It was previously shown that costimulation of CD8+ lymphocytes results in de novo expression of CD4. This study expanded on this observation to investigate the function of CD4 on CD8 cells. The ability of costimulated CD8 cells to respond to interleukin 16 (IL-16), a ligand that binds CD4 and induces cellular chemotaxis, was examined. IL-16–mediated ligation of CD4 expressed on CD8 T cells was found to induce an intracellular signal that directs migration of these cells in vitro. Thus, expression of CD4 on a CD8 lymphocyte has functional importance and may serve to control distribution of newly activated CD8 T cells in vivo.
Introduction
The CD4 protein has many different functions in the development and activity of a T cell. CD4 has an important role in T-helper (Th)–cell development and response to antigen in the context of major histocompatibility complex class II (MHC II). It also serves as an adhesion molecule and a chemotactic receptor, and it has a role in cellular activation. CD4, a 58-kd transmembrane glycoprotein, is a member of the immunoglobulin family of receptors.1 The extracellular 370–amino acid portion of CD4 is folded into 4 different domains, which are designated D1 to D4.1 These domains are involved in a variety of interactions with other proteins, such as the T-cell receptor, MHC II, interleukin-16 (IL-16), and human immunodeficiency virus (HIV) gp120. Engagement of CD4 plays an important role in the initiation of events that lead to activation of Th cells or recruitment of those cells to sites of inflammation. The cytoplasmic tail of the CD4 protein was shown to associate noncovalently with Lck, a Src-family protein tyrosine kinase.2,3 Cross-linking of CD4 with monoclonal antibodies (mAbs) or stimulation of CD4 by IL-16 results in Lck tyrosine phosphorylation and subsequent tyrosine phosphorylation of other cellular proteins.4-6 These tyrosine phosphorylation events lead to modulation of cellular activation and functional responses.7
CD4 expression is tightly regulated during the T-cell development process, and this control has an important role in T-cell maturation and function. The coordinate cell-surface expression of CD4 and CD8 and the subsequent down-regulation of either CD4 or CD8 are definitive markers of T-cell ontogeny. Furthermore, expression of one of these molecules on mature T cells is indicative of successful selection and commitment of these cells to either the Th or T-cytotoxic lineage.8 However, previous ideas on the terminal differentiation of a cell into either a CD4+CD8− Th cell or a CD4−CD8+ T-cytotoxic cell have been questioned. Several studies, including one of ours, found that activation of mature CD4−CD8+ (CD8 single-positive [SP]) T cells by costimulation9,10or superantigen stimulation11 results in expression of CD4, which renders these cells susceptible to infection by HIV-1.9-11 We previously found that CD45RA+CD8 SP cells respond to costimulation with greater expression of CD4 than do CD45RO+ CD8 SP cells.10 Thus, expression of CD4 can be modulated on T cells at more mature stages of development than was previously thought, suggesting that CD4 may be involved in mature CD8 T-cell function.
The CD4 molecule was previously shown to function as a chemotactic receptor for both IL-16 and the viral surface glycoprotein HIV gp120 on CD4+ Th cells. IL-16 specifically binds the D4 region, and gp120 binds the D1 region of CD4.12,13 IL-16 is a 14-kd molecule that forms homotetramers, which are required for biologic activity.14 This cytokine is produced by a variety of cells, including CD8 and CD4 T cells, eosinophils, mast cells, bronchial epithelial cells, synovial fibroblasts, and cells in the dermis and epidermis.15-18 Expression of IL-16 by the bronchial epithelium in patients with asthma, by the dermis and epidermis in patients with atopic dermatitis, and by synovial fibroblasts in patients with rheumatoid arthritis was found to correlate with CD4+ cell influx.15,17-19Furthermore, recombinant IL-16 induced chemotaxis of CD4+ T cells in vitro through interaction with CD4,13,14 and HIV-1 gp120 binding to CD4 increased CD4 T-cell activation and directed cell migration in vitro.20 21
In the current study, we expanded on our previous findings and determined that the level of CD4 cell-surface expression on CD8+ T cells is directly proportional to the level of costimulation with T-cell receptor and CD28. We also found that CD4 can direct migration of CD8+ T cells in response to IL-16 and that this migration depends on intracellular tyrosine phosphorylation signal-transduction events. Finally, we determined that HIV gp120 from CXC chemokine receptor 4 (CXCR 4)–tropic strains induces migration of these CD4+CD8+ cells in a CD4- and CXCR4-dependent manner. Thus, our data suggest that de novo expression of CD4 on CD8+ lymphocytes may function to control the trafficking and distribution of newly activated, previously naive CD8 T cells to sites of inflammation in vivo.
Materials and methods
Cell isolation and culture
Fresh peripheral blood was obtained from healthy human donors, and peripheral blood mononuclear cells (PBMC) were isolated using Ficoll-Hypaque separation (Sigma, St Louis, MO). CD8+ T cells were purified using negative selection columns (R&D Systems, Minneapolis, MN) to a purity of greater than 99% as determined by flow cytometry. After isolation, cells were cultured in RPMI 1640 medium containing penicillin (100 U/mL), streptomycin (100 μg/mL) (Sigma), and 10% human AB serum (Gemini Bioproducts, Calabasas, CA). Cells were stimulated by culture in either phytohemagglutinin (PHA; 1 μg/mL; Sigma), anti-CD3 mAb (OKT3; 50 μg/mL; Ortho Biotech, Raritan, NJ) immobilized on plates coated with goat antimouse antibody, or immobilized anti-CD3 mAb and soluble anti-CD28 in various concentrations.
Flow cytometry
Fluorescein isothiocyanate (FITC)–conjugated, phycoerythrin (PE)–conjugated, or allophycocyanin-conjugated mAbs specific for human CD4, CD8, CD25, CD69, and CD71 were obtained from Becton Dickinson (Mountain View, CA). FITC-conjugated and PE-conjugated mAbs specific for human CCR5 (clone 2D7) and CXCR4 (clone 12G5) were obtained from Pharmingen (San Diego, CA). PE-Texas Red (ECD)–conjugated and PE-cytochrome 5–conjugated mAbs specific for CD4 and CD8 were obtained from Coulter (Hialeah, FL). Four-color flow cytometry was done with a fluorescence-activated cell-sorter scanner flow cytometer (FACSCalibur; Becton Dickinson) or an EPICS flow cytometer (Coulter). Immunophenotypic analysis was done using Cellquest (Becton Dickinson) or FloJo (Treestar, San Mateo, CA) software. Instrument settings for flow cytometry were as described previously.10
Phosphotyrosine blotting assays
Purified CD8+ cells were costimulated for 3 days as described above. Cells (5 × 107/mL) were then either left untreated or pretreated with PP1 (10 μM) for 15 minutes. Alternatively, cells were incubated with antihuman CD4 (OKT4) for 30 minutes at 4°C. Then, 125 μg/mL goat antimouse mAb was added to cross-link OKT4 for 2 minutes. Subsequently, cells were washed and incubated with 1 μg/mL recombinant IL-16 or medium alone at 37°C for 2 minutes. Cells were lysed in buffer (50 mM Tris [pH 8.0], 1% Nonidet P-40, and 20 mM EDTA) containing protease inhibitors (10 μg/mL aprotinin,10 μg/mL leupeptin,1 mM phenylmethylsulfonyl fluoride, and 1mM sodium orthovanadate) for 30 minutes at 4°C.22 OKT4 (25 μg/mL) was added to all cell lysates in the presence of protein G–Sepharose (Pharmacia), and the lysates were incubated at 4°C overnight. Proteins were resolved by using sodium dodecyl sulfate–polyacrylamide gel electrophoresis and visualized with Western blotting. Membranes were blocked with 50 mM Tris (pH 8.0), 2.5 mM EDTA, 50 mM sodium chloride, 0.1% Tween 20, and 5% bovine serum albumin (BSA), and proteins were detected using 4G10 mAb (antiphosphotyrosine) or anti-CD4 mAb (R&D Systems) followed by iodine 125–protein A and autoradiography. Band intensity was quantified using phosphoimage analysis (Kodak, New Haven, CT).
Cell-migration assays
Purified CD8+ cells at a concentration of 1 × 106/mL were costimulated with anti-CD3 and anti-CD28 mAbs for 3 or 4 days as described previously.10 Cells were then either fluorescently labeled for rapid, sensitive enumeration purposes by culturing in the presence of 5 μg/mL Calcein-AM (Molecular Probes, Eugene, OR) for 30 minutes at 37°C23 or, in certain experiments, left unlabeled. Subsequently, cells (3.6 × 106 cells/mL) were placed in RPMI 1610 medium without phenol red (Irvine Scientific, Irvine, CA) but containing 1% BSA (Sigma). Labeled cells (90 000 cells; 25 μL) were placed on top of filters (8-μm pores) that were part of 96-well ChemoTX chemotaxis plates (Neuro Probe, Gaithersburg, MD). Recombinant human IL-16 (Pharmingen), rgp120 (National Institutes of Health Acquired Immunodeficiency Disease Research and Reference Reagent Program), recombinant stromal-derived factor 1α (SDF-1α), recombinant macrophage inflammatory protein 1α (MIP-1α; R&D Systems), or medium alone was added to the lower chambers at various concentrations in replicates of 6. Anti-CD4 mAb OKT4 (Ortho Biotech; 1 μg/mL), which competitively inhibits binding of IL-1613,14,24; anti-CD4 mAb Leu3a (Becton Dickinson; 1 μg/mL), which competitively inhibits binding of gp120 to CD412; and unconjugated 12G5 (1 μg/mL), which competitively inhibits gp120 interaction with CXCR4,25,26were used to control for migration specificity and were either placed in the bottom chamber with the appropriate chemotactic agent or used to prestain the cells. The Src-family kinase inhibitor PP127 was used at a concentration of 5 μM in certain experiments.
Migration from the top chamber to the bottom chamber was allowed to take place during a 2-hour incubation at 37°C. The filters were then removed and the chemotaxis chamber was placed in a multiwell fluorescent plate reader (Molecular Devices, Sunnyvale, CA) configured in a bottom-read position (excitation, 485 nm; emission, 530 nm).23 Cells that migrated to the bottom chamber were enumerated by comparing the Calcein fluorescence signal with standard numbers of labeled cells in the same plate. Migration of unlabeled cells was assessed by combining replicate wells and direct counting using a hemacytometer. Statistical analysis was done by using Wilcoxon rank sum testing (SAS software; SAS Institute, Cary, NC). When a single measurement deviated from the replicate measurements, rejection criteria were based on the Q-test.28 No more than one data point was rejected for each condition in each experiment (at least 5 replicates were used for each condition).
Results
Expression of CD4 on CD8+ T lymphocytes
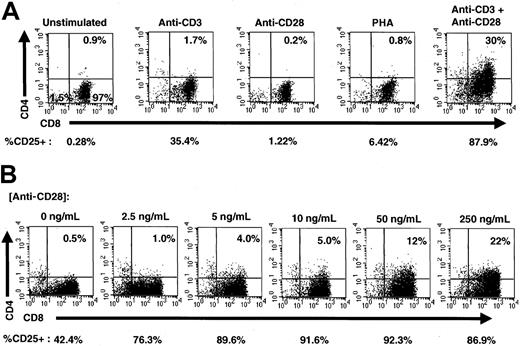
To determine activation signals that induce expression of CD4 on CD8 cells, highly purified CD8 SP T cells were isolated from fresh PBMC from adults and stimulated in the presence of either anti-CD3 mAb alone, anti-CD28 mAb alone, PHA, or anti-CD3 and anti-CD28 together. Three days after stimulation, cells were analyzed for expression of CD4 and CD8 using flow cytometry (Figure 1A). Consistent with our previous findings,10 costimulation by anti-CD3 and anti-CD28 mAbs resulted in de novo expression of CD4, whereas stimulation by either anti-CD3 alone, anti-CD28 alone, or PHA did not produce significant levels of CD4 expression on the surface of these cells. The costimulated population of cells also expressed higher levels of the activation markers CD25 (Figure 1A), CD69, and CD71 (data not shown). In fact, according to gating assessments, more than 99% of CD8+CD4+ cells expressed CD25 at this time. In contrast, approximately 88% of the total CD8+ population expressed CD25, indicating that the CD4+ population was highly activated.
Costimulation induces CD4 expression on CD8+ cells.
(A) Stimulation of purified CD8+ T lymphocytes. CD8+ T cells (> 99% purity) were left unstimulated or stimulated with anti-CD3 alone, anti-CD28 alone, PHA, or simultaneously with anti-CD3 and anti-CD28 for 3 days and analyzed for expression of CD4 (PE), CD8 (ECD), and CD25 (FITC). The percentage of CD8+ cells expressing CD4 is shown in the upper right-hand corner of each dot plot, and the percentage of total cells expressing CD25 is shown below each dot plot. (B) Costimulation of purified CD8+ T cells. Purified CD8+ T lymphocytes were costimulated with plate-bound anti-CD3 and increasing concentrations of anti-CD28 for 3 days and analyzed for expression of CD4 (PE), CD8 (ECD), and CD25 (FITC). The amount of soluble anti-CD28 added to each culture is shown above each dot plot, and the percentage of CD8+cells expressing CD4 is shown in the upper right-hand corner of each dot plot. The percentage of total cells expressing CD25 is shown below each dot plot.
Costimulation induces CD4 expression on CD8+ cells.
(A) Stimulation of purified CD8+ T lymphocytes. CD8+ T cells (> 99% purity) were left unstimulated or stimulated with anti-CD3 alone, anti-CD28 alone, PHA, or simultaneously with anti-CD3 and anti-CD28 for 3 days and analyzed for expression of CD4 (PE), CD8 (ECD), and CD25 (FITC). The percentage of CD8+ cells expressing CD4 is shown in the upper right-hand corner of each dot plot, and the percentage of total cells expressing CD25 is shown below each dot plot. (B) Costimulation of purified CD8+ T cells. Purified CD8+ T lymphocytes were costimulated with plate-bound anti-CD3 and increasing concentrations of anti-CD28 for 3 days and analyzed for expression of CD4 (PE), CD8 (ECD), and CD25 (FITC). The amount of soluble anti-CD28 added to each culture is shown above each dot plot, and the percentage of CD8+cells expressing CD4 is shown in the upper right-hand corner of each dot plot. The percentage of total cells expressing CD25 is shown below each dot plot.
To investigate whether the induction of cell-surface CD4 expression was proportional to the level of costimulation, we titrated anti-CD28 mAb in the presence of a constant level of anti-CD3 adherent to the plates. Purified CD8 T cells were stimulated for 3 days in increasing concentrations of anti-CD28 and analyzed for expression of CD4, CD8, and CD25 using flow cytometry. The level of CD4 expression was directly proportional to the level of anti-CD28 costimulatory signal, with increased costimulation producing increased CD4 expression (Figure 1B). CD25 expression also increased with costimulation, although maximal CD25 expression was observed before maximal CD4 expression. The coordinate expression of CD4 and CD25, especially at lower levels of anti-CD28 costimulatory signal, strongly suggests that any functional role of CD4 on CD8+ T cells is related to cellular activation events.
CD4 and chemotaxis of CD8+ T cells induced by IL-16
IL-16 was previously found to induce chemotaxis by binding the CD4 molecule.13,14 Therefore, we examined whether newly expressed CD4 could direct migration of a CD8+ cell in response to IL-16. Purified CD8+ T cells were costimulated with anti-CD3 and anti-CD28 mAbs for 3 days and then assessed for induction of chemotaxis in vitro by using a transwell assay. IL-16 induced significant chemotaxis of costimulated cells in a dose-dependent manner (Figure 2A). To determine whether the observed migration was due to specific interaction between IL-16 and CD4, the mAb OKT4, which competitively inhibits IL-16 binding, was placed in the bottom chamber with IL-16. OKT4 competitively inhibited IL-16–mediated chemotaxis, indicating that the IL-16 migration was specific for CD4. SDF-1α was used as a positive control for migration of these cells, which are positive for CXCR4.10 11 CXCR4-mediated migration by SDF-1α was not inhibited by OKT4; however, migration was competitively inhibited by the anti-CXCR4 mAb 12G5 (data not shown), which did not induce migration by itself or inhibit background migration levels. Placement of IL-16, SDF1α, OKT4, or 12G5 in the top and bottom chambers in equal concentrations did not induce chemokinesis (data not shown), indicating that the cellular migration observed in the earlier studies occurs in response to the chemokine gradient. Prestaining cells with OKT4 or 12G5 had the same effect of blocking migration induced by IL-16 or SDF, respectively (data not shown). Thus, expression of CD4 on the surface of a CD8+ T lymphocyte directs migration in response to IL-16.
Chemotaxis of CD8+ cells.
(A) IL-16–induced migration of costimulated CD8+ T cells. Purified CD8+ T cells were costimulated for 3 days. Cells were removed from culture and CD4 expression was verified (46% of cells were CD4+CD8+). Cells were then labeled with Calcein-AM, placed on 96-well chemotaxis chambers, and allowed to migrate in response to the indicated concentrations of IL-16 or SDF1α (250 ng/mL). Results are the percentage of cells migrating compared with medium-only controls (set at 100% migration). Anti-CD4 mAb (OKT4) was added to the lower chamber with the indicated samples. Asterisks indicate samples with results significantly different from those for medium-only controls (P < .05). (B) Migration of unstimulated and PHA-stimulated CD8+ T cells. Purified CD8+ T cells were left unstimulated or stimulated with PHA for 3 days. Cells were then removed from culture and CD4 expression was examined (≤ 1% of unstimulated cells or PHA-stimulated cells expressed CD4). Subsequently, cells were labeled with Calcein-AM, placed on 96-well chemotaxis chambers, and allowed to migrate in response to IL-16 (200 ng/mL) or SDF1α (250 ng/mL). Results are the percentage of cells migrating compared with unstimulated, medium-only controls (set at 100% migration). Asterisks indicate samples with results significantly different from those for medium-only controls (P ≤ .05). (C) Intensity of CD4 expression on migrating cells. Purified CD8+ T cells were costimulated for 3 days, placed on 96-well chemotaxis chambers, and allowed to migrate in response to IL-16 (200 ng/mL) or SDF1α (250 ng/mL). After 2 hours of incubation, cells that had migrated to the lower chamber were pooled, enumerated, stained for CD4 (allophycocyanin) and CD8 (ECD), and analyzed by flow cytometry. The graph shows the mean fluorescence intensity of CD4 expression on cells that migrated.
Chemotaxis of CD8+ cells.
(A) IL-16–induced migration of costimulated CD8+ T cells. Purified CD8+ T cells were costimulated for 3 days. Cells were removed from culture and CD4 expression was verified (46% of cells were CD4+CD8+). Cells were then labeled with Calcein-AM, placed on 96-well chemotaxis chambers, and allowed to migrate in response to the indicated concentrations of IL-16 or SDF1α (250 ng/mL). Results are the percentage of cells migrating compared with medium-only controls (set at 100% migration). Anti-CD4 mAb (OKT4) was added to the lower chamber with the indicated samples. Asterisks indicate samples with results significantly different from those for medium-only controls (P < .05). (B) Migration of unstimulated and PHA-stimulated CD8+ T cells. Purified CD8+ T cells were left unstimulated or stimulated with PHA for 3 days. Cells were then removed from culture and CD4 expression was examined (≤ 1% of unstimulated cells or PHA-stimulated cells expressed CD4). Subsequently, cells were labeled with Calcein-AM, placed on 96-well chemotaxis chambers, and allowed to migrate in response to IL-16 (200 ng/mL) or SDF1α (250 ng/mL). Results are the percentage of cells migrating compared with unstimulated, medium-only controls (set at 100% migration). Asterisks indicate samples with results significantly different from those for medium-only controls (P ≤ .05). (C) Intensity of CD4 expression on migrating cells. Purified CD8+ T cells were costimulated for 3 days, placed on 96-well chemotaxis chambers, and allowed to migrate in response to IL-16 (200 ng/mL) or SDF1α (250 ng/mL). After 2 hours of incubation, cells that had migrated to the lower chamber were pooled, enumerated, stained for CD4 (allophycocyanin) and CD8 (ECD), and analyzed by flow cytometry. The graph shows the mean fluorescence intensity of CD4 expression on cells that migrated.
To provide an additional control, we examined whether IL-16 could induce migration in unstimulated or PHA-stimulated CD8+ T cells, which do not express CD4. Unstimulated or PHA-stimulated cells cultured for 3 days were examined for their ability to migrate in response to IL-16 (200 ng/mL), SDF1α (250 ng/mL), or medium alone. SDF1α induced significant levels of migration in unstimulated CD8+ T cells, whereas IL-16 did not (Figure 2B). Stimulation of CD8+ T cells with PHA did not result in random migration that was significantly different from migration of unstimulated cells in medium only. In addition, IL-16 did not induce significant migration of PHA-stimulated CD8+ T cells. These data support the idea that expression of CD4 is required for migration in response to IL-16.
To determine the phenotype of cells that migrated in response to IL-16, purified CD8+ T cells were costimulated with anti-CD3 and anti-CD28 for 3 days and assessed for their ability to migrate in response IL-16 (200 ng/mL), SDF-1α (250 ng/mL), or medium alone. Cells that migrated into the lower chamber were then enumerated, stained with anti-CD4 and anti-CD8, and analyzed by flow cytometry. IL-16–induced migration in CD8+ T cells was 196% and SDF1α-induced migration was 336% that of medium-only controls. The mean fluorescence intensity of CD4 on cells migrating in response to IL-16 was significantly higher than that on cells migrating in response to SDF-1α and cells in medium only (Figure 2C). The selective migration in response to IL-16 of cells expressing higher levels of CD4 further supports the idea that this migration is mediated through CD4.
CD4 on bona fide CD4 T cells is known to associate with Lck, a member of the Src kinase family.4 To determine whether IL-16–induced migration of activated CD8 T cells was dependent on intracellular signaling through a Src-family kinase, we examined cellular migration in the presence and absence of the Src-family kinase–specific inhibitor PP1.27 Cells were preincubated in the presence of PP1, and migration in response to IL-16 and MIP1α was assessed. PP1 had no effect on total cell viability; however, inhibition of IL-16–induced migration was observed after treatment with PP1 (Figure 3). In contrast, migration in response to MIP1α was not significantly inhibited. In experiments using Western blotting, we also observed increased tyrosine phosphorylation of a high-molecular-weight, CD4–associated protein in costimulated CD8 T cells treated with medium containing IL-16 or medium containing cross-linking anti-CD4 mAbs, compared with results in costimulated cells treated with medium alone (data not shown). Furthermore, we found that pretreatment with PP1 blocked IL-16–induced tyrosine phosphorylation of this protein (data not shown). Together, these results indicate that Src-family kinases are involved in CD4-mediated signal transduction in CD8 cells and that a different signaling pathway is used to allow cellular migration in response to MIP1α.
Effect of the Src-family kinase inhibitor PP1 on migration of CD8+ T cells.
CD8+ T cells were costimulated for 3 days, stained with Calcein-AM, and left untreated or treated with 5 μM PP1. At this time, 32% of cells expressed CD4. Results are the percentage of cells migrating in response to IL-16 or MIP1α compared with medium-only (no inhibitor) controls. Asterisks indicate samples with levels of migration to the designated chemotactic factor after treatment with PP1 that were significantly different from levels of samples treated with the same chemotactic factor alone (P < .05).
Effect of the Src-family kinase inhibitor PP1 on migration of CD8+ T cells.
CD8+ T cells were costimulated for 3 days, stained with Calcein-AM, and left untreated or treated with 5 μM PP1. At this time, 32% of cells expressed CD4. Results are the percentage of cells migrating in response to IL-16 or MIP1α compared with medium-only (no inhibitor) controls. Asterisks indicate samples with levels of migration to the designated chemotactic factor after treatment with PP1 that were significantly different from levels of samples treated with the same chemotactic factor alone (P < .05).
CD4 and cell migration induced by HIV-1 gp120
To determine whether another ligand of CD4, HIV-1 gp120, induces migration of CD8+ T cells that express CD4, purified CD8+ T cells were costimulated for 3 days and examined for migration in response to gp120 from several different CXCR4-tropic viral isolates. HIV gp120 from HIV-1SF2 and HIV-1MN induced a significant chemotactic response (Figure4A). A nonglycosylated version of gp120 from HIV-1SF2, gp120 from HIV-1CM, and gp120 from a clade E isolate of HIV-1 induced migration levels that were consistently greater than control levels in 3 separate experiments, although the difference was not significant. When equal concentrations of gp120 from each isolate were placed in the top and bottom chambers, no cell chemokinesis was observed (data not shown).
Effect of HIV gp120 on CD8+ cell migration.
(A) HIV-1 gp120–induced migration of CD8+ T cells. CD8+ T cells were stimulated for 3 days and examined for their ability to migrate in response to IL-16 (200 ng/mL), SDF1α (250 ng/mL), MIP1α (500 ng/mL), and gp120 (2 μg/mL) from each of 5 different HIV-1 isolates. The HIV-1 gp120s were a glycosylated version from HIV-1SF2 (SF2 Glyc), a nonglycosylated version from HIV-1SF2 (SF2 Non-Glyc), HIV-1MN (MN), HIV-1CM (CM), and HIV-1clade E (clade E). Results are the percentage of cells migrating compared with medium-only controls. Asterisks indicate conditions that induced migration levels that were significantly different from those of medium-only controls (P < .05). (B) Inhibition of gp120-induced migration of CD8+ T cells. CD8+ T cells costimulated for 3 days were assessed for their ability to migrate to IL-16 (200 ng/mL) and HIV-1 gp120 (SF-2 Glyc). A mAb that inhibits gp120 interaction with CD4 (anti-CD4, Leu3a) and a mAb that inhibits gp120 interaction with CXCR4 (anti-CXCR4, 12G5) were placed in the lower chambers with gp120 in the indicated samples. The graph shows migration induced under the various conditions compared with that of medium-only controls, and asterisks indicate conditions that induced migration levels that were significantly different from those of controls (P < .05). Migration in response to IL-16 was not inhibited by 12G5 (data not shown).
Effect of HIV gp120 on CD8+ cell migration.
(A) HIV-1 gp120–induced migration of CD8+ T cells. CD8+ T cells were stimulated for 3 days and examined for their ability to migrate in response to IL-16 (200 ng/mL), SDF1α (250 ng/mL), MIP1α (500 ng/mL), and gp120 (2 μg/mL) from each of 5 different HIV-1 isolates. The HIV-1 gp120s were a glycosylated version from HIV-1SF2 (SF2 Glyc), a nonglycosylated version from HIV-1SF2 (SF2 Non-Glyc), HIV-1MN (MN), HIV-1CM (CM), and HIV-1clade E (clade E). Results are the percentage of cells migrating compared with medium-only controls. Asterisks indicate conditions that induced migration levels that were significantly different from those of medium-only controls (P < .05). (B) Inhibition of gp120-induced migration of CD8+ T cells. CD8+ T cells costimulated for 3 days were assessed for their ability to migrate to IL-16 (200 ng/mL) and HIV-1 gp120 (SF-2 Glyc). A mAb that inhibits gp120 interaction with CD4 (anti-CD4, Leu3a) and a mAb that inhibits gp120 interaction with CXCR4 (anti-CXCR4, 12G5) were placed in the lower chambers with gp120 in the indicated samples. The graph shows migration induced under the various conditions compared with that of medium-only controls, and asterisks indicate conditions that induced migration levels that were significantly different from those of controls (P < .05). Migration in response to IL-16 was not inhibited by 12G5 (data not shown).
To investigate whether the migratory response induced by gp120 was specific for CD4, anti-CD4 mAb (Leu3a), which binds the D1 region of CD4 and competitively inhibits gp120 interaction, was placed in the bottom chamber with gp120 from HIV-1SF2 (glycosylated). Leu3a completely inhibited gp120-induced migration (Figure 4B), thereby showing that migration in response to HIV gp120 is dependent on CD4. Moreover, blocking gp120 interaction with the viral entry coreceptor CXCR4 by using the CXCR4-specific mAb 12G5 also inhibited the migration induced by this CXCR4-tropic envelope protein. Similar results were observed with HIV-1 gp120 from HIV-1MN, HIV-1SF2, HIV-1CM, and gp120 from a clade E isolate. Prestaining cells with either 12G5 or Leu3a had the same effect of blocking migration induced by gp120. In control experiments (data not shown), we found that the 12G5 mAb did not inhibit IL-16–mediated migration of these cells. These results indicate that HIV gp120–induced migration of CD4+CD8 cells is dependent on interaction with both CD4 and the viral entry coreceptor CXCR4.
Discussion
The chemotactic response of CD8+CD4dimcells to IL-16 may have implications for cellular redistribution and localization of activated CD8 T cells in vivo. Our data suggest that because primarily naı̈ve CD8+ T cells express CD4 after activation, newly activated cells would be recruited to sites of infection or inflammation. Production of IL-16 by various cells at the site of inflammation could induce infiltration of these cells by means of chemoattraction. This could play an important part in the generation and maintenance of primary antiviral immunity. Furthermore, such a mechanism could affect the numbers or function of CD8+CD4dim cells at a site of inflammation, since IL-16 can modulate cellular activation.16 Because memory CD8 cells do not express detectable levels of CD4 in vivo, this protein may be lost as the cell enters the memory state. Alternatively, CD4 expression may be a characteristic of CD8 effector cells that are destined to die without ever becoming memory cells. Additional studies are needed to explore these ideas to determine the function of CD8+CD4dim cells.
We and others previously showed that expression of CD4 on CD8+ T lymphocytes renders those cells susceptible to infection by HIV-1.9-11 In the current study, we found that HIV-1 gp120 can also induce migration of these cells in a CD4- and CXCR4-dependent manner. Previous studies determined that HIV-1 gp120 can induce migration of bona fide CD4+ and CD8+ T cells.20,21 These studies showed that this migration can be induced in a CD4- or CXCR4-dependent manner and that migration of resting CD8+ T cells can be induced in a CXCR4-dependent manner. Our results suggest that interaction of gp120 with both CD4 and CXCR4 is required for migration of CD8+CD4dim cells. The gp120-induced migration is most likely mediated through interaction with CXCR4, since blocking this interaction with mAb 12G5 inhibited migration. It is known that structural changes in gp120 after interaction with CD4 allow sequential interaction with the chemokine coreceptor.29-32Interaction of gp120 with CD4 on CD8+ T cells likely results in exposure of epitopes in gp120 that interact with CXCR4 and induce the observed migration. Modification of recombinant gp120 proteins also appears to affect HIV-1 gp120–induced migration, as is observed of migration induced by a glycosylated protein compared with that induced by a nonglycosylated version of the same protein. The deglycosylated form of the HIV-1 gp120 (SF2) used in our studies was previously found to have more binding affinity toward CD4 and CXCR4 than its glycosylated counterpart.32 It is unclear how this would result in a protein less able to stimulate chemotaxis. The biologic outcome of gp120-induced migration of CD8+CD4dim T cells is unknown; however, it is possible that gp120 gradients recruit these cells to sites of HIV replication where they may become target cells for infection.
Our results show that CD4 expression on CD8+ T cells has functional importance. Engagement of CD4 with IL-16 or cross-linking with OKT4 resulted in initiation of intracellular tyrosine phosphorylation events, and the migration induced by IL-16, but not MIP1α, was found to be dependent on the activity of Src-family kinase or kinases. Increased tyrosine phosphorylation of Lck after treatment of cells with IL-16 was not observed. However, another high-molecular-weight cellular phosphoprotein that associated with CD4 was detected, and phosphorylation of this protein was inhibited by treatment with PP1 (data not shown). Our data suggest that IL-16 induces tyrosine phosphorylation events by means of a Src-family kinase, resulting in the chemotactic response of CD8+ T cells that express CD4. Lck remains a strong candidate for this signaling because of its known association with CD4 in this cell type.11 Our inability to observe increases in the tyrosine phosphorylation of this molecule could have been due to high background in this highly activated cell type. Our studies suggest that CD4 is an activation marker and a chemotactic receptor on a subset of CD8+ T cells and that the CD8+CD4dim cells in human peripheral blood may be cells that are actively responding to antigen.33 34
We thank Ted Sarafian for technical assistance with fluorometry.
Supported by National Institutes of Health grants AI 36554 and AI 36059 and the UCLA Center for AIDS Research. J.A.Z. is an Elizabeth Glaser scientist supported by the Pediatric AIDS Foundation. S.G.K. is a recipient of the UCLA Center for Clinical AIDS Research and Education HIV Pathogenesis Institutional Training Grant.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Jerome A. Zack, UCLA AIDS Institute/UCLA Dept of Medicine, Division of Hematology/Oncology, 10833 LeConte Ave, Rm 11-934 Louis Factor Bldg, Los Angeles, CA 90095; e-mail: jzack@ucla.edu.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal