Under conditions of impaired T-cell immunity, human cytomegalovirus (HCMV) can reactivate from lifelong latency, resulting in potentially fatal disease. A crucial role for CD8+ T cells has been demonstrated in control of viral replication, and high levels of HCMV-specific cytotoxic T-lymphocytes are seen in immunocompetent HCMV-seropositive individuals despite very low viral loads. Elucidation of the minimum portion of the anti-HCMV T-cell repertoire that is required to suppress viral replication requires further study of clonal composition. The ability of dendritic cells to take up and process exogenous viral antigen by constitutive macropinocytosis was used to study HCMV-specific T-cell memory in the absence of viral replication. The specificity and clonal composition of the CD8+ T-cell responses were evaluated using HLA tetrameric complexes and T-cell receptor β chain (TCRBV) spectratypic analyses. There was a skewed reactivity toward the matrix protein pp65, with up to 40-fold expansion of CD8+ T cells directed toward a single peptide-MHC combination. Individual expansions detected on TCRBV spectratype analysis were HCMV-specific and composed of single or highly restricted numbers of clones. There was preferential TCRBV gene usage (BV6.1/6.2, BV8, and BV13 in HLA-A*0201+ individuals) but lack of conservation of CDR3 length and junctional motifs between donors. While there was a spectrum of TCR repertoire diversity directed toward individual MHC-peptide combinations between donors, a relatively small number of clones appeared to predominate the response in each case. These data provide further insight into the range of anti-HCMV responses and will aid the design and monitoring of adoptive immunotherapy protocols.
Introduction
Human cytomegalovirus (HCMV) is a ubiquitous β-herpesvirus that infects up to 90% of individuals depending on the population studied. In common with other members of the herpesvirus group, HCMV is able to persist lifelong in a latent or persistent state following primary infection, remaining under the control of the immune system.1 HCMV remains a significant cause of morbidity and mortality following allogeneic bone marrow transplantation, due to reactivation of virus acquired prior to transplantation.2,3 Pressures to expand the potential donor pool to facilitate more widespread application of transplantation have led to the use of conditioning protocols or posttransplantation immunosuppressive regimens that result in more profound immune suppression and enhanced risk of viral infection or reactivation.4 The predisposition to viral reactivation appears to relate to the absence of a sufficient cytotoxic T-lymphocyte (CTL) response,5 because reconstitution of CD8+ HCMV-reactive T cells or supplementation by adoptive transfer of T-cell clones has been shown to correlate with a reduced risk of HCMV disease.5,6 While the development of sensitive surveillance methods to monitor for HCMV reactivation allows available therapies to be targeted to those at greatest risk for the development of HCMV disease,7,8 the limitations of currently available surveillance protocols and antiviral pharmacotherapies has led to the need to develop alternative treatment strategies, particularly because HCMV seropositivity of recipient or donor prior to transplantation remains a significant adverse prognostic factor in some studies.4
Adoptive transfer of unmanipulated T cells has been demonstrated to mediate both antiviral and antitumor immunity in humans.9 However, the cells' use has been constrained by the low frequency of antigen-specific T cells and by the presence of alloreactive T cells, which have the potential to induce graft versus host disease.9,10 Because of these considerations, the HCMV-specific CD8+ CTLs used for patient infusion in early studies by Riddell and colleagues were cloned from bulk cultures.6,11 These studies demonstrated the ability of infused cells to reconstitute short-term HCMV-specific CTL responses, which appeared sufficient to prevent the development of HCMV disease. In addition, they confirmed the requirement for CD4+ T-cell helper function to restore longer-term immune memory as had been suggested by earlier work in murine models.12 No adverse effects, including graft versus host disease, were documented. However, large numbers (up to 1 × 109/m2) of CD8+ cells were required at regular intervals to maintain in vivo anti-HCMV responses, and this required several weeks of cell culture and large-scale cloning that are both time-consuming and costly. These practical difficulties may partially explain why these studies have not led to the widespread use of adoptive immunotherapy for prevention and treatment of HCMV infection.
The human CTL response to CMV is dominated by structural protein pp65, targeted by 70% to 90% of HCMV-specific CTLs.13,14 Other immunogenic peptides accounting for a smaller part of the overall response include elements of the major immediate-early gene product (IE-1), the matrix protein pp150, and virion envelope glycoprotein B.15,16 Immunodominant nonapeptides from the pp65 matrix glycoprotein that are restricted to specific HLA molecules are being identified. In addition, the development of techniques to detect and quantify antigen-specific T cells now enable more detailed analysis of culture output cells and provide powerful tools for monitoring the fate of infused cells in immunotherapy protocols.17,18 As more interest focuses on the in vitro manipulation of immune responses in order to dissociate potentially beneficial from harmful effects, the elucidation of the “sufficient T-cell repertoire”19required to enable clinical efficacy (ie, the minimum portion of the overall T-cell response necessary to allow, for example, sustained viral clearance or regression of Epstein-Barr viris [EBV]–associated lymphoproliferative disorders) becomes increasingly important. Studies to more precisely document the overall clonal composition of T-cell responses and any differences or similarities in the responses of different individuals are required as a prelude to clinical studies based on selection of more restricted subpopulations of T cells. We have combined HCMV pp65–specific HLA tetrameric complex analysis with TCRBV spectratype analysis to evaluate HCMV-specific CD8+T-cell memory responses in HLA-A*0201+ and/or HLA-B*0702+ individuals following in vitro restimulation by dendritic cells (DCs) pulsed with exogenous viral antigen.
Materials and methods
Generation of human monocyte-derived DCs and CTL culture
Generation of human monocyte-derived DCs and CTL culture has been described previously.20 Briefly, fresh peripheral blood mononuclear cells from 8 HLA-A*0201+ and/or HLA-B*0702+ HCMV-seropositive donors were prepared by Ficoll-Paque density centrifugation (Pharmacia, St Albans, United Kingdom), suspended in X Vivo 20 medium (Biowhittaker, Wokingham, United Kingdom), and allowed to adhere to tissue culture flasks for 2 to 3 hours. Nonadherent cells were removed by washing 2 times with phosphate-buffered saline. Adherent cells were differentiated into immature DCs in X Vivo 20 medium supplemented with 10% autologous human serum, 100 ng/mL interleukin-4 (Insight Biotechnology, Wembley, United Kingdom), and 100 ng/mL granulocyte-macrophage colony-stimulating factor (Hoescht, Hounslow, United Kingdom) for 7 days in a 37°C 5% CO2 humidified incubator.
Autologous peripheral blood lymphocytes (PBLs) and monocyte-derived DCs were cocultured for 14 to 21 days in X Vivo 20 medium supplemented with 10% autologous human serum and 1 mg/mL CMV antigen (Dade Behring, Marburg, Germany) in tissue culture flasks. The cocultures were restimulated on day 7 with further autologous DCs and 0.5 mg/mL CMV antigen. From day 10, cocultures were supplemented with 20 U/mL interleukin-2 (Sigma, Poole, United Kingdom) every 2 days. For cocultures maintained longer than 14 days, there was an additional restimulation with autologous CMV antigen–pulsed DCs on day 14.
Characterization of the coculture system
The proliferative and cytolytic characteristics of the cocultures have been detailed elsewhere.20 Briefly, in 15 donor cocultures studied, T-cell proliferation (as detected in [3H]thymidine incorporation assays) in response to the control antigen was significantly less than in response to the HCMV antigen (P = .0007, Wilcoxon signed rank test) but often slightly greater than proliferation without any antigen (P = .01, Wilcoxon signed rank test). There was a median 2.1-fold expansion in total lymphocyte numbers over the 2-week culture period. Unseparated mononuclear cells obtained from the culture were used as effectors in cytotoxicity assays. Cells from 12 of 14 cocultures from HCMV-seropositive donors tested showed HCMV-specific cytotoxicity against HCMV-infected fibroblasts/DCs or antigen-pulsed DCs (median 14.7% specific lysis, range 6.1%-42% at maximal effector:target ratios of 20:1, vs median 0%, range 0%-5.1% for uninfected/unpulsed targets; P = .002, Wilcoxon signed rank test). In general, therefore, the culture conditions promoted the development of HCMV-specific cytotoxicity. In addition, killing was HLA-restricted because only autologous but not allogeneic HLA-mismatched target cells were lysed (median 0.8%, range 0%-5.1% specific lysis, P = .005, Wilcoxon signed rank test), and lysis was prevented by preincubation of targets with HLA class I blocking antibodies.
Tetrameric complex formation
Soluble MHC-peptide tetramers were produced using standard approaches.18 Briefly, recombinant HLA-A*0201 and HLA-B*0702 heavy chain and β2-microglobulin protein were produced in Escherichia coli cells transformed with the relevant expression vectors. Expression of the HLA heavy chain was limited to the extracellular domain, and the C-terminus of this domain was modified by the addition of a substrate sequence for the biotinylating enzyme BirA. The peptide epitopes were the HLA-A*0201–restricted NLVPMVATV epitope (amino acids 495-503 of the lower matrix protein pp65) and the HLA-B*0702 epitope, TPRVTGGGAM (amino acids 417-426 of the lower matrix protein pp65). Monomeric HLA-peptide complexes were folded in vitro by addition of HLA protein to β2-microglobulin in the presence of appropriate peptide. The MHC complexes were biotinylated using purified recombinant BirA enzyme and were then purified by gel filtration and anion exchange chromatography. HLA-peptide tetramers were made by mixing the biotinylated protein complex with streptavidin-phycoerythrin (PE) (Leinco, St Louis, MO) at a molar ratio of 4:1. Tetramers were purified by gel filtration on a Sephadex S-200 column (Amersham Pharmacia, St Albans, United Kingdom).
Flow cytometric analysis
Lymphocytes from day 14 or day 21 cocultures were both triple-stained with fluorescein isothiocyanate–conjugated anti-CD4 mAb (Dako, Ely, United Kingdom), PE-conjugated anti-CD8 mAb (Dako), and PE Cy5–conjugated anti-CD3 mAb (Dako) and dual-stained with the appropriate HLA-restricted HCMV-specific tetrameric complex and PE Cy5–conjugated anti-CD8 mAb (Dako) in 20 to 30 μL phosphate-buffered saline, 2% bovine serum albumin, and azide for 60 minutes on ice. Cells were washed twice in the same buffer and analyzed immediately in a flow cytometer (BeckmanCoulter, High Wycombe, United Kingdom). Between 4 × 106 and 1 × 107 cells were analyzed in the samples stained with tetrameric complex. Negative controls included cells from HCMV-seronegative individuals expressing the appropriate HLA antigen and from HCMV-seropositive individuals not expressing the appropriate HLA antigen. In addition, the HLA-A*0201 tetrameric complex provided a negative control for HLA-B*0702+ individuals and the HLA-B*0702 tetrameric complex for the HLA-A*0201+ individuals. These additional controls were performed for donors 01 to 04 but not thereafter in order to maximize the number of cells available for subsequent analysis. Cells demonstrating dual staining with CD8 mAb and tetrameric complex were electronically sorted on a BeckmanCoulter EPICS Elite flow cytometer.
TCR CDR3 spectratyping
RNA was extracted from preculture and postculture PBLs using Ultraspec RNA (BiotecX Laboratories, Houston, TX) according to the manufacturer's protocol. Complementary DNA (cDNA) was generated from 1 μg RNA in a 30 μL reaction using random hexanucleotide primers for reverse transcription with reverse transcriptase (Superscript, Gibco BRL, Paisley, United Kingdom). In the case of cells sorted according to dual staining with CD8 mAb and tetrameric complex (4000-17 000 cells), RNA was extracted in a similar manner but using glycogen (Boehringer Mannheim, Lewes, United Kingdom) as a carrier. The total RNA extracted was used to generate cDNA in a 15 μL reaction. Each of 22 functionally rearranged T-cell receptor (TCR) β chain variable (BV) gene subfamilies was amplified across the complementarity determining region 3 (CDR3)–encoding regions using the 24 BV subfamily-specific primers described previously by Maslanka et al21 and a fluorescent dye–conjugated (FAM, Perkin Elmer, Cambridge, United Kingdom) β chain constant (BC) region-specific primer (Table1). The polymerase chain reaction (PCR) product lengths using this technique reflect the CDR3 lengths of the input TCR RNA, being dependent upon joining (BJ) and diversity (BD) gene segment usage along with the balance of exonuclease activity and N nucleotide addition by terminal transferase at the junctional regions. Peaks corresponding to in-frame transcripts are detected at 3 nucleotide intervals. The appearance of a dominant peak suggests the presence of an oligoclonal or clonal T-cell population, while the absence of peaks or entire subfamily spectratypes suggests the absence of T cells of the given CDR3 length or BV subfamily, respectively. BV primers were combined in duplex PCR reactions as follows: BV 5.1 plus 1; BV2 plus 12; BV13 plus 3; BV4 plus 5.3; BV8 plus 7; BV9 plus 14; BV11 plus 20; BV17 plus 15; BV16 plus 21; BV18 plus 23; and BV24 plus 22. BV6.1 and BV6.2 were used unpaired.
Primer sequences of TCR BV and BC chains
| TCR primer . | Sequence . | Size (mer) . |
|---|---|---|
| BV1 | CAGTTCCCTGACTTGCACTC | 20 |
| BV2 | GCTTCTACATCTGCAGTGC | 19 |
| BV3 | GAGAGAAGAAGGAGCGCTTC | 20 |
| BV4 | GCAGCATATATCTCTGCAGC | 20 |
| BV5.1 | CTCGGCCCTTTATCTTTGCG | 20 |
| BV5.3 | CCCTAACTATAGCTCTGAGC | 20 |
| BV6.1 | GATCCAGCGCACACAGC | 17 |
| BV6.2 | GATCCAGCGCACAGAGC | 17 |
| BV7 | CCTGAATGCCCCAACAGC | 18 |
| BV8 | GAACCCAGGGACTCAGCTG | 19 |
| BV9 | GGAGCTTGGTGACTCTGCTG | 20 |
| BV11 | CAGGCCCTCACATACCTCTCA | 21 |
| BV12 | CAAAGACAGAGGATTTCCTCC | 21 |
| BV13 | GTCGGCTGCTCCCTCCC | 17 |
| BV14 | GTCTCTCGAAAAGAGAAGAGG | 21 |
| BV15 | GTCTCTCGACAGGCACAGGC | 20 |
| BV16 | GAACTGGAGGATTCTGGAGTT | 21 |
| BV17 | CCAAAAGAACCCGACAGCTTTC | 22 |
| BV18 | GTGCGAGGAGATTCGGCAGC | 20 |
| BV20 | CACACCCCAGGACCGGCAG | 19 |
| BV21 | GGCTCAAAGGAGTAGACTCC | 20 |
| BV22 | GTTGAAAGGCCTGATGGATC | 20 |
| BV23 | CAGTTCAGTGACTATCATTCTG | 22 |
| BV24 | GGGGACGCAGCCATGTACC | 19 |
| BC | CTGTGTTTGAGCCATCAGAAGC | 22 |
| TCR primer . | Sequence . | Size (mer) . |
|---|---|---|
| BV1 | CAGTTCCCTGACTTGCACTC | 20 |
| BV2 | GCTTCTACATCTGCAGTGC | 19 |
| BV3 | GAGAGAAGAAGGAGCGCTTC | 20 |
| BV4 | GCAGCATATATCTCTGCAGC | 20 |
| BV5.1 | CTCGGCCCTTTATCTTTGCG | 20 |
| BV5.3 | CCCTAACTATAGCTCTGAGC | 20 |
| BV6.1 | GATCCAGCGCACACAGC | 17 |
| BV6.2 | GATCCAGCGCACAGAGC | 17 |
| BV7 | CCTGAATGCCCCAACAGC | 18 |
| BV8 | GAACCCAGGGACTCAGCTG | 19 |
| BV9 | GGAGCTTGGTGACTCTGCTG | 20 |
| BV11 | CAGGCCCTCACATACCTCTCA | 21 |
| BV12 | CAAAGACAGAGGATTTCCTCC | 21 |
| BV13 | GTCGGCTGCTCCCTCCC | 17 |
| BV14 | GTCTCTCGAAAAGAGAAGAGG | 21 |
| BV15 | GTCTCTCGACAGGCACAGGC | 20 |
| BV16 | GAACTGGAGGATTCTGGAGTT | 21 |
| BV17 | CCAAAAGAACCCGACAGCTTTC | 22 |
| BV18 | GTGCGAGGAGATTCGGCAGC | 20 |
| BV20 | CACACCCCAGGACCGGCAG | 19 |
| BV21 | GGCTCAAAGGAGTAGACTCC | 20 |
| BV22 | GTTGAAAGGCCTGATGGATC | 20 |
| BV23 | CAGTTCAGTGACTATCATTCTG | 22 |
| BV24 | GGGGACGCAGCCATGTACC | 19 |
| BC | CTGTGTTTGAGCCATCAGAAGC | 22 |
The primers were as previously published by Maslanka et al.21
Hot-start PCR amplifications were performed in a total volume of 20 μL containing Genamp PCR buffer (Perkin Elmer), 2 mM MgCl2, 0.2 mM each dNTP, 1 mM of each primer, and 1 μL cDNA (equivalent to approximately 25 000 cells). In the case of the cells sorted according to dual staining with CD8 mAb and tetrameric complex, the total cDNA was divided equally between the PCR reactions. After a 5-minute denaturation step at 95°C, 0.5 U Amplitaq DNA polymerase (Perkin Elmer) was added. Optimal cycling conditions were 95°C for 30 seconds, 58°C for 30 seconds, and 72°C for 45 seconds, for 30 cycles, followed by a final extension at 72°C for 5 minutes. One microliter of PCR product was denatured in 12 μL formamide and electrophoresed through Performance Optimized Polymer 4 (Perkin Elmer) on an ABI 110 automated sequencer (Perkin Elmer) in the presence of Tamra 500 size standard (Perkin Elmer). Genescan software 2.1 (Perkin Elmer) was used to analyze the data.
TCR CDR3 cloning and sequencing
Fresh spectratype PCR products were cloned using TA Cloning Kit Dual Promoter Version B (pCRII vector) with TOP10F′ One Shot chemically competent E coli (Invitrogen, Groningen, The Netherlands) according to the manufacturer's instructions. Transformed cells were selected by ampicillin resistance and blue/white colony screening and then plucked and cultured in 96-well plates in the presence of 50 μg/mL ampicillin (Gibco BRL). Cells from each well were further screened by PCR amplification of TCR CDR3 sequence as previously described using primers appropriate for the expected BV family. Those expressing a product of the appropriate size as determined by agar gel electrophoresis with ethidium bromide staining were amplified in 50 μL PCR reactions and the amplicons purified using Wizard Plus Minipreps DNA Purification System (Promega, Southhampton, United Kingdom) according to the manufacturer's instructions. The DNA yield and purity were determined by spectrophotometry. A total of 90 ng of the purified PCR product was used as the template for sequencing using the ABI PRISM Dye Terminator Cycle Sequencing Ready Reaction Kit (Perkin Elmer). The primers used were the same as those used for the TCR CDR3 spectratype PCR reactions. Cycle sequencing was performed on the GeneAmp PCR Systems 2400 (Perkin Elmer) (96°C for 10 seconds, 50°C for 5 seconds, 60°C for 4 minutes for 25 cycles with rapid thermal ramping). Following ethanol precipitation of the extension products, the samples were resuspended in 25 μL Template Suppression Reagent (Perkin Elmer), denatured for 95°C for 2 minutes, and electrophoresed through Performance Optimized Polymer 6 (Perkin Elmer) on an ABI 110 automated sequencer (Perkin Elmer). The data were processed using ABI Prism DNA Sequencing Analysis software (Perkin Elmer).
Results
Phenotypic analysis of coculture output cells
The coculture output cells consisted of mixed populations of both CD4+ and CD8+ cells. The percentage of CD8-expressing cells varied between 10% and 47% (Table 1). HCMV-specific tetrameric complex staining of donor peripheral blood mononuclear cells prior to culture ranged from less than 0.1% to 0.6% of the CD8+ population. Following coculture, this increased to 0.3% to 8.0% of the CD8+ population, demonstrating a maximal 40-fold relative expansion. In 6 of 6 evaluable donors, there was at least a 10-fold expansion with 1 of the 2 HCMV-specific tetrameric complexes (Table 2 and Figure1). Donors 06 and 08 did not have preculture tetrameric complex studies performed and, thus, the degree of expansion in these donors was impossible to enumerate. The proliferation assay results for donor 08 were close to the median for the group (15 606 cpm; range, 11 875-56 574 cpm), suggesting that the low level of staining after the culture was not a result of a failure of the culture system to stimulate proliferation. Specificity of the tetrameric complexes was confirmed by the absence of staining of cells from HCMV-seronegative donors of the appropriate HLA type (both before the culture and after the culture) and the absence of staining of cells from HCMV-seropositive donors not expressing the appropriate HLA molecules (data not shown).
Phenotypic analysis of postculture T cells
| Donor . | Postculture CD8 positivity (% CD3*cells) . | Tetramer binding (% CD8* cells) . | |||
|---|---|---|---|---|---|
| HLA-A*0201 tetramer . | HLA-B*0702 tetramer . | ||||
| Preculture . | Postculture . | Preculture . | Postculture . | ||
| 01 | 25 | <0.1 | 0.3 | 0.6 | 6.5 |
| 02 | 10 | 0.2 | 8.0 | ||
| 03 | 12 | 0.1 | 1.6 | ||
| 04 | 35 | 0.1 | 5.4 | ||
| 05 | 26 | 0.1 | 1.3 | ||
| 06 | 47 | NA | 0.3 | NA | 7.9 |
| 07 | 21 | 0.1 | 1.1 | ||
| 08 | 39 | NA | 0.4 | ||
| Donor . | Postculture CD8 positivity (% CD3*cells) . | Tetramer binding (% CD8* cells) . | |||
|---|---|---|---|---|---|
| HLA-A*0201 tetramer . | HLA-B*0702 tetramer . | ||||
| Preculture . | Postculture . | Preculture . | Postculture . | ||
| 01 | 25 | <0.1 | 0.3 | 0.6 | 6.5 |
| 02 | 10 | 0.2 | 8.0 | ||
| 03 | 12 | 0.1 | 1.6 | ||
| 04 | 35 | 0.1 | 5.4 | ||
| 05 | 26 | 0.1 | 1.3 | ||
| 06 | 47 | NA | 0.3 | NA | 7.9 |
| 07 | 21 | 0.1 | 1.1 | ||
| 08 | 39 | NA | 0.4 | ||
NA indicates not available.
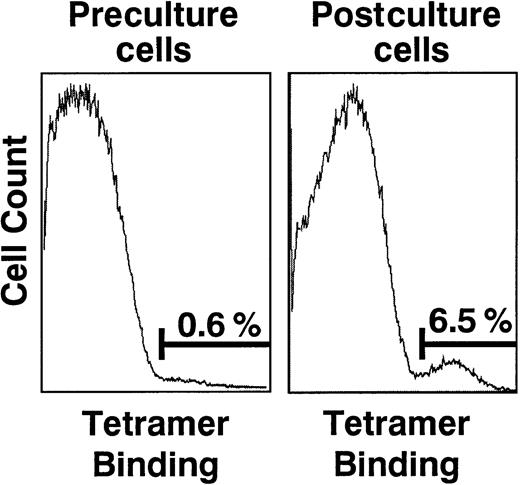
Phenotypic analyses of preculture and postculture cells from donor 01.
Cells were dual-stained with PE Cy5–conjugated anti-CD8 mAb and PE-conjugated HCMV pp65–specific HLA-B*0702 tetrameric complexes. The single parameter histograms are gated onto the CD8+ cell population and demonstrate an increase in the population of cells binding the tetrameric complexes over the period of culture.
Phenotypic analyses of preculture and postculture cells from donor 01.
Cells were dual-stained with PE Cy5–conjugated anti-CD8 mAb and PE-conjugated HCMV pp65–specific HLA-B*0702 tetrameric complexes. The single parameter histograms are gated onto the CD8+ cell population and demonstrate an increase in the population of cells binding the tetrameric complexes over the period of culture.
TCR CDR3 spectratyping
Preculture spectratypic analysis revealed typically complex appearances, with some skewing of a basic background Gaussian distribution of size classes evident in some of the BV subfamilies. Comparison of preculture and postculture unselected TCR CDR3 spectratypes showed that postculture T-cell repertoires were still polyclonal, ie, most BV spectratypes displayed a complete set of size class peaks. While most spectratypes were similar to the preculture spectratypes, some BV families contained one or more predominant size classes after the culture. When analysis was restricted to the CD8+ subset of cells, 7 of the 8 donors showed much more highly restricted postculture T-cell repertoires with fewer peaks and more irregular profiles (Figure 2). This became more pronounced when the culture period was prolonged to 21 days (Figure 3). The spectratypes of the cells sorted on the basis of dual staining with anti-CD8 mAb and HCMV-specific tetrameric complex showed even more highly restricted T-cell repertoires (Figures 3 and 4). In general, either no size classes were represented within a given BV subfamily (Figure 4, BV12, BV3, and BV9), or between 1 and 4 size classes were represented with a predominance of 1 or 2 members (Figure3, BV6.1, and Figure 4, BV13 and BV14). In 5 donors a minority of the BV subfamilies (range, 2-4; median, 3) demonstrated more than 5 peaks following tetrameric complex–guided sorting, but these were generally within a restricted subset of subfamilies (mainly BV6.1/6.2, BV13, and BV2), and again there was a predominance of 1 or 2 members (Figure 4, BV2 and BV6.1). The predominant peaks selected following tetrameric complex–guided sorting often corresponded to the expansions demonstrated in the unselected CD8+ subset, suggesting that the peptide specificity of the anti-HCMV response was skewed toward pp65 (Figure 3, BV6.1). However, examples were seen of both tetrameric complex–sorted cell size class peaks without correspondingly expanded size classes in the unselected CD8+ subset and expanded size classes in the unselected CD8+ subset, which were not present following tetrameric complex–guided sorting (Figure5). The former can be explained by selection of HCMV-specific CTLs that account quantitatively for less of the overall anti-HCMV immune response. The latter may represent expansions to other pp65 epitopes (associated with the same or different HLA molecules), to alternate HCMV proteins, or to antigens unrelated to HCMV.
TCRBV spectratypic profiles from donor 04.
Profiles are for both (A) the unselected preculture PBLs and (B) the postculture CD8+ population of cells. The postculture profile showed a more restricted repertoire, with prominent skewing evident in a number of families.
TCRBV spectratypic profiles from donor 04.
Profiles are for both (A) the unselected preculture PBLs and (B) the postculture CD8+ population of cells. The postculture profile showed a more restricted repertoire, with prominent skewing evident in a number of families.
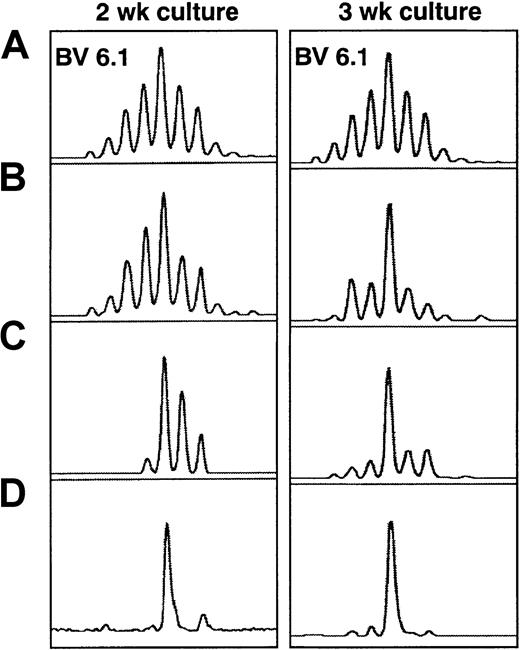
TCRBV spectratypic profiles from donor 02 at 2 and 3 weeks of culture.
BV6.1 profiles are shown for (A) preculture, (B) postculture unselected, (C) postculture CD8+ selected, and (D) postculture HCMV pp65–specific HLA-A*0201 tetrameric complex–selected cells. The tetramer-selected population showed a highly restricted repertoire. The major size class remained the same and became more apparent in the unselected population after 3 weeks of culture.
TCRBV spectratypic profiles from donor 02 at 2 and 3 weeks of culture.
BV6.1 profiles are shown for (A) preculture, (B) postculture unselected, (C) postculture CD8+ selected, and (D) postculture HCMV pp65–specific HLA-A*0201 tetrameric complex–selected cells. The tetramer-selected population showed a highly restricted repertoire. The major size class remained the same and became more apparent in the unselected population after 3 weeks of culture.
TCRBV spectratypic profiles from donor 05.
Profiles are for (A) the unselected preculture PBLs and (B) the postculture HCMV pp65–specific HLA-A*0201 tetrameric complex–selected cells. Many BV families had no representative members following tetramer-guided sorting. There was a predominance of 1 or 2 size classes in those families in which representatives were present.
TCRBV spectratypic profiles from donor 05.
Profiles are for (A) the unselected preculture PBLs and (B) the postculture HCMV pp65–specific HLA-A*0201 tetrameric complex–selected cells. Many BV families had no representative members following tetramer-guided sorting. There was a predominance of 1 or 2 size classes in those families in which representatives were present.
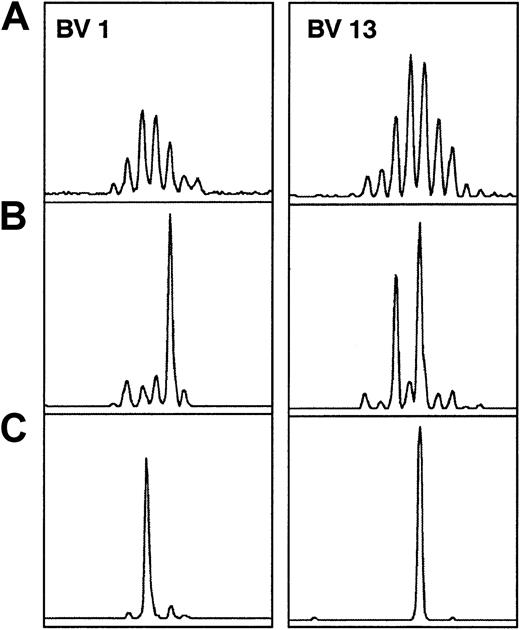
TCRBV spectratypic profiles from donor 04 for BV1 and BV13.
Profiles are for (A) preculture, (B) postculture CD8+selected, and (C) postculture HCMV pp65–specific HLA-A*0201 tetrameric complex–selected cells. Examples are shown of populations that were expanded after the culture that were selected based on tetramer-guided sorting (BV13—the larger of the 2 major size classes in panel B) along with those that were not selected (BV1 and BV13). In addition, the size class selected in panel C for BV1 showed no correspondingly expanded size class peak in the CD8+ selected population.
TCRBV spectratypic profiles from donor 04 for BV1 and BV13.
Profiles are for (A) preculture, (B) postculture CD8+selected, and (C) postculture HCMV pp65–specific HLA-A*0201 tetrameric complex–selected cells. Examples are shown of populations that were expanded after the culture that were selected based on tetramer-guided sorting (BV13—the larger of the 2 major size classes in panel B) along with those that were not selected (BV1 and BV13). In addition, the size class selected in panel C for BV1 showed no correspondingly expanded size class peak in the CD8+ selected population.
The number of BV subfamilies in which at least one size class was represented following HLA-A*0201 tetrameric complex–guided sorting was variable between donors (range, 5-20; median, 10). Due to variations in amplification efficiency between individual PCR reactions, strict quantitative comparison of the PCR products between subfamilies for any given individual is not valid. Allowing for this caveat, a normal spectratypic profile is composed of 180 to 190 size class peaks with no individual peak accounting for more than 5% of the total peak heights and/or area under the peaks. The spectratypes of all 8 donors whose postculture cells were sorted on the basis of dual staining with anti-CD8 mAb and HCMV-specific HLA-A*0201 tetrameric complex showed at least 3 (range, 3-5; median, 4) “major” peaks accounting for more than 5% of the total area under the peaks. In 5 of these donors (all with 4 major peaks), the combined area of these major peaks accounted for more than 50% of the total area under the peaks (51%-71%, median 61%), demonstrating a highly focused response to the single HCMV pp65 epitope. In these donors the combined area of the 10 largest peaks accounted for 63% to 91% of the total. In the other 3 donors, the combined area of the major peaks accounted for 31% to 42% and of the 10 largest peaks, 50% to 62% of the total area under the peaks. These donors therefore appeared to show a slightly less focused immune response, although overall a continuous spectrum of response was evident in terms of diversity of size class peaks represented.
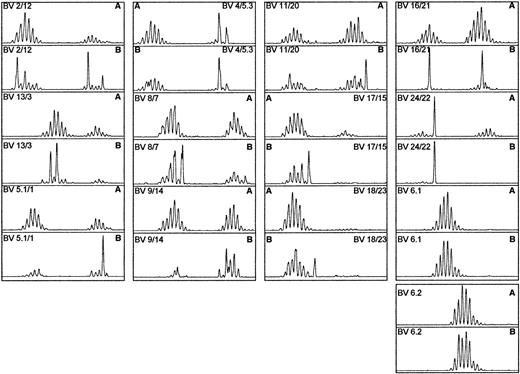
There was clear evidence of preferential BV gene usage among donors. Major peaks from HLA-A*0201 tetrameric complex–sorted cells were present in BV6.1/6.2 and BV8 in 6 of 8 donors and in BV13 in 4 of 8 donors (Figure 6). At least one size class peak was present in BV6.1/6.2 and BV8 in all 8 donors and in BV13 in 7 of 8 (Figure 7).
BV gene usage of the major size class peaks selected on the basis of HCMV pp65–specific HLA-A*0201 tetrameric complex staining.
A major peak was defined as accounting for more than 5% of the total area under the peaks in the TCRBV spectratypic profile of an individual donor.
BV gene usage of the major size class peaks selected on the basis of HCMV pp65–specific HLA-A*0201 tetrameric complex staining.
A major peak was defined as accounting for more than 5% of the total area under the peaks in the TCRBV spectratypic profile of an individual donor.
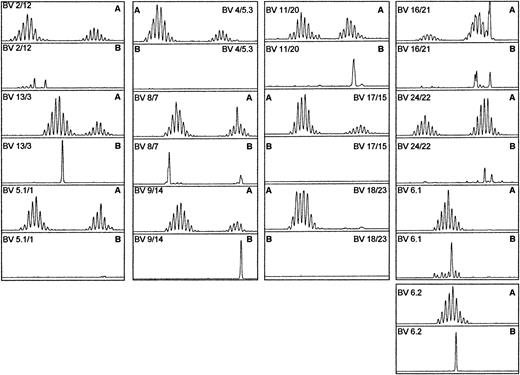
TCRBV spectratypic profiles for BV8 and BV13 of all 8 donors following HCMV pp65–specific HLA-A*0201 tetrameric complex–guided sorting.
The profiles are aligned according to PCR product size with a control profile for comparison and show a lack of conservation of a single TCR CDR3 size among the donors.
TCRBV spectratypic profiles for BV8 and BV13 of all 8 donors following HCMV pp65–specific HLA-A*0201 tetrameric complex–guided sorting.
The profiles are aligned according to PCR product size with a control profile for comparison and show a lack of conservation of a single TCR CDR3 size among the donors.
The results following sorting on the basis of dual staining with anti-CD8 mAb and HLA-B*0702 tetrameric complex in donors 01 and 06 were similar to those obtained with the HLA-A*0201 tetrameric complex. In both donors there were 20 to 30 times more CD8+ cells that bound the HLA-B*0702 tetrameric complex as compared with the HLA-A*0201 tetrameric complex following culture (Table2). Postculture spectratypes following tetrameric complex–guided sorting demonstrated 4 and 6 major size class peaks, respectively, which accounted for 78% and 46% of the total area under the peaks. Representatives were present in 8 BV subfamilies in donor 01 and 23 in donor 06, showing the same extremes of the spectrum of immune response focusing seen with the HLA-A*0201 tetrameric complexes. Major peaks were present in BV6.2 and BV14 in donor 01 (2 major peaks in each) and in BV1, BV4, BV9, BV14, BV6.1, and BV6.2 in donor 06.
Overall there appeared to be a lack of TCR CDR3 length conservation of the tetrameric complex–sorted T cells for a given BV family when compared between different donors (Figure 7).
Sorting based on HLA-A*0201 tetrameric complex binding was performed on 3 separate occasions on cells cultured from donor 02. Comparison of the spectratypic analysis from cells selected after 2 and 3 weeks of culture showed that the most abundant size classes remained identical (Figure 3). Analysis of cells recultured for 2 weeks from fresh peripheral blood mononuclear cells also demonstrated that the same size classes were selected. These data suggest that HCMV pp65 epitope-specific CTLs with the same CDR3 size are reproducibly expanded in the culture system and that the effects of stochastic events on the postsorting spectratype appearances remain minimal despite the relatively low numbers of cells used for RNA preparation following sorting.
Cloning and sequencing of CDR3 spectratype PCR products
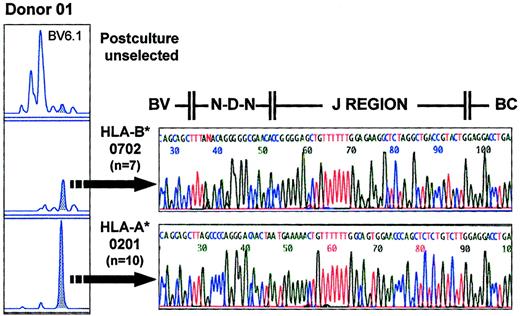
Individual size class peaks of the CDR3 spectratype analysis may represent the CDR3-encoding RNA of a single T-cell clone or may represent the CDR3-encoding RNA of multiple clones with identical CDR3 length. To test whether the immune responses to HCMV resulted in preferential expansions of multiple clones with a particular CDR3 length or of a single clone within a given BV family, we cloned the spectratype PCR products and sequenced the corresponding DNA regions. The postsorting spectratypes of donor 01 showed an identical size peak selected in the BV6.1 subfamily with both the HLA-A*0201 and HLA-B*0702 tetrameric complexes (Figure 8). The sequence of 7 clones from the HLA-B*0702–sorted population was identical. That of 10 clones from the HLA-A*0201–sorted population was also identical but differed from that of the HLA-B*0702–sorted cells in both J region usage and VDJ region sequence (Figure 8). These results confirm the specificity of the 2 distinct HLA tetrameric complexes and suggest that each size peak is composed of either a single clone or a very limited number of clones. Sequence comparison with the HLA-A*0201 tetrameric complex–sorted major size class peak from BV6.1 of donor 02 (5 of 5 clones with identical sequence) showed different J region usage and different CDR3 length (Figure9). Thus, T-cell clones sharing the same BV region and selected by the same HLA tetrameric complex in different individuals display markedly different CDR3 nucleotide and predicted amino acid sequences. Similarly, comparison of the sequences derived from size class peaks in the same or different BV families from a single donor selected with the same HLA-A*0201 tetrameric complex (BV6.1 and BV15 [10 of 10 clones with identical sequence] in Figure9) demonstrates that a single HLA molecule–nonapeptide combination can be recognized by a number of TCRs with differing TCR CDR3 lengths and differing predicted amino amino acid sequences within the same individual.
Sequence analysis of tetramer-selected populations.
The panel on the left shows the postculture TCRBV spectratypic profiles for BV6.1 of donor 01, including the unselected, the HLA-B*0702 tetramer–selected, and HLA-A*0201 tetramer–selected populations. Seven and 10 clones representing the major peaks (shaded) of the HLA-B*0702 tetramer–selected and HLA-A*0201 tetramer–selected populations, respectively, showed identical sequence, indicating that each size class peak is composed of a single or very limited number of clones. The sequence data confirm a lack of conservation of CDR3 motifs between the clones selected by the different tetrameric complexes.
Sequence analysis of tetramer-selected populations.
The panel on the left shows the postculture TCRBV spectratypic profiles for BV6.1 of donor 01, including the unselected, the HLA-B*0702 tetramer–selected, and HLA-A*0201 tetramer–selected populations. Seven and 10 clones representing the major peaks (shaded) of the HLA-B*0702 tetramer–selected and HLA-A*0201 tetramer–selected populations, respectively, showed identical sequence, indicating that each size class peak is composed of a single or very limited number of clones. The sequence data confirm a lack of conservation of CDR3 motifs between the clones selected by the different tetrameric complexes.
TCR CDR3 sequence data for clones corresponding to the size class peaks indicated for donor 01 and donor 02 (selected with the HLA-A*0201 tetrameric complex).
There was a lack of conservation of CDR3 length and no evidence of frequent usage of public clonotypes both between the same BV family in different donors (BV6.1 donor 01 and donor 02), different size class peaks from the same BV family in a single donor (BV6.1 donor 02), and different BV families in a single donor (BV6.1 and BV15 donor 02).
TCR CDR3 sequence data for clones corresponding to the size class peaks indicated for donor 01 and donor 02 (selected with the HLA-A*0201 tetrameric complex).
There was a lack of conservation of CDR3 length and no evidence of frequent usage of public clonotypes both between the same BV family in different donors (BV6.1 donor 01 and donor 02), different size class peaks from the same BV family in a single donor (BV6.1 donor 02), and different BV families in a single donor (BV6.1 and BV15 donor 02).
The sequences of the TCR CDR3 regions corresponding to the major peaks in BV15 and BV6.1 of donor 02 following culture and tetrameric complex–guided sorting on 2 separate occasions were also identical (Figure 10), confirming that the expanded T-cells with conserved TCR CDR3 lengths were indeed due to reproducible expansions of the same clones in these cases.
Reproducibility of clonal expansion and tetrameric complex–guided selection.
The spectratype appearances of BV15 and BV6.1 are shown following coculture and tetrameric complex–guided sorting on 2 separate occasions for donor 02. The major peaks selected were identical in size and sequence.
Reproducibility of clonal expansion and tetrameric complex–guided selection.
The spectratype appearances of BV15 and BV6.1 are shown following coculture and tetrameric complex–guided sorting on 2 separate occasions for donor 02. The major peaks selected were identical in size and sequence.
Discussion
We have studied immune responses to HCMV antigen in a culture system that is free of live HCMV virions and that uses monocyte-derived DCs as antigen-presenting cells (APCs). DCs are potent APCs that are able to process exogenously supplied antigen and present peptide fragments on both class I and class II HLA molecules, along with an array of costimulatory molecules.22,23 This in turn allows stimulation of lymphocytes expressing either CD8 or CD4 in combination with an appropriate TCR. We have previously demonstrated that cells derived from such a culture system that has been used to stimulate a recall response to HCMV antigen can be shown to possess HCMV-specific HLA-restricted cytotoxic activity, although the system has not allowed the expansion of HCMV-specific T cells from HCMV-naive donors.20 The development of techniques to allow identification of antigen-specific populations of T cells such as the enzyme-linked immunospot assay and HLA tetrameric complexes now allows more detailed analyses of immune responses.17,18 These developments are in part based on an increasing knowledge of the immunodominant epitopes involved in specific responses. Tetrameric complex analysis is particularly suited to cases in which restricted numbers of defined epitopes dominate the response, allowing direct quantification of the antigen-specific cells. Studies in healthy virus carriers of the immune response to HCMV pp65 have shown that in some individuals the peptide specificity of the pp65-specific CTLs is highly focused to a single peptide while in others multiple pp65 peptides are recognized.14 These studies have helped to define the minimal CD8+ T-cell epitopes of HCMV and their HLA restrictions,14,24 which in turn have allowed determination of which of these combinations can be formed into stable HLA tetrameric-peptide complexes. This study has therefore used 2 peptide-HLA combinations that have been shown to allow formation of such stable tetrameric complexes,25 which appear to be immunodominant in at least some individuals in a number of previous studies,14,24,26 and which allow examination of most individuals in view of the prevalence of these HLA alleles in our study population. However, evidence is also emerging that in some individuals the major immediate-early protein (IE-1) may be the major target for CD8+ T cells with only a minor or undetectable response to pp65.27
In the present study we show an increase in the number of HCMV pp65–specific T cells in donors expressing HLA-A*0201 and/or HLA-B*0702 over the time period of the culture. This confirms that the culture conditions result in expansion of a population of cells expressing CD8 that are able to recognize a single HLA molecule–peptide combination in agreement with earlier functional studies20 and adds to the previous literature suggesting that DCs derived from a variety of sources are able to present exogenous antigen constitutively on their MHC class I molecules following antigen capture by constitutive macropinocytosis.28 The relative importance of this mode of antigen presentation in vivo compared with classical MHC class I pathway presentation of pathogen-encoded proteins synthesized de novo within DCs remains unclear. Similarly, any differences in the antigenic profiles that are presented by these alternate pathways, as has been suggested for the EBV-encoded nuclear antigen EBNA1,29 are unknown. In the context of presentation of exogenous antigen, the results of this study suggest that none of the donors demonstrated a recall response completely polarized toward IE-1 to the exclusion of pp65, although a possible explanation for the relatively low level of tetrameric complex binding after the culture in donor 08 could relate to either a broader antigen or pp65 peptide–directed immune response in this donor. These findings may have implications for the mechanism of host control over viral replication in vivo. The persistently high levels of HCMV-specific CTLs in immunocompetent individuals suggest ongoing stimulation of the immune response.30 HCMV possesses numerous strategies to evade the immune response. These include resistance to natural killer cell–mediated lysis through surface expression of a MHC class I–like molecule31 and disruption of inducible MHC class I and class II expression resulting in reduced presentation of HCMV-associated antigens.32,33 However, nested PCR for HCMV DNA suggests very low viral loads in immunocompetent individuals (at least for the analytes studied, which include peripheral blood leucocytes, plasma, and serum).34 35 One possible explanation for the paradoxical existence of high circulating levels of HCMV-specific CTLs but low viral load could be that viral proteins expressed in mononuclear cells, the proposed “latent” reservoir of HCMV, continually stimulate CTLs. Our data show that oligoclonal pp65-specific CD8+ T-cell responses are restimulated following indirect presentation of CMV antigen and suggest that active viral replication may not be required to maintain the high level of CMV-specific CTLs seen in vivo.
An alternative explanation for those cases with low postculture tetrameric complex binding would be to invoke the possibility of greater in vitro deletion (ie, activation-induced cell death [AICD]) of lymphocytes in these cases. It has been demonstrated in animal models that peripheral T cells may be induced to die following high-intensity TCR signaling36 and that the result of viral infection in terms of viral clearance and induction of AICD may be determined by viral load alone.37 However, various costimulatory signals have been described that appear to reduce the potential for AICD. Probably the best described of these is CD28, which is activated by CD80 and CD86 (both expressed on APCs),38resulting in up-regulation of Bcl-xL39 and FLIP40 (both antiapoptotic) and down-regulation of FasL.41 Thus, DCs may be capable of inducing strong T-cell activation while protecting the cells from AICD. Subsequent antigen encounter (ie, in pathologically infected cells) and induction of apoptosis following appropriate effector function could provide one form of homeostatic control. We have previously published data showing that increasing antigen concentration in the DC/T-cell coculture system results in reducing levels of T-cell proliferation measured on day 6 of coculture and data on the variability in expansion of T cells between individuals under the same conditions.20 However, there was no evidence that those cases with the lowest postculture tetrameric complex binding exhibited either greater or lesser proliferation than those with higher levels of binding, nor that there was a greater degree of cell death as assessed by flow cytometry (data not shown) in these donors. Further studies into the degree of protection from AICD that is afforded by coculture and repeated restimulation with DCs and, in particular, comparison between differing approaches using alternate stimulants (eg, HCMV peptides versus proteins, mature versus immature DCs, and DCs versus infected fibroblasts) are warranted.
Analysis of TCRBV spectratypes gives a more detailed picture of total T-cell repertoire diversity than is available from phenotypic studies of either CD antigen or antigen-specific TCR (as assessed by tetramer complex binding) expression alone. While not strictly quantitative when compared across different primer pairs due to variations in amplification efficiency, the relative amounts of each size class PCR product within a given BV family do give some idea of the relative contributions of clones with differing CDR3 lengths to the diversity of the individual family. Attempts to ascribe clinical correlates to temporal alterations in the spectratypic profiles of PBLs have been strengthened by the demonstration of expansions of lymphocytes using the same BV genes and with identical CDR3 length within disease lesions (eg, tumor-infiltrating lymphocytes21 or those within the skin lesions of graft versus host disease42,43 or infiltrating the target organs in autoimmune disorders44). However, the lack of evidence that cells of a given CDR3 length have a defined antigen specificity has meant that the biological significance of alterations in such profiles detected in PBLs has remained speculative. In the environment of evolving immune reconstitution that prevails following allogeneic transplantation, changes in spectratypic patterns are to be expected. By combining a technique that allows isolation of antigen-specific T cells with TCRBV spectratypic analysis, we have been able to demonstrate the BV gene and CDR3 length usage of HCMV pp65 epitope-specific clones. This provides the strongest direct evidence to date that temporal alterations in spectratypic profiles relate to expansions of T cells of known antigen specificity, providing a tool for monitoring adoptive immunotherapy protocols.
Advances allowing more detailed analyses of the clonal composition of immune responses to defined antigens are giving increasing insight into the degree of diversity of T-cell responses, which in part reflect the degeneracy of the TCR–MHC-peptide interaction. It is well established that the interaction between TCR and MHC-peptide shows some degree of degeneracy in both directions, both for MHC class I–CD8 and MHC class II–CD4 interactions.45,46 A single TCR can recognize structurally distinct MHC-peptide complexes,47 and ontogenetically thymocytes bearing a specific TCR can be positively selected by a single MHC molecule in combination with a variety of peptides. Conversely, a single MHC-peptide combination can positively select thymocytes bearing many distinct TCRs,48-50 albeit based on interactions with relatively low avidity (because high-avidity interactions result in negative selection during this stage of development). It has been estimated in a murine model that these low-avidity interactions may result in a minimum of 105different BV rearrangements being selected by a single peptide-MHC class II complex.51 The diversity of the T-cell repertoire selected for the higher-avidity reactions required for antigen recognition and subsequent triggering of cellular response and proliferation as part of an immune response to exogenous antigen has been studied for a number of viral antigens and/or epitopes, including those from influenza A, HTLV-1, and EBV.52-56 Preferential usage of certain BV families by the TCR of T cells responding to specified peptides has been reported in these studies. Our study suggests that this is also true for responses to HCMV in HLA-A*0201+ donors, confirming the BV gene usage (BV8, BV6.1/6.2, and BV13) previously demonstrated by multiple independently derived epitope-specific CTL clones generated by formal single-cell cloning or from clonal CTL microculture.24 Although we have fewer results for HLA-B*0702+ donors, the possible preferential usage of BV14 and BV6.1/6.2 by CTL clones defined by binding of tetrameric complexes generated with the HLA-B*0702–restricted peptide similarly correlates with those documented in the previous study.24 Sequencing of clones selected according to their ability to lyse pp65-pulsed target cells following limiting dilution analysis has suggested that the memory CTL response to individual HCMV pp65 epitopes is highly focused.24 Tetrameric complex selection would not be predicted to select specifically for those clones with the greatest lytic capacity but rather to select all cells bearing TCRs that bind the complexes with sufficient avidity. Thus, while tetrameric complexes have been found to select for the highest-avidity T cells and would indeed be expected to select the clones with greatest lytic capacity, they would not do so selectively.57 The prediction that tetrameric complex–guided sorting would identify a larger number of T-cell clones than techniques based on assessment of lytic activity may in part explain the higher frequencies of antigen-specific T cells reported by this technique compared with previous studies using limiting dilution analysis.58,59 Indeed, T cells selected by single-specificity soluble HLA tetrameric-peptide complexes have been shown to be heterogeneous in terms of cell surface immunophenotype (CD11a, CD45RO/RA, and CD28 expression) and intracellular cytokine production,18,30,60-62suggesting some possible heterogeneity of function. In addition, the ability of tetramers to select a population of cells that are functionally unresponsive despite having many of the hallmarks of effector T cells (ie, cells that appear to have been selectively rendered anergic in vivo) has been demonstrated in the setting of a tumor-specific target.60 While our data do demonstrate a larger number of clonal T-cell expansions to a single pp65 epitope-MHC combination than the study by Weekes et al,24 they confirm that the response to specific HCMV pp65 epitopes is highly focused, apparently to a greater degree in some donors compared with others.
The general lack of conservation of other TCR gene segments and CDR3 length in HCMV pp65–specific T cells, both when comparing between different donors and between different CTL clones derived from the same donor, is also in agreement with the results of Weekes et al.24 These findings differ from those published for a dominant HLA-A2–restricted EBV epitope reported recently by Lim et al,56 where there was a frequent contribution of T-cell clonotypes with public TCR features (conserved CDR3 length and predicted TCR motifs) detected following selection with peptide-specific HLA tetrameric complexes. The suggestion that this feature may permit direct molecular follow-up of T-cell responses by, for example, use of primers specific for public clonotype junctions in BV spectratypic analysis (as elegantly demonstrated for EBV) may therefore not hold true for other antigenic responses such as that to HCMV. Analysis of HTLV-1 Tax-specific CTLs from different donors has also demonstrated largely unrelated TCR usage,55 and class II–restricted tetanus toxin peptide-specific T cells similarly have been shown to display highly heterogeneous CDR3 regions.63While analysis of larger numbers of donors would be required to fully exclude any common structural motifs within the CDR3 encoding region, our data do add to an the increasing literature suggesting that distinct TCRs may recognize a peptide-MHC complex by binding to it with differing primary TCR contact residues.14,24,55,61 63
In summary, we have combined the use of 2 techniques that are able to demonstrate differing facets of the immune response to gain further insight into the human immune response to HCMV in an in vitro culture system that has potential for application in immunotherapeutic protocols. We have shown that there is diversity between individuals in terms of the number of TCR clones that recognize single HLA-restricted viral epitopes, but that a small number of clones appear to predominate, and that there is preferential usage of certain BV gene segments within this restricted part of the overall immune response. Unlike the case for EBV, there does not appear to be conservation of CDR3 length or sequence (including J region usage) between donors for the HLA-peptide combinations we have studied. The ability to determine the BV gene usage and CDR3 size of the pp65 epitope-specific CTLs will allow comparison of in vitro culture systems with responses detected in vivo during periods of HCMV reactivation following allogeneic bone marrow transplantation and allow monitoring of the fate of infused cells during adoptive immunotherapy protocols. Correlation with quantitative techniques for evaluation of HCMV load will allow investigation of the relationships between immune response and viral dynamics in early infection. Attempts to further enrich infused cells for HCMV specificity (for example, by selection of cells expressing TCRs with a restricted range of BV genes or those binding specific HLA-tetramer-peptide combinations) may help to determine the “sufficient T-cell repertoire” for reestablishment of host control over viral replication, because the heterogeneity of responses (both in terms of the degree of focusing toward specific pp65 peptides and in terms of the clonal diversity directed toward individual HLA-tetramer-peptide combinations) suggests that any such enrichment based on positive selection is unlikely to encompass the entirety of the human anti-HCMV immune response.
Supported by the Leukaemia Research Fund, London, United Kingdom, and by a grant from the Golden Charitable Trust, London.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Stephen Mackinnon, Dept of Haematology, University College Hospital, 98 Chenies Mews, London WC1E 6HX, United Kingdom; e-mail: s.mackinnon@ucl.ac.uk.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal