The stem cell leukemia (SCL) gene encodes a basic helix-loop-helix transcription factor with a critical role in the development of both blood and endothelium. Loss-of-function studies have shown that SCL is essential for the formation of hematopoietic stem cells, for subsequent erythroid development and for yolk sac angiogenesis. SCL exhibits a highly conserved pattern of expression from mammals to teleost fish. Several murine SCLenhancers have been identified, each of which directs reporter gene expression in vivo to a subdomain of the normal SCL expression pattern. However, regulatory elements necessary for SCL expression in erythroid cells remain to be identified and the size of the chromosomal domain needed to support appropriate SCL transcription is unknown. Here we demonstrate that a 130-kilobase (kb) yeast artificial chromosome (YAC) containing the human SCL locus completely rescued the embryonic lethal phenotype ofscl−/− mice. Rescued YAC+scl−/− mice were born in appropriate Mendelian ratios, were healthy and fertile, and exhibited no detectable abnormality of yolk sac, fetal liver, or adult hematopoiesis. The human SCL protein can therefore substitute for its murine homologue. In addition, our results demonstrate that the human SCL YAC contains the chromosomal domain necessary to direct expression to the erythroid lineage and to all other tissues in which SCL performs a nonredundant essential function.
Introduction
The stem cell leukemia (SCL)gene (also known as TAL-1) encodes a basic helix-loop-helix (bHLH) transcription factor with an essential role in the development of both blood and endothelial cells.1 Mice lacking a functional SCL protein failed to develop yolk sac hematopoiesis and blastocyst reconstitution experiments have demonstrated that SCL is required for both definitive and primitive hematopoiesis.2,3 These data suggest that SCL plays a pivotal role in the formation or behavior of hematopoietic stem cells and are consistent with the observation that expression of antisense SCL suppressed the proliferation, cell cycle progression, and self-renewal of a multipotent hematopoietic cell line.4SCL is likely to perform additional functions following lineage commitment because enforced SCL expression enhanced erythroid differentiation of hematopoietic cell lines5,6 and increased erythroid and megakaryocytic differentiation of normal CD34+ progenitors.7 8
Several lines of evidence demonstrate that SCL is critical for normal endothelial development. scl knockout mice exhibit defective yolk sac angiogenesis thought to reflect an essential function for SCL during vessel formation.9 SCL is also likely to play an earlier role during the formation of endothelial cells. In zebrafish and in murine embryonic stem (ES) cell systems, SCL is expressed in hemangioblasts, bipotent progenitors of blood and endothelium.10,11 Moreover, SCL expression can partially rescue both blood and endothelial defects of the zebrafishcloche mutant12 and ectopic expression of SCL during early development alters the fate of mesodermal cells, resulting in excessive hemangioblast formation at the expense of several other cell types.10
Current evidence therefore suggests that SCL is essential for establishing the transcriptional program necessary for the formation of hemangioblasts and subsequently hematopoietic stem cells. It is also clear that transcriptional dysregulation of the sclgene has profound consequences. These observations emphasize the fundamental biologic significance of the mechanisms that regulateSCL transcription and our laboratory has therefore undertaken a systematic analysis of the transcriptional regulation of the murine scl locus.
Both human and murine SCL are transcribed from 2 lineage-specific promoters.13-17 A survey of the chromatin structure surrounding the murine scl gene has also revealed a panel of DNase I hypersensitive sites associated with enhancer or silencer activity in transfection assays.18 Transgenic reporter assays have so far identified 5 independent enhancers each of which targets expression to a specific subdomain of the normal SCL expression pattern.19-21 A 3′ enhancer is of particular interest because it targets expression to the vast majority of hematopoietic progenitors and long-term repopulating hematopoietic stem cells.19 22
However, additional elements remain to be identified and the size of the chromosomal domain that contains all of the scl gene regulatory elements remains unknown. This issue is particularly important given the mounting evidence for the existence of long-range regulatory elements at a number of mammalian loci. Yeast artificial chromosomes (YACs) spanning 120 kilobase (kb), 540 kb, or 625 kb of the Gata-3 locus were unable to completely rescue the pattern of endogenous Gata-3 expression23 and also failed to overcome embryonic lethality in Gata-3−/−mice.24,25 Similarly, large YAC transgenes resulted in only partial rescue of the Dazl and Wt1 null phenotypes.26,27 Studies of chromosome rearrangements have inferred the existence of c-kit regulatory regions at least 100 kb upstream of the coding region28 and more recently, Loots and colleagues have identified a regulatory element that coordinates expression of the interleukin 4, 13, and 5 genes over a region of 120 kb.29
In this paper we demonstrate that a 130-kb YAC containing the humanSCL locus completely rescues the lethal phenotype ofscl−/− mice and results in normal yolk sac, fetal liver, and adult hematopoiesis.
Materials and methods
Isolation and modification of a human SCL YAC
Three human YAC libraries were screened.30-32Filters were initially hybridized with a 750 bp BamHI fragment from the 3′ untranslated region (UTR) of SCL (3′ probe)13 and subsequently with a 2.2 kb XhoI fragment derived from the 5′ end of the SCL gene (5′ probe).13 Six clones came through 2 rounds of screening (4HD12 and 28EF2 from the ICI human YAC library; 48D10 and Y18C10 from the ICRF human YAC library; 638E10 and 781B6 from the CEPH megaYAC library). Preparation of high-molecular-weight yeast DNA plugs and total yeast/YAC DNA, long-range and short-range restriction mapping, and sizing of YACs were performed according to standard methods.33-35 Fluorescence in situ hybridization (FISH) was performed as previously described.36 YAC 4HD12 from the ICI human YAC library was subsequently used for further manipulation.
YAC 4HD12 was retrofitted by spheroplast transformation37to incorporate I-PpoI sites into each YAC arm firstly with plasmid pUC-OK and subsequently with pUC-WAN.38 The efficiency of transformation was 5% and 3%, respectively. After each transformation, high-molecular-weight yeast DNA plugs were prepared from at least 10 individual transformants. Pulsed field gel electrophoresis (PFGE) followed by Southern hybridization confirmed the size of the modified YAC was unchanged.
To monitor expression from the human SCL locus, an internal ribosomal entry site (IRES) (from encephalomyocarditis virus) nuclear localized (nls) lacZ reporter (kindly provided by E. Andermarcher) was cloned into the NcoI site within the 3′ UTR of humanSCL. This construct was cloned into the yeast integrating vector, pRS406 (Stratagene, La Jolla, CA), which contains the URA3 selectable marker, to form plasmid p5′lacZ3′. This plasmid was integrated into the modified YAC using Pop-In/Pop-Out technology.39 The YAC was retrofitted with linearized p5′lacZ3′ by spheroplast transformation (Pop-In). Total yeast/YAC DNA was prepared from 25 Ura+ Trp+ Lys+transformants, digested with BglII and HindIII and sequentially hybridized to the 3′ probe and the lacZ reporter gene by Southern analysis. The correct pattern was observed in 4 of 25 transformants. Integrity of the YAC was demonstrated by PFGE and Southern hybridization.
To generate transformants without vector sequence but containing 3′ UTR with IRES-nls-lacZ (Pop-Out), one transformant was grown in a selection medium containing uracil. The culture was plated onto agar supplemented with 1 mg/mL 5-fluoro-orotic acid (5-FOA) to select against colonies containing the URA3 gene. Fifty-seven of 78 Ura− Trp+ Lys+transformants40 were found to contain the lacZ reporter gene by hybridization. DNA was prepared from 18 of these, digested withBglII and HindIII and hybridized to the 3′ probe. Fourteen demonstrated the expected pattern and all were of the correct size, as determined by PFGE and Southern hybridization.
To determine the 5′ and 3′ limits of genomic DNA contained within clone 4HD12, the YAC ends were rescued by digesting 100 ng 4HD12 YAC DNA withHaeIII and self-ligating at a low concentration (1 ng/μL). Then, 2 ng of this ligated product was amplified by inverted polymerase chain reaction (PCR), using the following primers: YAC left arm; sense CGCAAGACTTTAATTTATCACTAC and antisense TAGTCGATAGTGGCTCCAAGTAGC; YAC right arm; sense TGGATCCTCTACGCCGGACGCATC and antisense AGTCGAACGCCCGATCTCAA. The resulting products were cloned into pGEM-T vector (Promega, Southampton, United Kingdom) and sequenced. Sequences were compared with sequences in GenBank using BLAST.41
Generation of transgenic mice
Purified YAC DNA was prepared for microinjection as previously described by 2 gel runs without exposure of the DNA to ethidium bromide or UV irradiation.42 YAC DNA in a gel slice was then equilibrated against TENPA (10 mM Tris-HCl, pH7.5, 1 mM EDTA, 100 mM NaCl, 30 μM spermine, 70 μM spermidine), melted at 68°C and the agarose digested with agarase (New England Biolabs, Hitchin, United Kingdom) at 42°C. YAC DNA was dialyzed against microinjection buffer (10 mM Tris, pH 7.4, and 0.25 mM EDTA) on a floating dialysis membrane (0.05 μm; Millipore, Bedford, MA) for 1.5 hours. and diluted to 1 μg/mL for microinjection.
Pronuclear injections were performed into CBA/C57.Bl6 fertilized mouse oocytes that were allowed to divide to 2 cells prior to implantation into the oviducts of pseudopregnant CD1 female mice.43 At 2 weeks of age tail DNA was prepared from founder mice and multiplex PCR analysis was performed to detect lacZ using myogenin as an internal control.44 Founder mice were subsequently backcrossed onto CBA/C57.Bl6 F1 mice to maintain the transgenic lines. In the case ofscl−/− rescue analysis, YAC transgenic lines were crossed onto the scl knockout line SV10245(kindly provided by L. Robb and C. G. Begley). Offspring were genotyped by PCR for the presence of the YAC transgene and thescl− allele as described previously.44 45 Mice heterozygous for the YAC and heterozygous for mutant scl were intercrossed and resulting offspring were genotyped as described above.
Southern analysis of transgenic mice
Copy number quantification of the YAC transgene was performed by digesting 10 μg tail DNA from F2 mice with EcoRI followed by Southern hybridization to equimolar concentrations of size-matched (1.9 kb) probes for the 3′UTR of the endogenous scl (mouse specific) and lacZ (transgene specific). Signal from the lacZ gene was quantified with respect to the 2 copy endogenous scl signal on a Molecular Dynamics PhosphorImager (Kemsing, Kent, United Kingdom). The 3′ UTR probe was a 1.9-kb KpnI/BglII fragment from the murine scl complementary DNA (cDNA)46and the lacZ probe was a 1.9-kb BamHI/SacI fragment from the IRESnlslacZ construct.
Single-cell suspensions were made from the spleens of YAC transgenic mice and were washed once in phosphate-buffered saline (PBS). Cells were resuspended at 2 × 107 cells/mL in PBS at room temperature. Equal volumes of cells and 2% agarose (made up in PBS and equilibrated to 40°C) were mixed and dispensed into chilled plug molds on ice. Plugs were lysed in the same way as for high-molecular-weight yeast plugs,33 washed twice with 24 mM EDTA, 0.5 mM Tris, 1.8 mM N-laurylsarcosine (pH 9.5) for 2 hours at room temperature and stored at 4°C in this buffer. High-molecular-weight DNA was digested in situ with I-PpoI (Promega) in the same manner as described above.33
Transgene integrity was determined by Southern blot analysis of tail DNA from F2 transgenic mice. Restriction digests and probes (as shown in Figure 1) were as follows: L-arm,EcoRI digest of genomic (various sizes detected as extends from L-arm into integration site) and hybridized with a 4-kbAvaI fragment from plasmid pUC-WAN spanning neoand TRP1; fragment 1, BamHI digest to yield a 15.7-kb fragment (29141-44906 in human SCL locus, GenBank accession no.AJ131016) when hybridized with a promoter 1a probe (44447-44647); fragment 2, BglII digest to yield an 11.6-kb fragment when hybridized with an intron 3 probe (49261-49281); fragment 3,BamHI digest to yield a 2.3- kb fragment when hybridized with an exon 4 probe (380-bp NotI/NaeI fragment from mouse scl cDNA); fragment 4, EcoRI digest to yield a 6.2-kb fragment (53539-lacZ gene) when hybridized with an intron 5 probe (54949-54967); fragment 5, EcoRI digest to yield a 3.6-kb fragment of the lacZ gene when hybridized with a lacZ probe (1.9-kb BamHI/SacI fragment); fragment 6,HindIII digest to yield a 16-kb fragment (lacZ-74500) when hybridized with a 3′UTR probe (600-bp fragment fromBglII/KpnI digest of human SCL cDNA); fragment 7, BglII digest to yield a 10-kb fragment (72347-82005) when hybridized with a downstream probe (73182-73524); R-arm, EcoRI digest to yield fragments of varying sizes (extending from R-arm into integration site) when hybridized with a 6.3-kb AatII/TthIII I fragment from plasmid pUC-OK spanning the LYS2 gene. The promoter 1a, intron 3, intron 5, and downstream probes were generated by PCR from human genomic DNA.
Structure and analysis of the human SCL YAC transgenes.
(A) Diagram of the 130-kb YAC containing the human SCL gene together with part of the SIL gene and the completeMAP17 gene. A fine detail map of the modified SCLlocus is shown together with the location of the IRES-nls-lacZ cassette (hatched box) within the 3′ UTR of exon 6 (open box). Numbered horizontal lines indicate the restriction fragments assessed by Southern analysis (Table 1). (B) Southern hybridization of tail DNA from 5 lines together with a nontransgenic control mouse using equimolar concentrations of size matched probes for lacZ (transgene specific) and murine scl 3′ UTR (mouse specific). Transgene copy numbers were calculated with respect to the endogenous mousescl gene.
Structure and analysis of the human SCL YAC transgenes.
(A) Diagram of the 130-kb YAC containing the human SCL gene together with part of the SIL gene and the completeMAP17 gene. A fine detail map of the modified SCLlocus is shown together with the location of the IRES-nls-lacZ cassette (hatched box) within the 3′ UTR of exon 6 (open box). Numbered horizontal lines indicate the restriction fragments assessed by Southern analysis (Table 1). (B) Southern hybridization of tail DNA from 5 lines together with a nontransgenic control mouse using equimolar concentrations of size matched probes for lacZ (transgene specific) and murine scl 3′ UTR (mouse specific). Transgene copy numbers were calculated with respect to the endogenous mousescl gene.
Northern blot analysis
Poly (A)+ RNAs were isolated from fetal liver as described.47 RNA samples (4 μg) were size fractionated by electrophoresis on a 1% agarose gel containing 0.6% formaldehyde in 1 times 3-[N-Morpholino]propanesulphonic acid (MOPS) (20 mM MOPS, 1 mM EDTA, 5 mM sodium acetate) running buffer, transferred to a nylon membrane (Hybond N+; Amersham Pharmacia Biotech, Bucks, United Kingdom) and hybridized with a 32P-labeled nlslacZ probe (1.9-kbBamHI/SacI fragment) by standard techniques.48 After hybridization and autoradiography with the nlslacZ probe, the filter was stripped and reprobed with a 3′UTR mouse scl probe (1.9 -b BglII/KpnI fragment) and a rat GAPDH probe (kindly provided by T. Enver).
Generation of SCL antisera, immunoprecipitation, and Western analysis
A peptide corresponding to the C-terminus of murine SCL was used to generate polyclonal SCL specific sheep antisera. The antisera obtained was affinity purified using the immunizing peptide and subsequently precleared using a protein extract from the SCL− negative cell line BW5147.
Cellular extracts were prepared by suspension in immunoprecipitation buffer (1% Triton X-100, 0.5% NP-40, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 10 mM Tris-Cl pH 8.0) for 10 minutes on ice, and subsequently passing several times through a 20-gauge needle. For immunoprecipitation, 5 μL rabbit antimouse SCL49 or rabbit serum was added to lysates obtained from a single fetal liver (YAC−scl+/−; YAC−scl+/+) or 2 fetal livers (YAC+scl−/−) and incubated overnight at 4°C. Immune complexes were recovered with protein A–Sepharose 4B (Amersham Pharmacia Biotech) and washed 3 times in PBS. Proteins and immune complexes were boiled in Laemmli buffer for 5 minutes, resolved using sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane. Equal transfer was confirmed by Ponceau S staining. Western analysis with sheep antimouse SCL antisera, followed by horseradish peroxidase–conjugated antisheep IgG (Sigma-Aldrich, Dorset, United Kingdom) was performed according to standard methods.50 Chemiluminescent visualization of proteins was performed using Supersignal West Femto substrate (Pierce, Rockford, IL) according to the manufacturer's instructions.
Analysis of blood
Blood was taken from age-matched (8-14 weeks old) control (2 males, 4 females) and rescued (1 male, 3 females) mice by cardiac puncture. Blood counts were performed using an Animal Blood Counter (ABX Hematologie, Montpellier, France). Cell morphology was assessed on May-Grünwald-Giemsa–stained blood smears.
Flow cytometry
Bone marrow, fetal liver, splenocytes, and thymocytes were harvested in PBS containing 5% fetal calf serum (FCS) and 0.01% sodium azide. Cells (1 × 106) were stained with the appropriate antibody for cell surface antigens on ice for 30 minutes prior to analysis on a FACSsort flow cytometer (Becton Dickinson, San Jose, CA). Dead cells were excluded by propidium iodide (PI) staining and by gating out cells with low forward and side scatter. Monoclonal antibodies were from Pharmingen (San Diego, CA) and included anti–c-kit (2B8), anti-CD34 (RAM34), anti–MAC-1 (M1/70), anti–Gr-1 (RB6-8C5), anti-Ter119 (TER119), anti-CD61 (2C9.G2), anti-B220 (RA3-6B2), anti-CD4 (H129.19), and anti-CD8 (53-6.7). In some instances incubation with streptavidin-phycoerythrin or streptavidin–fluorescein isothiocyanate (Pharmingen) was required for biotin-conjugated antibodies.
In vitro colony-forming assays for progenitors
Bone marrow cells were seeded at 5 × 104cells/plate containing 0.3% agar in Iscoves modified Dulbecco medium (IMDM; Gibco, Invitrogen, Paisley, United Kingdom) supplemented with 25% FCS and cytokines (interleukin 3 [IL-3], stem cell factor [SCF], thrombopoietin [Tpo]) for myeloid colony formation51 or in Methocult GF-M3434 methylcellulose (Stem Cell Technologies, Vancouver, BC, Canada) for erythroid and myeloid colony formation. Agar cultures were supplemented with conditioned medium from the BHK cell line (a kind gift from S. Tsai) containing SCF and WEHI 3B cell line conditioned medium as a source of IL-3,51 or with 2 ng/mL recombinant IL-3 and Tpo (R & D Systems, Abingdon, United Kingdom). Duplicate agar and triplicate Methocult cultures were incubated at 37°C in a humidified atmosphere of 5% CO2 and colonies were scored after 7 days of culture.
Yolk sacs from day 8.5 embryos were separated from the embryo proper, which was subsequently used for genotype analysis by PCR as described previously. The individual yolk sacs were placed in 1 mL PBS containing 20% FCS and passed through 26- and 30-gauge needles to obtain a single-cell suspension. All cells were centrifuged and resuspended in 100 μL IMDM and seeded into 1 mL Methocult GF-M3434 methylcellulose and were cultured in a fully humidified atmosphere of 5% CO2 for 7 days prior to scoring. To confirm that erythroid colonies were hemoglobinized 2,7-diaminofluorene (Sigma-Aldrich) staining was performed at 9 days of culture.52
Results
Isolation and characterization of a human SCLYAC
A YAC clone, 4HD12, containing the human SCL gene was isolated from the ICI human YAC library by colony hybridization. FISH with the YAC identified signals only at chromosome 1p32 consistent with the location of the SCL locus (data not shown). PFGE followed by Southern hybridization indicated the YAC was approximately 130 kb in size (Figure 1A and data not shown). Restriction digestion of YAC DNA with BamHI, EcoRI, HindIII, orSacI followed by Southern hybridization with 5′ and 3′SCL probes demonstrated that the SCL locus within the YAC was intact and long-range restriction mapping withNotI, SalI, and SfiI indicated that the SCL gene lay within the middle of the YAC (Figure 1A and data not shown). To determine the 5′ and 3′ limits of the clone, the end fragments were rescued by inverted PCR and sequenced. Comparison with the sequence of an SCL PAC clone (GenBank accession number AJ131016)21 demonstrated that the 3′ end of the YAC insert was located 36 kb downstream of the MAP17 transcriptional start site. The genomic sequence adjacent to the left YAC arm was found to be identical to exon 11 of human SIL (GenBank accession number AF349650). An EcoRI site lies 200 to 300 bp upstream of human SIL exon 1153 and the YAC library was generated by a partial EcoRI digest. These data indicate that this EcoRI site marks the 5′ end of the YAC insert and that the size of the insert is therefore 131 kb.
To allow rapid assessment of YAC-transgene integrity in transgenic mice, I-PpoI restriction sites were inserted into both arms of the YAC.38 To facilitate analysis of human SCL expression in transgenic mice, an IRES-lacZ cassette was inserted into the 3′ UTR of exon 6 of the human SCL gene (Figure 1A). Modified YAC clones were analyzed by PFGE and Southern hybridization at each stage of the modification. After modification and confirmation, purified high-molecular-weight YAC DNA was prepared and microinjected into oocytes.
Transgenic founders were screened by PCR for the presence of the transgene. From a total of 161 offspring, 5 transgenic founders were identified (2037, 2054, 2074, 2083, and 2133). Southern blot analysis was performed to assess transgene copy number. Line 2083 was found to contain approximately 13 copies of the human SCL YAC, whereas all other lines contained approximately 2 copies of the transgene (Figure 1B).
Pulsed field gel electrophoresis and Southern hybridization were performed to assess the structure of the integrated YAC in each transgenic line (data not shown). Lines 2054, 2083, and 2133 each gave rise to a band of the expected size (130 kb) demonstrating an absence of major genomic rearrangements. A band of approximately 90 kb was observed in line 2037 suggesting the presence of a 40-kb deletion. Line 2074 gave rise to a band that comigrated with high-molecular-weight genomic DNA, consistent with loss of one or both I-PpoI sites.
A more detailed Southern analysis was then performed of a 57-kb region surrounding and including the human SCL gene (Figure 1A, Table 1, and data not shown). Consistent with the PFGE results, line 2037 contained a large deletion, which removed SCL exons 1a to 5. However, the human SCLexon/intron structure was intact in the remaining 4 transgenic lines and all of these contained unrearranged flanking sequence extending 16 kb upstream and 20 kb downstream of the human SCL gene.
Integrity of human SCL/lacZ YAC within transgenic lines
| Transgenic line . | Transgene copy no. . | Restriction fragments . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Left arm . | 1 . | 2 . | 3 . | 4 . | 5 . | 6 . | 7 . | Right arm . | ||
| 2037 | 2.2 | + | − | − | − | + | + | + | + | +a |
| 2054 | 2.4 | R | + | + | + | + | + | + | + | + |
| 2074 | 2.3 | R | + | + | + | + | + | + | + | + |
| 2083 | 13.3 | + | + | + | + | + | + | +b | + | + |
| 2133 | 2.3 | + | + | + | + | + | + | + | + | R |
| Transgenic line . | Transgene copy no. . | Restriction fragments . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Left arm . | 1 . | 2 . | 3 . | 4 . | 5 . | 6 . | 7 . | Right arm . | ||
| 2037 | 2.2 | + | − | − | − | + | + | + | + | +a |
| 2054 | 2.4 | R | + | + | + | + | + | + | + | + |
| 2074 | 2.3 | R | + | + | + | + | + | + | + | + |
| 2083 | 13.3 | + | + | + | + | + | + | +b | + | + |
| 2133 | 2.3 | + | + | + | + | + | + | + | + | R |
Southern analysis was performed to assess the copy number and integrity of YAC transgenes. The integrity of 7 restriction fragments (1-7, Figure 1) was assessed, together with two additional fragments containing the left and right YAC arms.
+ indicates fragment of correct size detected; −, fragment absent; a, weak signal indicating deletion of the arm from one of the two YACs present in this line; b, two fragments detected, one of the correct size (10 copies) and one of a larger size (3 copies); R, fragment rearranged.
Expression of the human SCL transgene
To assess expression of the human SCL gene, X-gal staining was performed on E12.5 and E13.5 embryos from all YAC transgenic lines. At these time points SCL is known to be expressed in the yolk sac, fetal liver, and central nervous system.54,55 Control transgenic embryos carrying a −7E3/lacZ murine scl transgene20 gave appropriate X-gal staining. However, no staining was observed in embryos from any of the YAC transgenic lines, even after 48 hours of incubation at 37°C.
To determine whether the fusion gene was transcribed, Northern blot analysis was performed on E14.5 fetal liver, which is known to express readily detectable levels of murine scl messenger RNA (mRNA)54 (Figure 2). No expression of the transgene was detected in line 2037 consistent with the Southern blot data demonstrating deletion of SCL 5′ exons in this line. Human SCL transcripts were detected in the 4 remaining transgenic lines. These data therefore further suggest that the failure to detect lacZ expression by X-gal staining was likely to reflect a defect in lacZ translation, probably related to the specific IRES sequence that was used. Lines 2074 and 2133 both contained 2 copies of the transgene but exhibited markedly different levels of transgene expression, suggesting that the humanSCL gene is subject to stable or variegating position effects.56 Line 2083 contained 13 copies of the transgene but displayed a level of transgene expression similar to line 2133, which only contained 2 copies, thus demonstrating an absence of copy number-dependent expression. These results suggest that either the human SCL locus does not have a locus control region (LCR)–like element or that such an element exists but is not contained within the 130-kb transgene. It is also possible that the presence of lacZ may have rendered the transgene particularly susceptible to position effects.57
Expression of the human
SCL gene. Northern analysis of poly(A)+ RNA from E14.5 fetal liver hybridized with a lacZ probe to detect expression of the human SCL/lacZ transgene, a 3′ UTR probe for endogenous mouse scl, and a GAPDH probe as a control for mRNA loading. RNA was obtained from the indicated transgenic lines as well as from a wild-type control (WT).
Expression of the human
SCL gene. Northern analysis of poly(A)+ RNA from E14.5 fetal liver hybridized with a lacZ probe to detect expression of the human SCL/lacZ transgene, a 3′ UTR probe for endogenous mouse scl, and a GAPDH probe as a control for mRNA loading. RNA was obtained from the indicated transgenic lines as well as from a wild-type control (WT).
The human SCL YAC rescues the scl−/−phenotype
The scl−/− embryos die at approximately E9.0 as a consequence of defective yolk sac hematopoiesis and angiogenesis.9,45,55 58 To assess whether the humanSCL YAC could rescue the scl−/−phenotype, YAC transgenic lines 2133, 2054, and 2074 were bred withscl+/− mice. Offspring were typed by PCR for the presence of the human SCL transgene and for the murinescl− and scl+ alleles. Compound heterozygotes (YAC+scl+/−) were intercrossed and viable offspring were typed by PCR (Figure3 and Table2). As expected, noscl−/− offspring lacking the humanSCL YAC were identified. By contrast all 3 YAC transgenic lines gave rise to viable scl−/− offspring that carried the human SCL YAC (Figure 3 and Table 2). These YAC+scl−/− offspring were found in the expected Mendelian frequency, were indistinguishable from their littermates (up to 8 months of age) and were fertile. The total number of YAC+scl−/− mice followed for 2 to 8 months were 37 (line 2054), 25 (line 2074), and 12 (line 2133). In addition, a further 23 (line 2054), 4 (line 2074), and 17 (line 2133) were followed for 8 to 16 months.
The human SCL YAC completely rescues the
scl−/− embryonic lethal phenotype.PCR genotype analysis of offspring derived from interbreeding YAC+/−scl+/− male and YAC+/−scl+/− female mice from line 2074. Lanes 1 and 2, YAC+/−scl+/−male and female parents; lanes 3 to 9, offspring with the following genotypes: 3, (YAC−scl+/−); 4, (YAC+scl+/+); 5, (YAC+scl+/−); 6-9, (YAC+scl−/−); +ve, positive control (YAC+scl+/−); −ve, no DNA control.
The human SCL YAC completely rescues the
scl−/− embryonic lethal phenotype.PCR genotype analysis of offspring derived from interbreeding YAC+/−scl+/− male and YAC+/−scl+/− female mice from line 2074. Lanes 1 and 2, YAC+/−scl+/−male and female parents; lanes 3 to 9, offspring with the following genotypes: 3, (YAC−scl+/−); 4, (YAC+scl+/+); 5, (YAC+scl+/−); 6-9, (YAC+scl−/−); +ve, positive control (YAC+scl+/−); −ve, no DNA control.
Rescue of scl−/− phenotype by the human SCL YAC
| Transgenic line . | No. of mice . | Human SCLYAC . | scl genotype . | ||
|---|---|---|---|---|---|
| +/+ . | +/− . | −/− . | |||
| 2133 | 104 | − | 7 | 21 | 0 |
| + | 19 | 42 | 15 | ||
| 2054 | 86 | − | 8 | 13 | 0 |
| + | 11 | 35 | 19 | ||
| 2074 | 75 | − | 8 | 12 | 0 |
| + | 11 | 28 | 16 | ||
| Transgenic line . | No. of mice . | Human SCLYAC . | scl genotype . | ||
|---|---|---|---|---|---|
| +/+ . | +/− . | −/− . | |||
| 2133 | 104 | − | 7 | 21 | 0 |
| + | 19 | 42 | 15 | ||
| 2054 | 86 | − | 8 | 13 | 0 |
| + | 11 | 35 | 19 | ||
| 2074 | 75 | − | 8 | 12 | 0 |
| + | 11 | 28 | 16 | ||
Genotypes of offspring resulting from matings between YAC+/−scl+/− parents from 3 transgenic lines. Note that in the presence of the human SCLYAC, all the transgenic lines produce viablescl−/− offspring in the expected mendelian ratios.
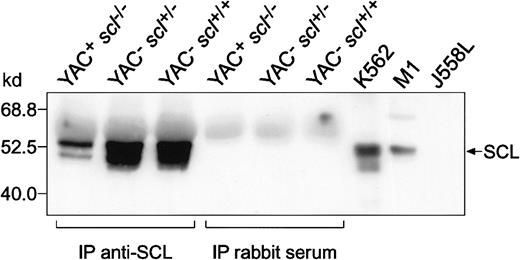
To assess the level of human SCL protein in rescued YAC+scl−/− mice, we performed Western analysis on E14.5 fetal livers from lines 2054 and 2074. Low levels of human SCL protein were observed in YAC+scl−/− livers relative to YAC−scl+/+ and YAC−scl+/−controls (data not shown). The level of human SCL protein was demonstrated more clearly by immunoprecipitation with a rabbit anti-SCL antibody followed by Western analysis with a sheep anti-SCL antibody (Figure4). These results demonstrate that low levels of human SCL transcript are accompanied by a correspondingly low level of SCL protein and that the latter is sufficient for rescue of the lethal scl−/−phenotype.
Detection of human SCL protein in rescued YAC+
scl−/− E14.5 fetal liver. Cell lysates were prepared from E14.5 fetal livers obtained from transgenic (line 2054) and nontransgenic mice. These were immunoprecipitated (IP) with rabbit antimouse SCL antisera or preimmune rabbit serum as indicated. The immunoprecipitates were subsequently Western blotted using an antimouse SCL antibody raised in sheep. Lysates from SCL-expressing cell lines (K562, human erythroleukemia; M1, mouse myeloid leukemia) and an SCL-nonexpressing cell line (J558L, mouse plasmacytoma) were included on the immunoblot as controls.
Detection of human SCL protein in rescued YAC+
scl−/− E14.5 fetal liver. Cell lysates were prepared from E14.5 fetal livers obtained from transgenic (line 2054) and nontransgenic mice. These were immunoprecipitated (IP) with rabbit antimouse SCL antisera or preimmune rabbit serum as indicated. The immunoprecipitates were subsequently Western blotted using an antimouse SCL antibody raised in sheep. Lysates from SCL-expressing cell lines (K562, human erythroleukemia; M1, mouse myeloid leukemia) and an SCL-nonexpressing cell line (J558L, mouse plasmacytoma) were included on the immunoblot as controls.
Rescued scl−/−mice have normal adult, fetal liver, and yolk sac hematopoiesis
The major phenotypic defect in scl−/−embryos comprises a complete absence of hematopoiesis. A detailed analysis of adult and embryonic hematopoiesis in rescued YAC+scl−/− mice was therefore performed. Analysis of peripheral blood demonstrated that adult YAC+scl−/− mice had normal hemoglobin levels together with normal platelet, white cell, and differential counts (Table 3). Inspection of peripheral blood smears did not reveal any morphologic abnormalities (data not shown).
Comparison of full blood counts of control and rescuedscl−/− mice
| Blood count . | Control YAC−scl+/+ . | Rescued YAC+scl−/− . |
|---|---|---|
| White cell, × 109/mm3 | 1.6 ± 1.0 | 2.6 ± 1.1 |
| Hemoglobin, g/dL | 14.5 ± 0.5 | 13.5 ± 0.5 |
| Platelets, × 106/mm3 | 896 ± 149 | 879 ± 69 |
| Hematocrit | 0.54 ± 0.02 | 0.52 ± 0.02 |
| Lymphocytes, % | 73.2 ± 10.8 | 78.3 ± 3.7 |
| Granulocytes, % | 21.5 ± 9.0 | 16.9 ± 3.4 |
| Monocytes, % | 5.4 ± 2.0 | 4.9 ± 1.0 |
| Blood count . | Control YAC−scl+/+ . | Rescued YAC+scl−/− . |
|---|---|---|
| White cell, × 109/mm3 | 1.6 ± 1.0 | 2.6 ± 1.1 |
| Hemoglobin, g/dL | 14.5 ± 0.5 | 13.5 ± 0.5 |
| Platelets, × 106/mm3 | 896 ± 149 | 879 ± 69 |
| Hematocrit | 0.54 ± 0.02 | 0.52 ± 0.02 |
| Lymphocytes, % | 73.2 ± 10.8 | 78.3 ± 3.7 |
| Granulocytes, % | 21.5 ± 9.0 | 16.9 ± 3.4 |
| Monocytes, % | 5.4 ± 2.0 | 4.9 ± 1.0 |
Blood was obtained from age-matched mice (8-14 weeks) by cardiac puncture. Numbers indicate means ± SD for mice of the indicated genotype from line 2074. Means were derived from the results for 4 mice (rescued data, control lymphocyte, granulocyte and monocyte percentages) or from 6 mice (remaining control data).
Flow cytometry was used to assess the spectrum of hematopoietic cells present in adult bone marrow, spleen, thymus, and fetal liver (Table 4). A panel of antibodies recognizing markers present on progenitors (c-kit, CD34), monocytes (Mac1), granulocytes (Gr-1), erythroid cells (Ter119), megakaryocytes (CD61), mast cells (IgE/anti-IgE), B cells (B220), and T cells (CD4, CD8) was used. Percentages of double-negative and double-positive thymocytes were also assessed, as were percentages of c-kit+ and Ter119+ cells in E11.5 and E12.5 fetal livers. No significant differences were obtained between control (YAC+scl+/−) and rescued (YAC+scl−/−) mice.
FACS analysis of hematopoietic tissues from rescuedscl−/− mice
| Tissue . | Surface marker . | Control YAC+scl+/− . | Rescue YAC+scl−/− . |
|---|---|---|---|
| Bone marrow | c-kit | 15.0 ± 2.44-150 | 12.8 ± 1.14-150 |
| CD34 | 6.7 ± 1.8 | 6.1 ± 0.9 | |
| Mac-1 | 30.8 ± 10.3 | 35.2 ± 3.6 | |
| Gr-1 | 33.0 ± 12.1 | 35.0 ± 2.6 | |
| Ter119 | 51.2 ± 10.2 | 42.9 ± 9.0 | |
| CD61 | 32.9 ± 11.5 | 36.1 ± 1.7 | |
| IgE/αIgE | 0.8 ± 0.34-150 | 0.9 ± 0.04-150 | |
| Spleen | B220 | 25.4 ± 10.7 | 23.2 ± 10.7 |
| CD4 | 5.2 ± 0.6 | 4.4 ± 1.4 | |
| CD8 | 4.6 ± 1.3 | 4.7 ± 1.3 | |
| Thymus | CD4−CD8− | 15.0 ± 8.6 | 12.6 ± 6.8 |
| CD4+CD8+ | 73.9 ± 7.4 | 76.1 ± 5.1 | |
| CD4+CD8− | 8.4 ± 2.6 | 8.9 ± 1.7 | |
| CD4−CD8+ | 2.8 ± 1.4 | 2.5 ± 0.2 | |
| Fetal liver | |||
| E11.5 | c-kit | 64.3 ± 18.9 | 72.0 ± 8.5 |
| E11.5 | Ter119 | 28.3 ± 24.8 | 20.5 ± 7.8 |
| E12.5 | c-kit | 1.7 ± 0.6 | 2.9 ± 0.9 |
| E12.5 | Ter119 | 85.0 ± 2.0 | 84.5 ± 2.9 |
| Tissue . | Surface marker . | Control YAC+scl+/− . | Rescue YAC+scl−/− . |
|---|---|---|---|
| Bone marrow | c-kit | 15.0 ± 2.44-150 | 12.8 ± 1.14-150 |
| CD34 | 6.7 ± 1.8 | 6.1 ± 0.9 | |
| Mac-1 | 30.8 ± 10.3 | 35.2 ± 3.6 | |
| Gr-1 | 33.0 ± 12.1 | 35.0 ± 2.6 | |
| Ter119 | 51.2 ± 10.2 | 42.9 ± 9.0 | |
| CD61 | 32.9 ± 11.5 | 36.1 ± 1.7 | |
| IgE/αIgE | 0.8 ± 0.34-150 | 0.9 ± 0.04-150 | |
| Spleen | B220 | 25.4 ± 10.7 | 23.2 ± 10.7 |
| CD4 | 5.2 ± 0.6 | 4.4 ± 1.4 | |
| CD8 | 4.6 ± 1.3 | 4.7 ± 1.3 | |
| Thymus | CD4−CD8− | 15.0 ± 8.6 | 12.6 ± 6.8 |
| CD4+CD8+ | 73.9 ± 7.4 | 76.1 ± 5.1 | |
| CD4+CD8− | 8.4 ± 2.6 | 8.9 ± 1.7 | |
| CD4−CD8+ | 2.8 ± 1.4 | 2.5 ± 0.2 | |
| Fetal liver | |||
| E11.5 | c-kit | 64.3 ± 18.9 | 72.0 ± 8.5 |
| E11.5 | Ter119 | 28.3 ± 24.8 | 20.5 ± 7.8 |
| E12.5 | c-kit | 1.7 ± 0.6 | 2.9 ± 0.9 |
| E12.5 | Ter119 | 85.0 ± 2.0 | 84.5 ± 2.9 |
Numbers indicate percentage of positive cells (± SD) following analysis of 4 adult mice or a minimum of 3 fetal livers for each genotype from line 2054.
Two animals analyzed.
Colony assays were used to investigate whether more subtle defects were present in the progenitor compartment. The numbers of myeloid and erythroid colonies present in adult bone marrow were the same in rescued YAC+scl−/− and control YAC+scl+/− littermates from line 2054 (Figure 5A). Subdivision of the myeloid colonies into granulocyte, monocyte, granulocyte-monocyte, and megakaryocyte did not reveal any differences between the rescued YAC+scl−/− mice and controls (Figure 5B). To assess yolk sac hematopoiesis, colony assays were performed using cells from E9.5 yolk sacs (Figure 5C). As previously described,45 58 no hematopoietic colonies were obtained from scl−/− yolk sacs (data not shown). By contrast, there was no significant difference between the numbers of erythroid and myeloid colonies obtained from rescued YAC+scl−/− or control YAC+scl+/− yolk sacs.
Analysis of hematopoietic progenitors and yolk sac angiogenesis in rescued YAC+
scl−/− mice. Hematopoietic progenitor assays were performed using adult bone marrow (A,B) or yolk sac (C) from mice of the indicated genotype derived from line 2054. In each case, histograms represent the mean of 6 cultures from 2 mice. (A) Myeloid and erythroid (erythroid burst-forming unit [BFU-E]) colonies obtained from adult bone marrow from line 2054. Myeloid colonies were obtained by growth either in IL-3 and SCF or IL-3 and Tpo. (B) Myeloid colony types obtained by growth in IL-3 and SCF. (C) Yolk sac BFU-Es obtained from line 2133 or 2054. (D) Yolk sac vasculature from E9.5 embryos of the indicated genotype from line 2074. Note the large vitelline vessels present in both control and rescued yolk sacs.
Analysis of hematopoietic progenitors and yolk sac angiogenesis in rescued YAC+
scl−/− mice. Hematopoietic progenitor assays were performed using adult bone marrow (A,B) or yolk sac (C) from mice of the indicated genotype derived from line 2054. In each case, histograms represent the mean of 6 cultures from 2 mice. (A) Myeloid and erythroid (erythroid burst-forming unit [BFU-E]) colonies obtained from adult bone marrow from line 2054. Myeloid colonies were obtained by growth either in IL-3 and SCF or IL-3 and Tpo. (B) Myeloid colony types obtained by growth in IL-3 and SCF. (C) Yolk sac BFU-Es obtained from line 2133 or 2054. (D) Yolk sac vasculature from E9.5 embryos of the indicated genotype from line 2074. Note the large vitelline vessels present in both control and rescued yolk sacs.
In addition to an absence of hematopoiesis,scl−/− embryos have also been reported to exhibit defects in yolk sac angiogenesis. In particular large vitelline vessels are absent in E9.5 scl−/− yolk sacs as a consequence of a cell autonomous defect in endothelial development.9 By contrast, the vitelline vessels present in rescued YAC+scl−/− yolk sacs were indistinguishable from those present in control YAC+scl+/− embryos (Figure 5D).
These data therefore demonstrate that the human SCL YAC completely rescues the lethal scl−/− phenotype and in particular is able to support normal yolk sac, fetal liver, and adult hematopoiesis together with yolk sac angiogenesis.
Discussion
We demonstrate here that the human SCL locus can rescue the lethal phenotype of scl−/− mice. The human SCL protein can therefore function in place of the murine SCL protein. More importantly, our results demonstrate that a 130-kb region spanning the human SCL locus contains the chromosomal domain necessary to direct SCL expression to erythroid cells and to other tissues in which SCL performs a nonredundant essential function. Identification of a human SCL domain capable of complementing the scl−/− phenotype opens up the possibility of systematically deleting individual regulatory elements and thereby examining the consequences of ablating SCL expression in specific cell types.
The SCL gene is expressed in a subset of blood cells (progenitors, erythroid, mast, and megakaryocytic lineages), endothelial cells, and specific regions of the brain and spinal cord. This pattern of expression is highly conserved throughout vertebrate evolution from zebrafish to mammals.1,10,12,20,55 Systematic analysis of the murinescl locus has identified a series of independent enhancers, each of which directs reporter gene expression to a subdomain of the normal SCL expression pattern.18-21 Of particular interest is a 3′ enhancer that directs expression to blood and endothelial progenitors throughout ontogeny19 and also to long-term repopulating hematopoietic stem cells.22 However additional elements remain to be identified because this 3′ enhancer is the only hematopoietic element identified so far and it does not direct expression to Ter119+ erythroid cells.19 By contrast the endogenous SCL gene is expressed in erythroid cells,54,55,59,60 thus suggesting that maintenance of SCL expression during erythroid differentiation is mediated by an as yet unidentified enhancer. Comparative genomic sequence analysis of the murine and human SCL loci have identified a number of peaks of homology that do not correspond to known enhancers and that therefore represent candidates for additional regulatory elements.61
Previous attempts to rescue the scl−/−phenotype in mice have met with only partial success probably because the transgenes were not subject to appropriate transcriptional regulation. An SCL cDNA under the control of murine stem cell virus regulatory elements has been used to rescuescl−/− ES cells.3 However germ line transmission of the ES cells was not obtained and the hematopoietic potential of rescued scl−/− ES cells was studied in chimeras by assessing the ES cell contribution to pooled populations of blood cells. This precluded any assessment of whether the rescued hematopoietic cells were quantitatively or qualitatively normal. We have recently demonstrated that anSCL cDNA controlled by the SCL gene 3′ enhancer results in selective rescue of early hematopoietic progenitors and yolk sac angiogenesis in scl−/−embryos.22 However, consistent with the idea of a distinctSCL erythroid enhancer, this construct was unable to support normal erythropoiesis and the embryos died of severe anemia. By contrast, the human SCL YAC reported here rescued normal levels of yolk sac, fetal liver, and adult hematopoiesis including erythropoiesis thus demonstrating that the YAC contains the missing erythroid regulatory elements.
SCL is known to be essential for the formation of hematopoietic stem cells,2,3 for subsequent erythroid differentiation,22 and for yolk sac angiogenesis.9 SCL may also play an essential role in other tissues including midbrain, hindbrain, spinal cord, and intraembryonic endothelium, but analysis has been precluded by the early embryonic lethality of scl−/− mice. Our results suggest that the human SCL YAC contains all the regulatory elements necessary to direct appropriate SCL expression to the full complement of tissues in which SCL provides essential nonredundant functions.
This notion is consistent with comparative synteny data. We have previously characterized and sequenced the SCL genomic loci from human, mouse, chicken, and pufferfish and have found that the genes immediately flanking pufferfish SCL were unrelated to those known to flank both avian and mammalian SCLgenes.21,61,62 In view of the conserved pattern of SCL expression between mammals and teleost fish, these results implied thatSCL regulatory elements might be confined to the region between the upstream and downstream flanking genes. Moreover, a 10.4-kb fragment of pufferfish genomic DNA, containing the SCL gene and extending to the 5′ and 3′ flanking genes, directed appropriate expression to hematopoietic and neural tissue in transgenic zebrafish embryos.62 Our current results accord with these data because the human SCL YAC contains the entire SCLlocus extending to both upstream (SIL) and downstream(MAP17) genes.
Interestingly, the human SCL YAC did not give rise to copy number–dependent expression and the levels of transgene expression also varied between transgenic lines carrying the same transgene copy number. These phenomena are well recognized in transgenic mice and are usually attributed to stable or variegating position effects (for a review, see Martin and Whitelaw56). Although we cannot completely exclude the possibility that different YAC transgenic lines have acquired distinct mutations or deletions of regulatory elements, the use of I-PpoI restriction sites allowed us to confirm the size of the integrated YAC in each transgenic line. Moreover, higher resolution conventional electrophoresis and Southern hybridization did not detect any rearrangements in a region of 57 kb spanning the SCL gene and including 16 kb and 20 kb of upstream and downstream sequence, respectively. Instead our results suggest that the SCL YAC does not exhibit LCR-like activity. It is possible that an SCL LCR element lies outside the YAC, but the comparative synteny data discussed above argue against this possibility. Instead, our results are consistent with the concept that the complement of regulatory elements sufficient to ensure appropriate expression of the SCL gene in its endogenous location may be inadequate to protect the gene from transcriptional constraints operating elsewhere in the genome.
We thank Dr Clare Huxley for advice concerning YAC modification and transfer and for the plasmids pUC-OK, pUC-WAN, and pRS406. We are grateful to Prof Glenn Begley and Dr Lorraine Robb for sending us the scl knockout line, SV102, and for help with hematopoietic colony assays. We thank Dr E. Andermarcher for the IRES-nls-lacZ construct, Dr T. Enver for the rat GAPDH probe, and Dr S. Tsai for BHK-conditioned medium. We also thank Drs Andreas Schedl, Wendy Dean, and Wolf Reik for advice and assistance regarding the generation of transgenic mice.
Supported by the Wellcome Trust, the Leukaemia Research Fund, the Medical Research Council, and the Pre-Leukaemia Society.
A.M.S. and A.J.B. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Anthony R. Green, University of Cambridge, Department of Haematology, Cambridge Institute for Medical Research, Hills Rd, Cambridge, CB2 2XY, United Kingdom; e-mail:arg1000@cam.ac.uk.




![Fig. 5. Analysis of hematopoietic progenitors and yolk sac angiogenesis in rescued YAC+. / scl−/− mice. Hematopoietic progenitor assays were performed using adult bone marrow (A,B) or yolk sac (C) from mice of the indicated genotype derived from line 2054. In each case, histograms represent the mean of 6 cultures from 2 mice. (A) Myeloid and erythroid (erythroid burst-forming unit [BFU-E]) colonies obtained from adult bone marrow from line 2054. Myeloid colonies were obtained by growth either in IL-3 and SCF or IL-3 and Tpo. (B) Myeloid colony types obtained by growth in IL-3 and SCF. (C) Yolk sac BFU-Es obtained from line 2133 or 2054. (D) Yolk sac vasculature from E9.5 embryos of the indicated genotype from line 2074. Note the large vitelline vessels present in both control and rescued yolk sacs.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/99/11/10.1182_blood.v99.11.3931/6/m_h81122606005.jpeg?Expires=1765892862&Signature=U836iddIY8R-AdPYQDBBuJATAfxN0uSYsH2LyeiRdEgUZk-ChIbuubiRe78o2IIKuTmUe6hxLdcuel6Ln6m3MgolsU9pIRh0Z9s5lw-vfUGMX-6Xpi8cNwf6qlajIGxTJdS3ebtgLni4BBZgtkBcANHbVNOXeqwcEzECz1Em~FA8oiTwUEnfvEKjfjd2h0RdkWOkUmX0G8PEqjGLiSHBBgalUpsAdfoAmy2oznEtBO0oIljIBknCHfcvkx6ods2TfRJFmjme18TVpQK89unKurhE96eFZSGTiKvf-VE7k-fjQer0VL2QHBeOLCaDLI2zTZPX8zUr764Ci44aApiE-A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal