 75). Identities to the closest germline sequence V4-30.4 are indicated by dashes, and each mutation is shown by the appropriate nucleotide. Stop codons are marked by asterisks. The numbering refers to codons counted from the beginning of VH according to Chothia et al.58
Deletions within the rearranged IgVH gene of case 3.
Two deletions (indicated by dots and gray background) are detectable: a 19-bp deletion is found at the border between FR1 and CDR1 involving codons 26 through 31a of the corresponding V4-30.4 germline sequence, and an 8-bp deletion is detectable within FR3 (codons 72 through 75). Identities to the closest germline sequence V4-30.4 are indicated by dashes, and each mutation is shown by the appropriate nucleotide. Stop codons are marked by asterisks. The numbering refers to codons counted from the beginning of VH according to Chothia et al.58
Distribution of mutations
To determine whether mutations were selected for a functional antibody, we calculated whether the detected R/S ratios were significantly lower than the R/S to be expected in the case of mutations introduced in a random fashion. As pointed out by Klein et al,11 mutational patterns within the CDR are difficult to interpret. Depending on the affected amino acid, some R mutations in the CDR may have little effect on the antibody affinity while others occurring within an already favorably mutated IgVH gene could dramatically reduce the binding affinity and thus even be counterselected. Therefore, we concentrated on the analysis of the FR and did not consider the CDR. In 12 cases, the VDJ rearrangements were in frame and potentially functional. The only nonfunctional VDJ rearrangement was found in case 3, which was out of frame and displayed 2 stop codons. In 10 of the 12 cases (83%) with potentially functional rearrangements, replacement mutations within the frame regions occurred significantly less frequently (P < .05) than would be expected by chance. Moreover, the mean value of observed R/S ratios was 1.4 (range, 0.7 to 3.4) and therefore within the range of magnitude found in normal memory B cells and plasma cells that can be assumed to have undergone affinity selection.11 These results strongly suggest that antibody-mediated selective forces maintained the overall structure of the B-cell receptor in PMBL.
Ongoing mutation
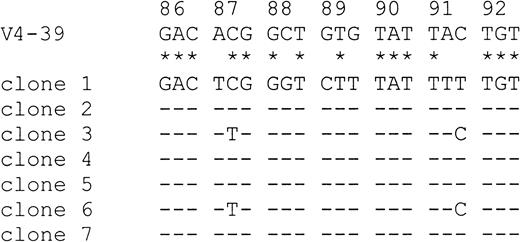
To ensure that base exchanges were not introduced by errors of the Taq polymerase (Taq error in our laboratory, 0.13%), mutations were only counted as such if they were present in at least 2 clones.28 With 2 exceptions (cases 9 and 12), the number of lymphoma-specific PCR clones sequenced in the present study (Table2) was within the range analyzed by other groups addressing the issue of ongoing mutation.29-31 No intraclonal variation was observed in any of the lymphoma samples, and there was thus no evidence of ongoing mutational activity. Identical VDJ sequences were also amplified from sequentially obtained samples of one patient (case 13a and 13b). In contrast, MedB-1 cells displayed a low intraclonal diversity, with 2 base exchanges in codon 87 and 91, respectively (Figure 3).
Evidence for ongoing mutation in MedB-1 cells.
Seven PCR clones amplified from MedB-1 cells were aligned and compared with the most homologous germline IgVH sequence V4-39 from codons 86 through 92. Sequence homologies between V4-39 and the consensus sequence of the PCR clones are highlighted by asterisks. Dashes designate identities among PCR clones. Clones 3 and 6 show deviations from the consensus sequence in codons 87 and 91, suggesting ongoing somatic mutation.
Evidence for ongoing mutation in MedB-1 cells.
Seven PCR clones amplified from MedB-1 cells were aligned and compared with the most homologous germline IgVH sequence V4-39 from codons 86 through 92. Sequence homologies between V4-39 and the consensus sequence of the PCR clones are highlighted by asterisks. Dashes designate identities among PCR clones. Clones 3 and 6 show deviations from the consensus sequence in codons 87 and 91, suggesting ongoing somatic mutation.
Use of VH families
In 6 of 12 potentially functional VDJ sequences (50.0%), members of the VH3 family were rearranged. VH4 was found in 2 cases (16.7%), of which V4-34 was amplified from one sample. VH1 could also be detected twice (16.7%), and VH2 and VH5 were each found once (8.3%). Thus, no bias toward use of the VH4 family was obvious in comparison with normal peripheral B cells.32
Expression of isotype mRNA
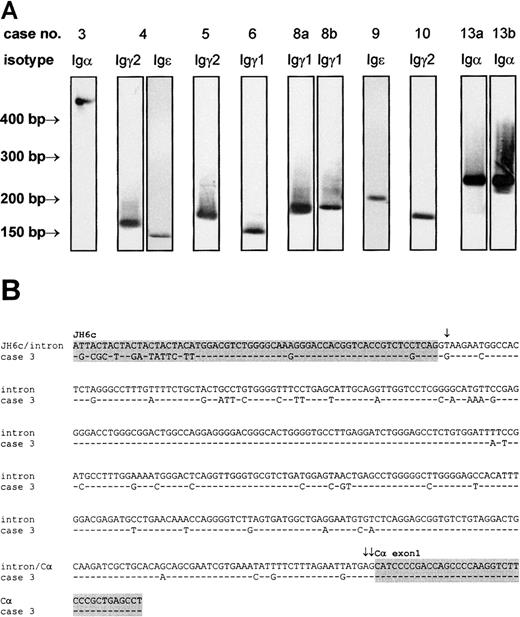
We amplified IgH mRNA composing the 3′ part of the variable chain and the 5′ end of the first exon of the constant chain of all 5 immunoglobulin isotypes by RT-PCR with lymphoma-specific primers. Following Southern blotting and hybridization to an isotype-specific probe, PCR products yielding positive signals (Figure4A) were cloned and sequenced (Table3). In 8 cases (61.5%), immunoglobulin transcripts were detected that, with the use of sequence analysis, could be unambiguously assigned to the corresponding lymphoma cell clone. No Igμ or Igδ mRNA was found. This was not due to low sensitivity of the RT-PCR or Southern blot assay, since positive control samples gave strong PCR bands and hybridization signals, respectively (data not shown). Igγ transcripts were detected in 5 cases, among which Igγ1 was observed twice and Igγ2 was observed 3 times. Igα was detectable in 2 cases, and 2 cases were shown to express Igε. A simultaneous expression of Igγ2 and Igε mRNA was observed in case 4. The coexpression of constant region transcripts from 2 different isotypes by one lymphoma has been previously described in follicular lymphoma and DLBCL.33 Possible mechanisms accounting for this phenomenon are differential deletional switch recombination in subpopulations of the lymphoma or switching without DNA recombination by alternative splicing or transsplicing.34 In case 3, Igα was not directly connected to VHDHJH6c but joined 373 bp downstream into the intron adjacent to JH6c. The intronic sequence displayed a high level of somatic hypermutation, which was more pronounced in the 5′ half (13.9%) than in the 3′ half (8.7%). No mutations were found in the Cα exon 1 covered by the sequence analysis. As illustrated in Figure 4B, somatic mutation generated a T>G transversion at position 2 of the intron that led to a destruction of the regular donor splice site and use of a cryptic donor splice sequence at position 373 (AG373GTGCGC) (Senapathy score35 of the cryptic donor site: 79.4).
Detection of VH-CH transcripts of the IgH gene.
VH-CH transcripts were amplified by RT-PCR with lymphoma-specific 5′ primers and isotype-specific 3′ primers, transferred to nylon membranes, and labeled by isotype-specific probes (A). The identity of the PCR products was confirmed by cloning and sequencing. In case 3, an abnormally large Igα transcript was observed. Sequence analysis of this transcript (B) revealed an aberrant VH-Cα splicing, caused by a T>G transversion within the regular donor splice site at the 3′ end of VHDHJH6 (single arrow) and use of a cryptic donor splice sequence (double arrows mark the first 2 bases of cryptic donor site). As a result, VHDHJH6 and Cα exon 1 (both indicated by gray background) are separated by a 373-bp intronic sequence. Note the high rate of somatic hypermutation carried far into the intron. This sequence has been submitted to the EMBL/GenBank/DDBJ database under accession number AJ292071.
Detection of VH-CH transcripts of the IgH gene.
VH-CH transcripts were amplified by RT-PCR with lymphoma-specific 5′ primers and isotype-specific 3′ primers, transferred to nylon membranes, and labeled by isotype-specific probes (A). The identity of the PCR products was confirmed by cloning and sequencing. In case 3, an abnormally large Igα transcript was observed. Sequence analysis of this transcript (B) revealed an aberrant VH-Cα splicing, caused by a T>G transversion within the regular donor splice site at the 3′ end of VHDHJH6 (single arrow) and use of a cryptic donor splice sequence (double arrows mark the first 2 bases of cryptic donor site). As a result, VHDHJH6 and Cα exon 1 (both indicated by gray background) are separated by a 373-bp intronic sequence. Note the high rate of somatic hypermutation carried far into the intron. This sequence has been submitted to the EMBL/GenBank/DDBJ database under accession number AJ292071.
Immunophenotype and immunoglobulin gene expression
| Case no. . | CD20 . | J chain . | IgM . | IgD . | IgG . | IgA . | IgE . | Igλ . | Igκ . | Ig mRNA . |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | + | − | − | − | − | − | − | − | − | — |
| 2 | + | +/− | − | − | − | − | − | − | − | — |
| 3 | + | − | − | − | − | − | − | − | − | Igα |
| 4 | + | − | − | − | − | − | − | − | − | Igγ2, Igε |
| 5 | + | − | − | − | − | − | − | − | − | Igγ2 |
| 6 | + | NA | − | − | − | − | NA | NA | NA | Igγ1 |
| 7 | + | − | − | − | − | − | − | − | − | — |
| 8a | + | − | − | − | − | − | − | − | − | Igγ1 |
| 8b | + | − | − | − | +≪− | − | − | − | +≪− | Igγ1 |
| 9 | + | NA | − | − | − | − | NA | NA | NA | Igε |
| 10 | + | − | − | − | − | − | − | − | − | Igγ2 |
| 11 | + | − | − | − | − | − | − | − | − | — |
| 12 | + | NA | − | − | − | − | NA | NA | NA | — |
| 13a | + | +<− | − | − | − | +<− | − | − | +<− | Igα |
| 13b | + | − | − | − | − | − | − | − | − | Igα |
| Case no. . | CD20 . | J chain . | IgM . | IgD . | IgG . | IgA . | IgE . | Igλ . | Igκ . | Ig mRNA . |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | + | − | − | − | − | − | − | − | − | — |
| 2 | + | +/− | − | − | − | − | − | − | − | — |
| 3 | + | − | − | − | − | − | − | − | − | Igα |
| 4 | + | − | − | − | − | − | − | − | − | Igγ2, Igε |
| 5 | + | − | − | − | − | − | − | − | − | Igγ2 |
| 6 | + | NA | − | − | − | − | NA | NA | NA | Igγ1 |
| 7 | + | − | − | − | − | − | − | − | − | — |
| 8a | + | − | − | − | − | − | − | − | − | Igγ1 |
| 8b | + | − | − | − | +≪− | − | − | − | +≪− | Igγ1 |
| 9 | + | NA | − | − | − | − | NA | NA | NA | Igε |
| 10 | + | − | − | − | − | − | − | − | − | Igγ2 |
| 11 | + | − | − | − | − | − | − | − | − | — |
| 12 | + | NA | − | − | − | − | NA | NA | NA | — |
| 13a | + | +<− | − | − | − | +<− | − | − | +<− | Igα |
| 13b | + | − | − | − | − | − | − | − | − | Igα |
See Table 1 footnote for descriptions of cases 8b and 13b.
− indicates no positive cells; +/−, 50% positive cells; +≪−, fewer than 10% positive cells; +<−, fewer than 50% positive cells; NA, not analyzed.
Immunohistochemistry
All cases were intensely CD20+ on immunohistology (Table 3). J-chain expression was detectable in subpopulations of neoplastic B cells in 2 cases (cases 2 and 13a) and negative in all other cases. In one case (13a), a coexpression of IgA and J chain was observed. In this case, the expression of IgA/κ protein correlated with the detection of Igα transcripts. It is of note that lymphoma tissue taken at autopsy of the same patient 1 year later was negative for IgA/κ and J chain but still expressed Igα transcripts (Table 3; Figure 4A). In no other case was there an expression of light-chain or heavy-chain isotypes.
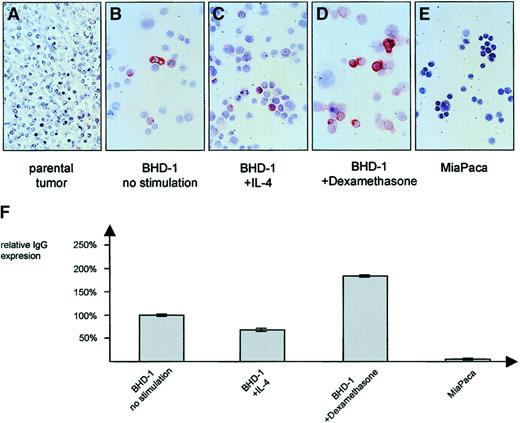
Stimulation of the MedB-1 cell line
In contrast to the IgG− parental lymphoma tissue of case 8 (Figure 5A), the MedB-1 cell line derived from it was found to express IgG/κ protein in the cytoplasm of a small fraction composing fewer than 10% of the cells (Figure 5B; Table 3). IgG expression was moderately influenced upon addition of stimulating substances. Whereas IL-4 (100 U/mL) led to a down-regulation by 32% (Figure 5C), there was a 1.8-fold increase of IgG expression in the presence of 10 μg/mL dexamethasone (Figure 5D) when compared with nonstimulated cells. Immunocytology confirmed the ELISA results (Figure 5F) and demonstrated that the modulatory effect was restricted largely to the constitutively IgG+ cell fraction while the number of IgG-expressing cells remained essentially unaltered. No IgG was found in the epithelial control cell line MiaPaca (Figure 5E).
IgG protein expression of MedB-1 cells.
In comparison with the IgG− parental tumor (A), a small subset of MedB-1 cells showed an expression of IgG protein, detectable by immunocytochemistry (B) and ELISA (F). Incubation for 48 hours in the presence of 100 U/mL IL-4 resulted in a down-regulation of IgG (C, F) whereas 10 μg/mL dexamethasone induced a moderate increase (D, F). ELISA data are given as mean value (no. = 5) and SD. Panel E shows negative control pancreatic carcinoma cell line MiaPaca. Original magnification ×50 (A), and ×100 (B-E).
IgG protein expression of MedB-1 cells.
In comparison with the IgG− parental tumor (A), a small subset of MedB-1 cells showed an expression of IgG protein, detectable by immunocytochemistry (B) and ELISA (F). Incubation for 48 hours in the presence of 100 U/mL IL-4 resulted in a down-regulation of IgG (C, F) whereas 10 μg/mL dexamethasone induced a moderate increase (D, F). ELISA data are given as mean value (no. = 5) and SD. Panel E shows negative control pancreatic carcinoma cell line MiaPaca. Original magnification ×50 (A), and ×100 (B-E).
Discussion
The PMBL analyzed in the present study carried a high load of somatic mutation. The mean base exchange rate of 13.0% in our lymphoma panel exceeds the mutation level found in normal germinal center and memory B-cell populations36 and is slightly higher than the average mutation frequency of DLBCL, ranging from 8.5%37 to 9.9%.13 This raises the question as to whether ongoing mutation within the neoplastic clone contributed to the high mutational load in PMBL as has been demonstrated for follicular center lymphoma.38 Applying stringent criteria for the definition of intraclonal diversity,28 we found no differences among the individual PMBL cases. Moreover, identical VDJ sequences were amplified from a biopsy sample and from lymphoma tissue taken 1 year later at autopsy of the same patient (cases 13a and 13b). Reports about ongoing mutation in DLBCL are inconsistent; while some groups demonstrated ongoing mutation in DLBCL,33,39 others did not.37,40 Data published by Ottensmeier et al33 demonstrate that antitumor chemotherapy or disease progression is related to a loss of intraclonal variation in DLBCL that had initially displayed ongoing mutation at primary diagnosis. However, this mechanism does not explain the absence of ongoing mutation in the present study as, with only one exception (case 8), lymphoma tissue from untreated patients was analyzed. In a recent survey, it was shown that DLBCLs are heterogeneous with respect to ongoing mutation.28 Using microarrays to generate expression profiles, the same group reported that ongoing somatic mutation was limited to germinal center B-cell–like lymphomas, whereas lymphomas resembling activated peripheral B cells in their expression pattern generally lacked intraclonal variation.41 If this correlation applies to all subentities of DLBCL, PMBL has to be assigned to the activated B-cell rather than to the germinal center B-cell category. Although an origin from a germinal center B cell that lost the ability to mutate its immunoglobulin gene by neoplastic transformation cannot be excluded, it is also possible that PMBL derives from a B cell at a postgerminal-center differentiation stage.41 During the multistep process of lymphomagenesis, early transforming events could lead to a retention within the germinal center, thereby facilitating the accumulation of an abnormally high mutational load. Such a cell might still be able to withdraw from the germinal center environment or the site of somatic mutation,12 switch off the mutational machinery, and proceed with differentiation before additional hits eventually complete the neoplastic transformation. With respect to PMBL, this notion is supported by the constitutive lack of the germinal-center marker CD10 in PMBL6,15 42 and the fact that the normal thymus as the primary site of PMBL does not usually harbor lymphoid follicles.
Our sequencing data provide evidence for a counterselection of replacement mutations within FR in the majority of PMBLs. This finding indicates that selective forces have favored mutations of the rearranged IgVH gene to conserve the antibody structure and maintain the potential for a functional B-cell receptor. Evidence for selection of a functional antibody in DLBCL has been previously reported.13 However, selection for a functional antibody is particularly noteworthy in the case of PMBL since this subentity of DLBCL is generally devoid of immunoglobulin protein expression and thus lacks the structure subject to selective constraint. One possible explanation for this apparently contradictory situation is that the loss of immunoglobulin protein occurred at a late stage of tumor development when somatic mutation had already taken place and tumor survival had become independent of B-cell receptor–mediated signals.43 The distribution of IgVH gene use by PMBL largely reflects the repertoire of normal peripheral B cells.32 In contrast to previous studies reporting an overrepresentation of VH4DJ rearrangements in DLBCL,37 44 we found VH4 to be used only twice among 12 potentially functional rearrangements (16.7%). Thus, there is no evidence that immunoglobulin binding specificity for autoantigens or superantigens plays a role in the lymphomagenesis of PMBL.
Analysis of immunoglobulin mRNA expression in PMBL supplemented our data on the hypermutation of the VDJ rearrangement. Using RT-PCR, we detected immunoglobulin transcripts in 8 of 13 cases (61.5%). All of these contained the switched isotypes Igγ, Igα, and Igε, while Igμ or Igδ were never observed. The exclusive finding of switched isotypes further differentiates PMBL from DLBCL, which predominantly,11,45 but not entirely,33exhibits an IgM+ phenotype. Although isotype switching can occur without somatic hypermutation in the T-cell areas of secondary lymphatic organs, it occurs mainly within the germinal center after somatic hypermutation has been triggered.46 The detection of switched immunoglobulin transcripts therefore confirms that the progenitor B cell that gives rise to PMBL had been under the influence of the germinal-center environment at some stage of lymphoma development.
The lack of surface and cytoplasmic immunoglobulin protein6,16 is a characteristic but enigmatic feature of PMBL. The immunohistologic data of the present study (Table 3) confirm the lack of immunoglobulin protein with only one exception. The reason for the consistently absent immunoglobulin protein in PMBL is not clear. Classical Hodgkin lymphoma, which is now generally accepted to be of B-cell origin in the vast majority of cases,47,48 is another lymphoma entity regularly lacking immunoglobulin expression.49 An additional link between Hodgkin disease and PMBL has lately been demonstrated by the identification of recurrent amplifications on chromosome 9p in both lymphoma entities.7,8,50 Recently, it was shown that Hodgkin and Reed-Sternberg cells, which represent the neoplastic cell population of classical Hodgkin lymphoma, frequently harbor crippling mutations in their rearranged IgV genes, creating stop codons and thereby preventing expression of an immunoglobulin protein.51 Our data suggest that crippling mutations are not likely to be a major cause of the absence of immunoglobulin in PMBL, since rearrangements of the IgVH gene were potentially functional in 12 of 13 cases. We cannot exclude the possibility that faulty IgVLrearrangements or aberrations located within the untranslated regions of the immunoglobulin gene52 might prevent antibody expression in PMBL. In fact, in one case (case 3) we did detect aberrant splicing owing to a mutated regular splice site (Figure 4). However, in this case, an out-of-frame VDJ rearrangement and 2 stop codons would have prevented the translation into an immunoglobulin protein even in the case of a correctly processed pre-mRNA. Rather than nonfunctioning cis-acting elements, a defective transcription machinery was recently identified as a competing mechanism for down-regulation of immunoglobulin expression in classical Hodgkin disease.49,53 In the present study, we demonstrate that MedB-1, a cell line generated from an IgG− parental tumor, constitutively expresses IgG in a small subset of cells. Interestingly, we also found a low degree of intraclonal variation in MedB-1 cells (Figure 3), whereas there was no evidence for ongoing mutation in the parental tumor. It remains to be clarified whether the reactivation of the mutational machinery is limited to the IgG+ cell subset. Moreover, IgG expression could, albeit to a moderate extent, be downregulated in vitro by IL-4 (which also has the propensity to inhibit immunoglobulin production in normal human B cells54) and increased by dexamethasone, in accordance with the inductive effect of corticosteroid hormones on immunoglobulin expression at the late stage of human B-cell differentiation55 (Figure 5). We therefore provide suggestive evidence that immunoglobulin protein expression in PMBL is reversibly down-regulated in vivo by extrinsic signals, one of which might be IL-4.
Considering the presumably thymic origin of PMBL, it is a plausible and attractive hypothesis that this form of lymphoma originates from a specific subpopulation of B cells resident in the thymic medulla.3 Thymic B cells display a CD20+CD21−, IgM+ but IgG− immunophenotype.56 To date, the molecular analysis of the VDJ rearrangement of human thymic B cells has been the subject of only one study,57 which demonstrated that most rearrangements were in the germline configuration and that fewer than 20% showed a moderate rate of somatic hypermutation. Hence, PMBL diverges from its putative normal progenitor cell by means of the prevalence of class switching, the high degree of somatic hypermutation, and the lack of immunoglobulin protein expression. While immunoglobulin expression has probably been down-regulated after neoplastic transformation, it is unlikely that isotype switching and somatic hypermutation of the IgVH gene occurred after neoplastic transformation had been fully completed. With the data presented in this study, a more precise identification of the thymic B-cell subset that gives rise to PMBL appears to be warranted. The lack of ongoing mutation and the prevalence of class switching further emphasize the distinct and rather homogeneous character of PMBL, providing additional justification for separating this lymphoma from other DLBCLs.
Our thanks to Beate Wotschke, Simone Westenfelder, and Beate Dörr for their superb technical assistance. We are indebted to Dr Paolo Casali and Dr Hong Zan, Division of Molecular Immunology, Department of Pathology, Cornell University Medical College, New York, NY, for providing their data analysis software.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Frank Leithäuser, Department of Pathology, University of Ulm, Albert Einstein Allee 11, 89081 Ulm, Germany; e-mail: frank.leithaeuser@medizin.uni-ulm.de.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal