The messenger RNA (mRNA) from 5 of 69 patients with severe hemophilia A did not support amplification of complementary DNA containing the first few exons of the factor VIII (F8) gene but supported amplification of mRNA containing exon 1 ofF8 plus exons of the VBP1 gene. This chimeric mRNA signals an inversion breaking intron 1 of the F8 gene. Using an inversion patient, one deleted for F8 exons 1 to 6, and cosmids mapped 70 to 100 kb telomeric of the F8gene, this study shows that this break strictly affects a sequence (int1h-1) repeated (int1h-2) about 140 kb more telomerically, between the C6.1A andVBP1 genes. The 1041-base pair repeats differ at a single nucleotide (although int1h-2 also showed one polymorphism) and are in opposite orientation. The results demonstrate that they cause inversions by intrachromosome or intrachromatid homologous recombination. The genomic structure of the inversion region shows that transcription traverses intergenic spaces to produce the 2 chimeric mRNAs containing the F8 sequences and characteristic of the inversion. This observation prompts the suggestion that nature may use such extended transcription to test whether the addition of novel domains from neighboring genes creates desirable new genes. A rapid polymerase chain reaction test was developed for the inversion in both patients and carriers. This has identified 10 inversions, affectingF8 genes with 5 different haplotypes for the BclI, introns 13 and 22 VNTR polymorphism, among 209 unrelated families with severe hemophilia A. This indicates a prevalence of 4.8% and frequent recurrence of the inversion. This should result in absence ofF8, and one inversion patient is known to have inhibitors.
Introduction
Higuchi et al1,2 observed that thorough screening of all the exons of the factor VIII (F8) gene was efficient in detecting the mutations of patients with mild and moderate hemophilia A and yet failed in 50% of patients with severe disease. Traces of factor VIII messenger RNA (mRNA) from peripheral lymphocytes soon revealed that this high failure rate was largely due to mutations affecting internal regions of intron 22 of theF8 gene.3 These mutations were later shown to be inversions resulting from homologous intrachromatid or intrachromosome (ie, intranemic) recombination between a 9503-base pair (bp) sequence (int22h-1) in intron 22 of theF8 gene and one or other of 2 inverted copies of this sequence (int22h-2, int22h-3) located, respectively, 500 and 600 kb more telomeric.4-6
The int22h-related inversions appeared to be sufficiently frequent to account for the shortfall in mutation detection experienced using methods based on the screening of all exons of the F8gene. However, analysis of factor VIII mRNA continued to detect further mutations that escape detection by this approach. Base substitutions deep inside introns were found that generate novel exons disrupting the factor VIII coding sequence7 (R. D. Bagnall, unpublished observations, January 2001). Moreover, an inversion was identified that broke the F8 gene at intron 1 and resulted in the production of 2 chimeric mRNAs.8 One of these mRNAs, presumably under the control of the factor VIII promoter, contains the first exon of the F8 gene followed by facultative exons and then exons 2 to 6 of a gene (VBP1) coding for subunit 3 of prefoldin.9-11 The other mRNA, transcribed under the control of the C6.1A promoter, contains all but the last exon of the C6.1A gene plus a number of facultative exons followed by exons 2 to 26 of the F8gene.8
During the construction of a confidential United Kingdom database of hemophilia A mutations and pedigrees, destined to optimize the genetic service provided nationally for this disease, we have discovered that the above inversion occurs repeatedly and accounts for about 5% of severe cases of hemophilia A. We have also characterized the breakpoint regions, elucidated the mode of origin of the inversions, and developed a rapid polymerase chain reaction (PCR) procedure for the detection of these mutations in both patients and carriers.
Materials and methods
Blood samples
A 20-mL sample of fresh blood was used for extraction of both DNA, using Puregene Kit (Gentra, Minneapolis, MN), and RNA according to the procedure described by Waseem et al.12
Factor VIII mRNA amplification and mutation analysis
Factor VIII mRNA was amplified by reverse transcription-PCR (RT-PCR) and screened for mutations using the method of solid-phase fluorescent chemical cleavage of mismatch previously described in detail.12
Vectorette library construction
Using the method of Riley et al,13 100 ng cosmid DNA was restricted with 10 U RsaI in a final volume of 20 μL for 12 hours, followed by heat inactivation at 65°C for 2 minutes. This digestion reaction (15 μL) was ligated to 4 μM of anRsaI vectorette cassette using 6 U DNA ligase at 4°C for 12 hours. The vectorette library was diluted with an equal volume of Tris (10 mM) plus EDTA (0.1 mM) buffer (pH 8) and stored at −20°C. An aliquot of this library was used to amplify between anint1h-specific primer and the vectorette-specific primer 224 (Table 1).
Primer sequences used in PCR amplifications
| Primer . | Sequence . |
|---|---|
| Vectorette | |
| 224 | CGAATCGTAACCGTTCGTACGAGAATCGCT |
| Factor VIII intron 1 primers | |
| 1F | GGGAGCTAAAGATATTTTAGAGAA |
| 1R | AAATAAATCCAATAGCCAACAAG |
| 2F | TTGTCATATGGCATGAGAGAGGGAT |
| 2R | CAATTCATTTGATAAAGCCAAC |
| 3F | GCAGCTGGTATCATCTGAATTG |
| 3R | ATGCACCATTGCTGGTAGAGG |
| 4F | AGGGCTGCACTCCTTAGTGA |
| 4R | GAACTTGAGGTGGGCAAAGATTTC |
| 5F | GAAATCTTTGCCCACCTCAA |
| 5R | GGTTTTCCCATGCTGTTCAC |
| 6F | AATCATGGTGGAAGGCAAAG |
| 6R | GGAACACTCATACTTTGGTAGTG |
| 7F | TTGGATTGTTTCCCCTTTTG |
| 7R | CCAACATGAAGAAACCCTGTCTAC |
| 8F | GCTCTTGATCCTGTTCCCTCTCTG |
| 8R | GATCTGAGGCGGACAGTTTC |
| 9F | GTTGTTGGGAATGGTTACGG |
| 9R | CACCACATCCAAACTCATGC |
| 10F | GAGGCAAGTTGTCTGGACCTATATAG |
| 10R | CCATCCTGGCTAACATGGTGAAACCC |
| 11F | TTTGTTTGTTCGTTTGCATCT |
| 11R | GCATTTGTCTTTTCATGACTGG |
| 12F | CATGCTACGACATAGATGAACC |
| 12R | TGGCAGCTGCACTTTTTAACTGC |
| Factor VIII intron 1 segment 9 primers | |
| 9aF | TTCCTAACAGGCCACAGACC |
| 9aR | CACCCGAGAAAGCACGTAGT |
| 9bF | GGAGTGAAATGCAGGGCTAC |
| 9bR | TCAGGTTGCAGACAAAGCAG |
| 9cF | GCGTTTTCCCTCAGTTACCA |
| 9cR | CTAGCTTGAGCTCCCTGTGG |
| 9dF | GAGGCAAGTTGTCTGGACCT |
| 9dR | ATGGCTTACAAAGGGCATGA |
| Int1h-2 primers | |
| Int1h-2F | GGCAGGGATCTTGTTGGTAAA |
| Int1h-2R | TGGGTGATATAAGCTGCTGAGCTA |
| Int1h repeat primers | |
| Int1hF | GCCCTCAGCCAGTACAGAAAGTCA |
| Int1hR | CCTTTGGGAGAATTCATTGCCAGCT |
| Factor VIII-VBP1 chimeric mRNA primers | |
| FVIII 1A | TGGGAGCTAAAGATATTTTAGAGAA |
| VBP1 PRIMEX1 | ACTTATACTTCTGGTACTGTTCATCCAGC |
| FVIII 1C (nested) | GAATTAACCTTTTGCTTCTCCAGTTGAAC |
| VBP1 3-M (nested) | TGTTTCATGAAGGAATCTAC |
| Primer . | Sequence . |
|---|---|
| Vectorette | |
| 224 | CGAATCGTAACCGTTCGTACGAGAATCGCT |
| Factor VIII intron 1 primers | |
| 1F | GGGAGCTAAAGATATTTTAGAGAA |
| 1R | AAATAAATCCAATAGCCAACAAG |
| 2F | TTGTCATATGGCATGAGAGAGGGAT |
| 2R | CAATTCATTTGATAAAGCCAAC |
| 3F | GCAGCTGGTATCATCTGAATTG |
| 3R | ATGCACCATTGCTGGTAGAGG |
| 4F | AGGGCTGCACTCCTTAGTGA |
| 4R | GAACTTGAGGTGGGCAAAGATTTC |
| 5F | GAAATCTTTGCCCACCTCAA |
| 5R | GGTTTTCCCATGCTGTTCAC |
| 6F | AATCATGGTGGAAGGCAAAG |
| 6R | GGAACACTCATACTTTGGTAGTG |
| 7F | TTGGATTGTTTCCCCTTTTG |
| 7R | CCAACATGAAGAAACCCTGTCTAC |
| 8F | GCTCTTGATCCTGTTCCCTCTCTG |
| 8R | GATCTGAGGCGGACAGTTTC |
| 9F | GTTGTTGGGAATGGTTACGG |
| 9R | CACCACATCCAAACTCATGC |
| 10F | GAGGCAAGTTGTCTGGACCTATATAG |
| 10R | CCATCCTGGCTAACATGGTGAAACCC |
| 11F | TTTGTTTGTTCGTTTGCATCT |
| 11R | GCATTTGTCTTTTCATGACTGG |
| 12F | CATGCTACGACATAGATGAACC |
| 12R | TGGCAGCTGCACTTTTTAACTGC |
| Factor VIII intron 1 segment 9 primers | |
| 9aF | TTCCTAACAGGCCACAGACC |
| 9aR | CACCCGAGAAAGCACGTAGT |
| 9bF | GGAGTGAAATGCAGGGCTAC |
| 9bR | TCAGGTTGCAGACAAAGCAG |
| 9cF | GCGTTTTCCCTCAGTTACCA |
| 9cR | CTAGCTTGAGCTCCCTGTGG |
| 9dF | GAGGCAAGTTGTCTGGACCT |
| 9dR | ATGGCTTACAAAGGGCATGA |
| Int1h-2 primers | |
| Int1h-2F | GGCAGGGATCTTGTTGGTAAA |
| Int1h-2R | TGGGTGATATAAGCTGCTGAGCTA |
| Int1h repeat primers | |
| Int1hF | GCCCTCAGCCAGTACAGAAAGTCA |
| Int1hR | CCTTTGGGAGAATTCATTGCCAGCT |
| Factor VIII-VBP1 chimeric mRNA primers | |
| FVIII 1A | TGGGAGCTAAAGATATTTTAGAGAA |
| VBP1 PRIMEX1 | ACTTATACTTCTGGTACTGTTCATCCAGC |
| FVIII 1C (nested) | GAATTAACCTTTTGCTTCTCCAGTTGAAC |
| VBP1 3-M (nested) | TGTTTCATGAAGGAATCTAC |
PCR amplification
The PCR amplifications of factor VIII intron 1 regions andint1h repeats were performed on 100 ng genomic DNA or 10 ng cosmid DNA with 2.5 μL 10 × Amplitaq Reaction Buffer (Perkin-Elmer, Warrington, United Kingdom), 1.5 mM MgSO4, 200 ng of each oligonucleotide primer, 0.5 mM of each dNTP, 5% dimethyl sulfoxide, and 2.5 U Amplitaq DNA polymerase (Perkin-Elmer). Thirty cycles of PCR were carried out at 94°C for 30 seconds, 65°C for 30 seconds, and 72°C for 2 minutes. The primers used for PCR assays are shown in Table 1.
Fluorescent sequencing
Sequencing was performed on Genecleaned PCR products using thed-Rhodamine dye terminator kit according to the manufacturer's instructions (ABI Perkin-Elmer, Warrington, United Kingdom). Products were analyzed on an ABI 377 DNA sequencer using ABI Sequence Analysis software.
Detection of int22h-related inversions
These inversions were detected using a PCR-based method described by Liu et al,14 modified by reducing the concentrations of both primer A and B to 20 ng and performing the PCR annealing step at 67°C.
Results
The intron 1 breaking inversion is a recurring cause of severe hemophilia A
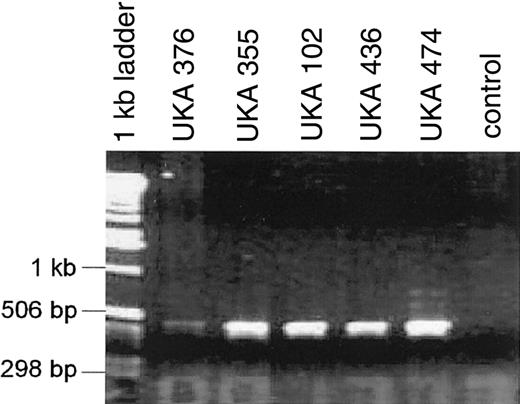
Our laboratory has received blood samples from 209 unrelated patients with severe disease in the course of constructing a United Kingdom confidential database of hemophilia A mutations and pedigrees. Ninety-four (45%) of these patients were found to have an intron 22 breaking inversion, whereas 115 patients, negative for these inversions, required full mutation screening. Analysis of factor VIII mRNA extracted from lymphocytes of 69 of these 115 patients showed that the mRNA of 5 patients did not support amplification of the segment containing exons 1 to 9, whereas the other segments containing the rest of the coding sequence readily amplified and contained no mutations. This was reminiscent of findings in 2 monozygotic twins with severe hemophilia A who were found to have an approximate 100-kb inversion breaking intron 1 of the F8 gene.8 Because this inversion leads to the production of a hybrid transcript that comprises exon 1 of the F8 gene and exons of the VBP1 gene, we used an RT-PCR reaction specific for this abnormal transcript and obtained a positive result in each of the above 5 patients (Figure1). They therefore appear to have the intron 1 breaking inversion previously observed by Brinke et al.8 The families of the 5 patients were not related to each other or to the family of the monozygotic twins examined by Brinke et al,8 and analysis of the F8 gene haplotypes for variations at the BclI and intron 13 and 22 VNTR polymorphisms15-17 demonstrated that the intron 1 breaking inversion has occurred independently more than once because the 6 patients show 3 different haplotypes.
RT-PCR amplification products from factor VIII-VBP1 chimeric mRNA transcripts in 5 severe hemophilia A patients.
Nested primers in factor VIII exon 1 combined with primers in VBP1 exon 2 produced a 500-bp band. The control lane RT-PCR contained normal lymphocyte RNA. Sequence analysis (not shown) revealed factor VIII exon 1 and 2 facultative exons followed by VBP1 exon 2.
RT-PCR amplification products from factor VIII-VBP1 chimeric mRNA transcripts in 5 severe hemophilia A patients.
Nested primers in factor VIII exon 1 combined with primers in VBP1 exon 2 produced a 500-bp band. The control lane RT-PCR contained normal lymphocyte RNA. Sequence analysis (not shown) revealed factor VIII exon 1 and 2 facultative exons followed by VBP1 exon 2.
The intron 1 breaking inversion appears to be due to intranemic homologous recombination involving repeats in inverted orientation
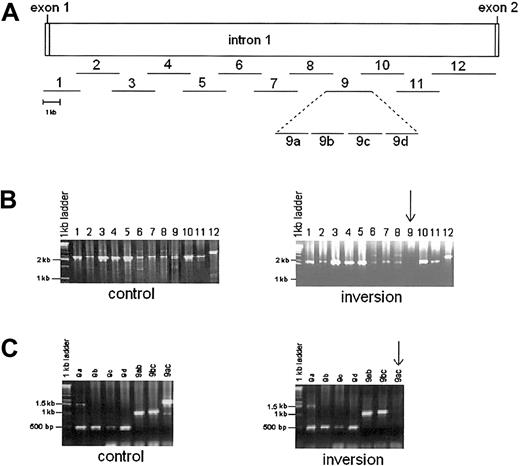
To discover the mode of origin of the above inversions, the region broken in intron 1 of the F8 gene was sought by subdividing the intron 1 sequence (GenBank accession no. AL390881) into 12 amplifiable overlapping segments of 2 to 3.5 kb (Figure2). The PCR reactions for all 12 segments were successful in wild-type DNA but the DNA from one of the monozygotic twins with the intron 1 inversion (UKA29) consistently failed to support amplification of segment 9. This suggested that the 2 kb of segment 9 comprises an inversion junction. To locate this junction more precisely, segment 9 was divided into 4 adjacent sections (a-d, shown in Figure 2) but, surprisingly, PCR reactions for all 4 sections were successful in the DNA from UKA29. However, the 1.5-kb PCR stretching from segments a to c consistently failed, whereas 2 overlapping segments containing a + b and b + c consistently amplified (Figure 2). This suggested that the inversion break involved the a to c region in such a way that the sequences of the individual segments a, b, and c are not disrupted. This could have occurred if the inversions had arisen by homologous recombination within a repeated sequence, as previously proposed and demonstrated for the intron 22 inversions.4-6 This presupposes the existence of repeated sequences in inverted orientation spanning segment 9b as illustrated in Figure 3.
PCR amplification of factor VIII intron 1 in control and intron 1 inversion patient (UKA 29).
(A) Amplification strategy. Lines with numbers show position and length of each segment amplified. Segment 9 has been expanded to show 4 adjacent reactions, 9a to 9d. (B). PCR products from control (left) and UKA29 (right) genomic DNA. Segment 9 consistently failed to amplify in UKA29 (arrow). (C) PCR amplification of sections of segment 9 in control (left) and UKA29 DNA (right). The 1.5 kb comprising sections a to c (9ac, arrow) could not be amplified from the DNA of UKA29. However, the 1-kb overlapping segments comprising sections a + b and b + c (9ab and 9bc) were amplified as readily as the individual sections 9a to 9d.
PCR amplification of factor VIII intron 1 in control and intron 1 inversion patient (UKA 29).
(A) Amplification strategy. Lines with numbers show position and length of each segment amplified. Segment 9 has been expanded to show 4 adjacent reactions, 9a to 9d. (B). PCR products from control (left) and UKA29 (right) genomic DNA. Segment 9 consistently failed to amplify in UKA29 (arrow). (C) PCR amplification of sections of segment 9 in control (left) and UKA29 DNA (right). The 1.5 kb comprising sections a to c (9ac, arrow) could not be amplified from the DNA of UKA29. However, the 1-kb overlapping segments comprising sections a + b and b + c (9ab and 9bc) were amplified as readily as the individual sections 9a to 9d.
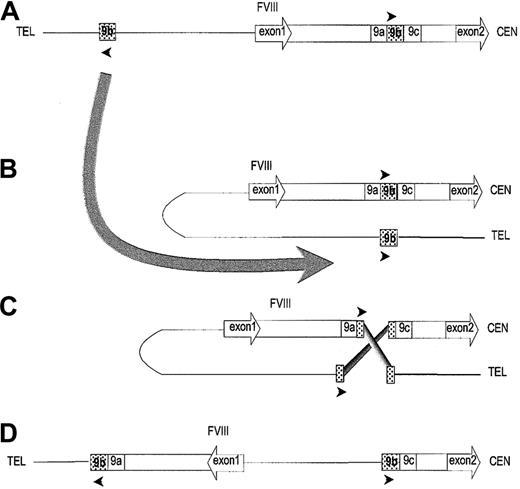
Diagram of factor VIII intron 1 and proposed model for inversion.
(A) Bar shows factor VIII intron 1, flanked by exons (drawn to indicate the direction of transcription), and containing a repeated sequence 9b (shaded) flanked by unique sequences 9a and 9c. The line shows DNA outside the F8 gene with the repeated sequence as a shaded box. Arrowheads indicate orientation of repeated sequences. The large curved arrow indicates the folding required for homologous recombination. (B,C) Homologous recombination between the two 9b repeats proposed to explain origin of inversion. (D) Inversion resulting from recombination. This shows why in the DNA of UKA29 PCR amplification succeeds for fragments 9ab and 9bc but not for 9ac. CEN indicates centromeric; TEL, telomeric.
Diagram of factor VIII intron 1 and proposed model for inversion.
(A) Bar shows factor VIII intron 1, flanked by exons (drawn to indicate the direction of transcription), and containing a repeated sequence 9b (shaded) flanked by unique sequences 9a and 9c. The line shows DNA outside the F8 gene with the repeated sequence as a shaded box. Arrowheads indicate orientation of repeated sequences. The large curved arrow indicates the folding required for homologous recombination. (B,C) Homologous recombination between the two 9b repeats proposed to explain origin of inversion. (D) Inversion resulting from recombination. This shows why in the DNA of UKA29 PCR amplification succeeds for fragments 9ab and 9bc but not for 9ac. CEN indicates centromeric; TEL, telomeric.
A proof of this hypothesis required the definition of the repeats and their flanking sequences. The existence of a repeat sequence was established using the DNA from a United Kingdom hemophilia A patient with a deletion extending from the promoter region to intron 6 of theF8 gene. This DNA sustained the amplification of a 500-bp sequence identical to segment 9b. Moreover, this sequence was also amplified from 3 cosmids (183 B4, 21B11, 240D11) containing no segments of the F8 gene. These were obtained from a 49,XXXXX library18 and mapped to a region 70 to 100 kb telomeric to the F8 gene.19 The cosmids containedC6.1A gene sequences that were shown to be placed 5′ of the second exon of the F8 gene in one of the hybrid mRNAs detected in the lymphocytes of UKA29.8
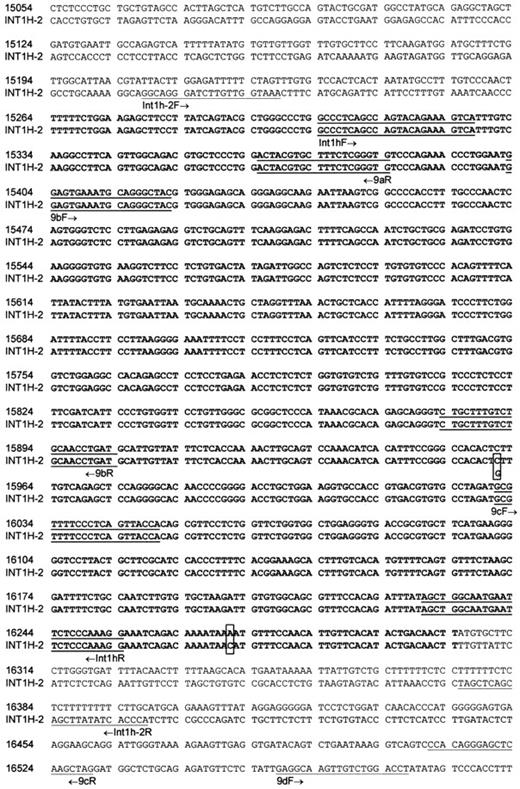
To determine the length of the repeat and identify its flanking sequences, an RsaI vectorette library of cosmid 183B4 was prepared. This library supported PCR reactions driven by primer 9bF or 9bR paired with the vectorette primer 224. The 9b primers were oriented outwardly and in opposite direction and were located in the 500 bp of known repeated sequence. The reaction containing primers 9bF and 9bR, respectively, yielded a 1-kb and an 0.8-kb product. The former comprised 901 bp identical to nucleotide number 15 403 to 16 304 of intron 1 of the F8 gene, except at nucleotide 16 271 where C was found rather than A, plus 58 bp of novel sequence. The latter PCR product was completely contained within the repeated sequence. Therefore, the second boundary of the repeat and the adjacent flanking sequence were sought by searching the GenBank database with the 500 bp of known repeated sequence. This showed that the 500-bp sequence bridged a gap between 2 independent sequence entries of 11.5 and 8.7 kb (GenBank accession no. AC016977), which contained, respectively, 216 and 160 bp of the 500-bp repeat. With the help of these sequences, a PCR was designed that amplified both wild-type genomic DNA and cosmid 183B4. This revealed that the repeated sequence extended for 1041 bp and was flanked in the PCR product by unique sequences of 56 and 94 bp (Figure 4). We call the 2 repeatsint1h (from intron 1 homology) and use -1 and -2 to specify, respectively, the copy in the F8 gene and the one more telomeric. A search of the current data on the human genome sequence does not show any other copy of int1h.
Nucleotide sequence alignment of
int1h-1 and int1h-2 and flanking sequences. Bold letters show the 1041-bp repeats. Nucleotide numbers are specific to intron 1 of the F8 gene where nt 1 is the G residue at the start of the intron. Primer binding sites are underlined with arrows to indicate orientation of primers. Polymorphic site in int1h-2 is indicated by alternative nucleotides in box. Difference between int1h-1 and int1h-2 is shown boxed.
Nucleotide sequence alignment of
int1h-1 and int1h-2 and flanking sequences. Bold letters show the 1041-bp repeats. Nucleotide numbers are specific to intron 1 of the F8 gene where nt 1 is the G residue at the start of the intron. Primer binding sites are underlined with arrows to indicate orientation of primers. Polymorphic site in int1h-2 is indicated by alternative nucleotides in box. Difference between int1h-1 and int1h-2 is shown boxed.
The int1h repeat sequence (nucleotides 15 264-16 304 of intron 1) and flanking regions were searched for both sequences of possible biologic relevance and repetitive elements using NIX.20 No biologically significant sequences were identified within the repeat region; however, int1h-1 was flanked telomerically by a 316-bp LTR/ERVL (ERVL) repeat extending from intron 1 nucleotide 14 963 to 15 252 and on the other side by a 345-bp LTR/ERVL (LTR 16A) repeat extending from nucleotide 16 397 to 16 742. The int1h-2 repeat extended on the telomeric side into a 1026-bp LINE L2 repetitive element.
According to the hypothesis illustrated in Figure 3, theint1h repeats of patient UKA29 should have recombined to yield repeats with novel arrangements of flanking sequence so that the repeat near exon 1 of the F8 gene should be flanked by the unique sequence centromeric of normal int1h-1 and the unique sequence centromeric to the normal int1h-2. Conversely, the repeat near to the C6.1A gene should be flanked by the sequences telomeric to normal int1h-1 andint1h-2. This was confirmed using the primer pairs 9aF + int1h-2R and int1h-2F + 9cR, because these yield no products in wild-type DNA while they amplify segments of 1.5 and 1.3 kb in DNA from UKA29, which show the arrangement of flanking sequences predicted above. This is in keeping with the hypothesis that the inversion of UKA29 results from homologous intranemic recombination within theint1h repeats.
Location of int1h-2 and verification of its orientation
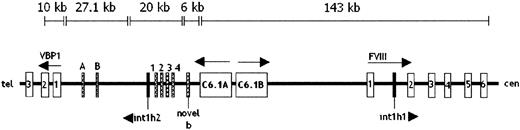
Using information from the GenBank genome database, we constructed a continuous sequence extending from intron 6 of the F8 gene to the 3′ untranslated region (UTR) of the C6.1A gene. This 143-kb sequence contains 6 overlapping sequence entries that were aligned using the Sequencher program. Unfortunately this sequence did not join up with the 20 kb contig comprising int1h-2. Therefore, to verify the orientation of int1h-2 we searched this 20-kb contig for the novel exon sequences found by Brinke et al8 in the chimeric mRNA of patient UKA29 containing all but the last exon of C6.1A and exons 2 to 26 of theF8 gene. Four of the 5 novel exons found spliced between the sequences of the 2 above genes were contained in the 20-kb contig. They were flanked by GT, AG splice consensuses and were in the order proposed by Brinke et al.8 They were separated by introns ranging in size from 440 to 3027 bp. The nucleotide analysis software, NIX,20 indicates that these exons may be part of an incompletely characterized gene encoding a protein homologous to the clathrin assembly protein. The novel 94-bp exon nearest to theC6.1A gene, called novel B, was not in the 20-kb contig but in a 6-kb sequence that should map to the gap between this contig and that of 143 kb. The order of the novel exons relative to theF8 and C6.1A genes indicates thatint1h-2 is oriented with its first residue most centromeric, whereas int1h-1 has its first residue most telomeric. A map of this region showing the relevant features is shown in Figure5.
Genomic map of the region surrounding the
int1h repeats in Xq28.Int1h repeats are shown as black boxes. Only the first 6 exons of factor VIII and first 3 exons of VBP1 are shown, with arrows indicating direction of transcription. Hatched boxes indicate facultative exons spliced into chimeric mRNAs of patients with int1h-related inversions. Exons A, B, 1, 2, 3, and 4 represent seq 29A, seq 29B, 45 bp, 30 bp, 100 bp, and 142 bp exons, respectively.8 28 Size bars indicate the location and extent of DNA sequence contigs constructed from entries in GenBank human DNA sequence database.
Genomic map of the region surrounding the
int1h repeats in Xq28.Int1h repeats are shown as black boxes. Only the first 6 exons of factor VIII and first 3 exons of VBP1 are shown, with arrows indicating direction of transcription. Hatched boxes indicate facultative exons spliced into chimeric mRNAs of patients with int1h-related inversions. Exons A, B, 1, 2, 3, and 4 represent seq 29A, seq 29B, 45 bp, 30 bp, 100 bp, and 142 bp exons, respectively.8 28 Size bars indicate the location and extent of DNA sequence contigs constructed from entries in GenBank human DNA sequence database.
Because current knowledge of the human genome sequence does not indicate the size of the gaps between the 3 genomic sequences considered above, the precise distance between int1h-1 andint1h-2 cannot be determined, but the available data suggest a minimum of 136 kb.
Similarity and conservation of int1h-1 andint1h-2
As mentioned earlier, only one difference was found between theint1h-1 sequences in GenBank AL390881 and theint1h-2 sequence from cosmid 183B4. Sequence variation inint1h was further investigated by screening the DNA of 43 United Kingdom men and 7 women using solid-phase fluorescent cleavage of mismatch.21 This showed that int1h-1 was identical in all 57 X chromosomes thus examined, whereasint1h-2 showed a polymorphism due to a C>G change at nucleotide 15 961 in 9 X chromosomes. In these 9 X chromosomes there are 2 nucleotide differences between int1h-1 andint1h-2.
PCR test for the detection of theint1h-related inversion and prevalence of this mutation
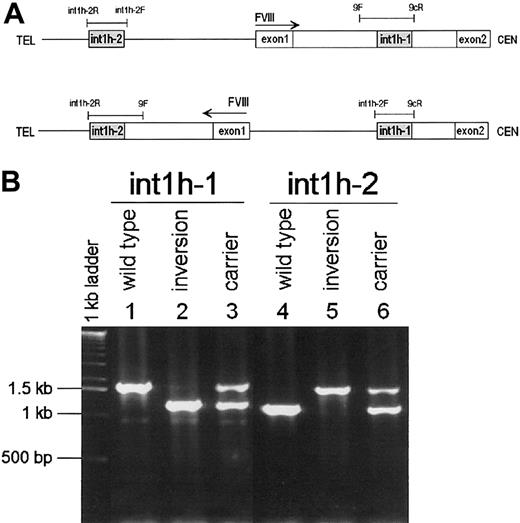
Because the inversion results in a reassortment of sequences flanking each int1h repeat, these flanking sequences were used to design 2 PCR reactions capable of detecting the inversion in patients and carriers. One reaction contains the primers specific forint1h-1 plus the primer specific for the sequence flankingint1h-2 on the centromeric side (9F, 9cR,int1h-2F); the other reaction contains the primers specific for int1h-2 plus that specific for the flanking sequence at the telomeric side of int1h-1 (int1h-2F,int1h-2R, 9F). The former reaction yields a 1.5-kb product from normal DNA and a 1.0-kb product from a patient with the inversion, whereas a female carrier's DNA yields both products. The latter reaction correspondingly yields 1, 1.5, 1 + 1.5-kb products from normal, inversion patient, and carrier DNA (Figure6).
PCR test for the FVIII
int1h-related inversion. (A) Diagram of normal (top) and inversion (bottom) DNA showing the location and orientation of relevant sequences as well as PCR primers and reactions (thin bars). (B) Amplification of int1h-1 using 9cR, 9F, andint1h-2F primers (lanes 1, 2, and 3) and int1h-2using int1h-2F, int1h-2R, and 9F primers (lanes 4, 5, and 6). Control and inversion are males; carrier is female.
PCR test for the FVIII
int1h-related inversion. (A) Diagram of normal (top) and inversion (bottom) DNA showing the location and orientation of relevant sequences as well as PCR primers and reactions (thin bars). (B) Amplification of int1h-1 using 9cR, 9F, andint1h-2F primers (lanes 1, 2, and 3) and int1h-2using int1h-2F, int1h-2R, and 9F primers (lanes 4, 5, and 6). Control and inversion are males; carrier is female.
DNA from UKA29 and the other 5 patients with the inversion were tested with the above reactions and the expected results were obtained. Thus, we used this PCR diagnostic test on the 46 most recently referred severely affected patients whose hemophilia A mutation had not yet been sought. The test was positive in 4 patients so that a total of 10int1h-related inversions was found to occur among the 209 unrelated patients with severe hemophilia A referred to our laboratory. This suggests a prevalence of 4.8% (95% CI = 2.4-8.7) in severe hemophilia A. The 9 inversion patients who could be analyzed with regard to haplotype for the BclI and intron 13 and 22 microsatellite polymorphisms of the F8 gene showed 5 different haplotypes. Among these patients one was reported to have developed inhibitors.
Formal proof that the int1h-related inversions are due to intranemic homologous recombination
All the results presented so far support the mode of origin for the int1h-related inversion proposed in Figure 3. However, formal proof of the hypothesis can only be obtained by sequencing the inversion breakpoint regions in patients. This was done for the 9 inversion patients whose DNA was available for this investigation. PCR products containing the inversion break regions were obtained as described above and sequenced. Int1h-1 andint1h-2 were found to be intact in 8 patients, whereas in one the A at nucleotide 16 271 of int1h-1 appears to have been converted to C as in int1h-2. Moreover, the repeats were precisely flanked on one side by the sequences present in wild-type DNA and on the other by the sequence normally flanking the alternative int1h repeat and expected to be part of the inverted DNA segment. These data formally prove that theint1h-related inversions are due to homologous intranemic recombination and because int1h-1 and int1h-2differ at residue 16 271, the observation that theint1h-2–specific residue was associated with the wild-typeint1h-2 flanking sequence indicates that the recombination breakpoint occurred between nucleotides 15 264 and 16 271 in 7 patients, and in 1 patient the presence of the rarer allele at the polymorphic site in int1h-2 further restricts the position of the recombination breakpoint to the segment between nt 15 264 and 15 961.
Discussion
As stated by Brinke et al,8 the intron 1 breaking inversion causes severe hemophilia A because the coding sequence for coagulation factor VIII, although transcribed, lies downstream of the translation stop codon of the C6.1A gene and is separated from the sequence coding for the factor VIII signal peptide. Thus, even if the chimeric mRNAs discovered in the patient's peripheral lymphocytes were sufficiently stable and produced in the liver in adequate amounts, no factor VIII protein should be present in the patient's blood unless unexpected internal translation of the mRNA occurred. This suggests that the patient should be predisposed to the inhibitor complication. Among the 10 patients we have identified, 1 is known to have developed inhibitors. Now a larger series of patients with this inversion is required better to estimate their empirical risk of developing inhibitors.
Our results demonstrate that Brinke et al8,10 were right in saying that the inversion breakpoint external to the F8gene does not disrupt either the C6.1A or theVBP1 gene (Figure 5) even though sequences of these genes are found spliced into mRNA with, respectively, exons 2 to 26 and exon 1 of the F8 gene. A number of facultative exons separated the factor VIII exons from those of the C6.1A andVBP1 genes in the chimeric mRNA of the inversion patients. We find that the order of the facultative exons proposed by Brinke et al8 on the basis of the structure of chimeric mRNA amplified from the lymphocytes of UKA29 agrees with the information we could obtain on their genomic location and we also find that these exons are flanked by conventional splice signals. In addition, we show that int1h-2 is at least 16 kb downstream and telomeric to the polyadenylation site of the C6.1A gene and about 37 kb upstream and centromeric to the major transcription start site of theVBP1 gene. Therefore, our patients are expected to produce normal C6.1A and VBP1 mRNA as well as the chimeric mRNA mentioned above.
The observation that in the inversion patients the intact transcription units of the C6.1A and VBP1 genes were also used to produce chimeric mRNA suggests that rearrangements may provide the opportunity to explore the value of gene products acquiring novel domains from new neighbors without loss of the original functional units. This is supported by several reports of intergenic splicing in humans.22-26
The results of Naylor et al6 and those presented above demonstrate that the same mechanism—homologous, intranemic recombination—is responsible for both the inversions breaking intron 1 and intron 22 of the F8 gene. Our data also present one suggestive instance of the conversion of nucleotide differences (A>C at nucleotide 16 271 of int1h-1) expected to occur in the hybrid DNA that forms at the sites of homologous recombination events. The int1h-related inversions occur at about one tenth the frequency of the int22h-related ones. This may largely be due to the size of the int1h repeats that are 9-fold smaller than int22h (1041 versus 9503 bp). In addition there is only one extragenic int1h repeat, whereas there are 2int22h repeats of which, however, the most telomeric is involved in the inversion 5 times more frequently than the other.22 The similarity between the copies ofint1h (as between those of int22h) is very high (ie, 99.9%) and therefore difference in the degree of similarity should not be responsible for the difference in occurrence of the inversions associated with these different repeats.
The int22h-related inversions originate mainly in the male germline26 and it was suggested that monosomy of the X chromosome may favor folding of the X chromosome onto itself and hence the occurrence of intrachromatid recombination at male meiosis. A similar male bias may characterize the int1h-related inversion, but more families with this inversion are required to verify this expectation.
Internemic recombination between int1h-1 andint1h-2, that is, recombination between these repeats from sister chromatids or homologous chromatids and chromosomes, would result in dicentric chromosomes and acentric fragments and hence should not lead to viable embryos.
The inversions causing severe hemophilia A are part of a varied group of rearrangements involving duplicons in the human genome. Ji et al27 suggest that this group of abnormalities causes diseases with a combined incidence of 0.7 to 1/1000 births. However, the inversions causing hemophilia A dramatically demonstrate how some mutations may seriously disrupt gene function and yet go undetected by strategies directed to the screening of exons in genomic sequences. Thus, the relevance of duplicon-related rearrangements to human disease may well be higher than suggested by Ji et al.27
The analysis of mRNA has allowed the detection of common inversions causing hemophilia A. This approach should be equally effective in other X-linked disorders. Once the inversions are identified, it is possible to develop rapid diagnostic tests based on the analysis of genomic DNA as we have illustrated for the intron 1 breaking inversion. This test will now allow the rapid identification of patients with severe hemophilia due to the int1h-related inversions and will become a very useful addition to the methods available to provide genetic service in hemophilia A. In addition, it will serve to identify a further group of patients who may be at high risk of developing inhibitors.
We are grateful to the hemophilia A patients and the hemophilia centers in the United Kingdom that have collaborated in the construction of the United Kingdom database of hemophilia A mutations and pedigrees. In particular, we are grateful to Dr E. A. Chalmers, Dr P. W. Collins, Dr B. T. Colvin, Professor P. L. F. Giangrande, Dr M. Laffan, Professor C. A. Lee, Dr G. L. Scott, and Dr R. F. Stevens, whose centers have referred patients and carriers with the int1h-related inversions to us. We acknowledge the secretarial help of Adrienne Knight.
Supported by Medical Research Council grant G9500698 Mb and the Guy's and St Thomas' Charitable Foundation.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Francesco Giannelli, Division of Medical and Molecular Genetics, GKT School of Medicine, 8th Fl Guy's Hospital Tower, London Bridge, London SE1 9RT, United Kingdom; e-mail:adrienne.knight@kcl.ac.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal