Caspase 10 (Mch4/FLICE2) is a caspase homologous to caspase 8. A recent report described that inherited CASP10 gene mutations underlie defective lymphocyte and dendritic cell apoptosis in autoimmune lymphoproliferative syndrome (ALPS). In this study, to explore the possibility that mutation of this gene might be involved in the development of non-Hodgkin lymphoma (NHL), we have analyzed the entire coding region and all splice sites of the CASP10gene for the detection of somatic mutations in 117 human NHLs. Overall, 17 NHLs (14.5%) were found to have CASP10mutations, which were identified in the coding regions of the prodomain (n = 3), the p17 large protease subunit (n = 11), and the p12 small protease subunit (n = 3). We expressed the tumor-derived caspase 10 mutants in 293 cells and found that apoptosis was suppressed. These data suggest that the inactivating mutations of theCASP10 gene might lead to the loss of its apoptotic function and contribute to the pathogenesis of some human NHLs.
Introduction
Apoptosis is a fundamental biochemical cell death pathway essential for normal tissue homeostasis, cellular differentiation, and development.1,2 Dysregulation of apoptosis may be directly involved in several human diseases, including degenerative and autoimmune diseases, neoplasia, and acquired immunodeficiency syndrome.1,2 The death signals originating from death receptors, such as tumor necrosis factor (TNF) receptors, Fas, and TNF-related apoptosis-inducing ligand (TRAIL) receptors, are transduced through the recruitment of procaspase 8 and 10 to the death-induced signaling complex (DISC) by the adaptor molecule Fas-associated death domain protein (FADD).1 The local aggregation of the procaspases 8 and 10 is sufficient to allow autoprocessing or transprocessing to produce active caspases 8 and 10, which can subsequently activate downstream executioners, such as caspases 3 and 7.3,4 The death receptors are widely expressed in normal and neoplastic cells,1,5 but the expression of these proteins does not necessarily predict susceptibility to killing.6 This can reflect the presence of inhibiting mechanisms of death receptor–mediated apoptosis. The death receptor–mediated apoptosis can be blocked by several mechanisms, including the mutation of the primary structure of death receptors and their ligand.7-10
The consequences of naturally occurring mutants of Fas/Fas ligand (FasL) have been well demonstrated in both mice and humans. BothFas gene–deficient mice (lpr) andFasL gene–deficient mice (gld) have an abnormality of mature T-cell deletion in the peripheral tissues, resulting in lymphadenopathy and systemic autoimmune disease.11 Autoimmune lymphoproliferative syndrome (ALPS), the human counterpart of lpr and gldmice, is characterized by an increase in double-negative T cells and profound lymphadenopathy.4,12,13 Most of the ALPS patients (ALPS type Ia) have inherited mutations of theFas gene.12 In ALPS type Ib, systemic lupus erythematosus and lymphadenopathy are associated with germ line mutation of the FasL gene.13 ALPS type II is manifested by a related clinical phenotype and apoptosis defects in the absence of either Fas or FasL mutations, indicating that other modifying factors besides Fas orFasL mutations affect cell death signaling pathways in these patients.4,14 15
Recently, Wang and coworkers traced along the Fas-signaling pathway of 2 ALPS type II families.4 They found 2 germ line mutations of the CASP10 gene, but not of other death-related genes such as Fas, FasL, TRAILR1, TRAILR2, CASP8, orFADD.4 These caspase 10 mutants had significantly reduced enzymatic and autoprocessing activities, and when transfected into mammalian cells, these mutants had a reduced ability to induce apoptosis, indicating that caspase 10 is defective in patients with ALPS type II.4
Unlike the embryonic lethality of FADD and caspase 8 deficiencies reported in knock-out mice,11,16 the CASP10mutations in ALPS patients exert consequences limited to the control of cell death in the immune system.4 Because theCASP10 gene mutation results in abnormal lymphocyte proliferation as in ALPS,4 it is possible that this gene mutation may be involved in the development of non-Hodgkin lymphoma (NHL). In addition, the previous loss of heterozygosity (LOH) studies have suggested that loss of one or more putative tumor suppressor genes at chromosome 2q33-34, where the CASP10 gene resides,17 may be involved in the development of NHL.18 In the present study, we analyzed a series of 117 NHLs for the detection of somatic mutations of theCASP10 gene, which might lead to a longer survival of affected tumor cells and contribute to the tumorigenesis of NHL.
Materials and methods
Tissue samples and microdissection
Paraffin-embedded tissues of human NHL were obtained from 117 patients. These samples were stained with hematoxylin and eosin, examined by immunohistology, and then classified according to the Revised European-American Lymphoma (REAL) classification.19 The NHLs analyzed consisted of 7 B-cell small lymphocytic lymphomas, 3 mantle cell lymphomas, 4 follicular lymphomas, 23 mucosa-associated lymphoid tissue (MALT)–type lymphomas, 46 diffuse large B-cell lymphomas, 4 precursor T-lymphoblastic lymphomas, 1 T-cell chronic lymphocytic leukemia, 14 peripheral T-cell lymphomas (unclassified), 14 angiocentric lymphomas, and 1 intestinal T-cell lymphoma. Approval was obtained from the Catholic University of Korea, College of Medicine's institutional review board for these studies. Informed consent was provided according to the Declaration of Helsinki.
Malignant cells and normal cells were selectively procured from hematoxylin and eosin-stained slides using a 30G1/2 hypodermic needle (Becton Dickinson, Franklin Lakes, NJ) affixed to a micromanipulator, as described previously.20
Single-strand conformation polymorphism analysis
Genomic DNAs from tumor cells and corresponding normal cells were amplified with 15 primer pairs covering the entire coding region (10 exons) of the CASP10 gene (Table1). Oligonucleotide primers were designed with the program Oligo (National Biosciences, Plymouth, MN) using sequences obtained from GenBank (accession no. M67454). Numbering of complementary DNA (cDNA) of CASP10 was done with respect to the ATG start codon of CASP10/b (FLICE 2)according to Ng et al.21 Each polymerase chain reaction (PCR) was performed under standard conditions in a 10-μL reaction mixture containing 1 μL template DNA (usually 500 cells/PCR), 0.5 μM of each primer, 0.2 μM of each deoxynucleotide triphosphate, 1.5 mM MgCl2, 0.4 U AmpliTaq gold polymerase (Perkin-Elmer, Foster City, CA), 0.5 μCi of [32P]dCTP (Amersham, Buckinghamshire, United Kingdom), and 1 μL 10X buffer. The reaction mixture was denatured for 1 minute at 94°C and incubated for 30 cycles (denaturing for 40 seconds at 94°C, annealing for 40 seconds at 59°-68°C, and extending for 40 seconds at 72°C). Final extension was continued for 5 minutes at 72°C. After amplification, PCR products were denatured for 5 minutes at 95°C at a 1:1 dilution of sample buffer containing 98% formamide-5 mmol/L NaOH and were loaded onto a single-strand conformation polymorphism (SSCP) gel (FMC Mutation Detection Enhancement system; Intermountain Scientific, Kaysville, UT) with 10% glycerol. Samples were electrophoresed at 8 W at room temperature overnight. After electrophoresis, the gels were transferred to 3-mm Whatman paper and dried, and autoradiography was performed with Kodak X-OMAT film (Eastman Kodak, Rochester, NY). For the detection of mutations, DNAs showing mobility shifts were cut out from the dried gel, and reamplified for 30 cycles using the same primer set. Sequencing of the PCR products was carried out using the cyclic sequencing kit (Perkin-Elmer) according to the manufacturer's recommendation.
Primer sequences used for PCR amplification of individual exons of CASP10
| Name . | Exon . | Sense primer sequence (5′-3′)* . | Antisense primer sequence (5′-3′) . | Product length (bp) . |
|---|---|---|---|---|
| E2A | 2 | gggccatatgtcctcactctc | AAACTTGAGGTTCTCCACATCTTG | 184 |
| E2B | 2 | CTTTCGTGAGAAGCTTCTGATTAT | TTCTGCCGTATGATATAGAGGAGT | 210 |
| E2C | 2 | TGCTGAGTGAGGAAGACCCTTTCT | ctcccatctccaccacagacc | 169 |
| E3 | 3 | cttacaagtgtaaggctttattt | cattgattaagacagtgctcaca | 191 |
| E4 | 4 | tgagtggataatcaataggcaagt | ctccaagttagcaatcacaagc | 217 |
| E5 | 5 | actgcaacctccgcctcctg | cattgaccagcacaccactgaacc | 224 |
| E6 | 6 | gtccttccctgcatcaagtc | ccctaccataccgatctaagttgt | 173 |
| E7 | 7 | tggggaagatatttggagtctgag | gcccctaaagaaaccgtcctt | 213 |
| E8 | 8 | Aaggattcctactaagtggctcta | gcttttgataaactgttccaga | 177 |
| E9A | 9 | tgtgcccggccttgtttcag | GGGCTGGATTGCACTTCTGCTTCT | 214 |
| E9B | 9 | CGAAAGTGGAAATGGAGATGGT | CAGGCCTGGATGAAAAAGAGTTTA | 220 |
| E9C | 9 | GGGAGATCATGTCTCACTTCACA | CCACATGCCGAAAGGATACA | 234 |
| E9D | 9 | CTACTTGGTCTGGCCACTGT | taccaaaggtgttgaatgagagta | 189 |
| E10 | 10 | aaattttgttttcttctttgttgc | caatgattcgtttgaggtctaag | 222 |
| E11 | 11 | ttccccttttatttctctttgtgc | gtcaatctcaggcgatgtgg | 237 |
| Name . | Exon . | Sense primer sequence (5′-3′)* . | Antisense primer sequence (5′-3′) . | Product length (bp) . |
|---|---|---|---|---|
| E2A | 2 | gggccatatgtcctcactctc | AAACTTGAGGTTCTCCACATCTTG | 184 |
| E2B | 2 | CTTTCGTGAGAAGCTTCTGATTAT | TTCTGCCGTATGATATAGAGGAGT | 210 |
| E2C | 2 | TGCTGAGTGAGGAAGACCCTTTCT | ctcccatctccaccacagacc | 169 |
| E3 | 3 | cttacaagtgtaaggctttattt | cattgattaagacagtgctcaca | 191 |
| E4 | 4 | tgagtggataatcaataggcaagt | ctccaagttagcaatcacaagc | 217 |
| E5 | 5 | actgcaacctccgcctcctg | cattgaccagcacaccactgaacc | 224 |
| E6 | 6 | gtccttccctgcatcaagtc | ccctaccataccgatctaagttgt | 173 |
| E7 | 7 | tggggaagatatttggagtctgag | gcccctaaagaaaccgtcctt | 213 |
| E8 | 8 | Aaggattcctactaagtggctcta | gcttttgataaactgttccaga | 177 |
| E9A | 9 | tgtgcccggccttgtttcag | GGGCTGGATTGCACTTCTGCTTCT | 214 |
| E9B | 9 | CGAAAGTGGAAATGGAGATGGT | CAGGCCTGGATGAAAAAGAGTTTA | 220 |
| E9C | 9 | GGGAGATCATGTCTCACTTCACA | CCACATGCCGAAAGGATACA | 234 |
| E9D | 9 | CTACTTGGTCTGGCCACTGT | taccaaaggtgttgaatgagagta | 189 |
| E10 | 10 | aaattttgttttcttctttgttgc | caatgattcgtttgaggtctaag | 222 |
| E11 | 11 | ttccccttttatttctctttgtgc | gtcaatctcaggcgatgtgg | 237 |
Upper case letters correspond to exons and lower case letters correspond to introns.
Because a biallelic polymorphism at nucleotide position 1337 A/G (exon 9) has been found by previous studies4 and additional biallelic polymorphism at nucleotide position 177 A/G (exon 2) has been detected in the present study, SSCP analysis at these polymorphic sites was used for the detection of LOH as well as for the detection of mutations. The PCR and SSCP conditions of LOH study were the same as the conditions described above. Complete or nearly complete absence of one allele in tumor DNA of informative cases, as defined by direct visualization, was considered as LOH.
Site-directed mutagenesis
Site-directed mutagenesis was performed using a Quick Change Site-Directed Mutagensis kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions. To change a base, a plasmid that contained the CASP10 gene in pcDNA3.1-Myc (Invitrogen, Carlsbad, CA) was used as a template. The sequences of the mutagenized plasmids were confirmed.
Coimmunoprecipitations and immunoblotting assay
Human embryonic kidney 293 cells were cultured at 37°C in 5% CO2 in Dulbecco modified Eagle medium with 10% heat-inactivated fetal bovine serum. Cells in log phase were transfected in 60-mm diameter dishes with expression plasmids using Superfect transfection reagent (Qiagen, Valencia, CA) according to the manufacturer's recommendations. Cells were harvested 2 days later and lysed in ice-cold Nonidet P-40 lysis buffer (10 mM Hepes, pH 7.4, 142.5 mM KCl, 0.2% Nonidet P-40, 5 mM EGTA) supplemented with 1 mM glycerophosphate, 1 μM Na3VO4, 1 mM phenylmethylsulfonyl fluoride, and 1 times protease inhibitor mix (Roche Molecular Biochemicals, Mannheim, Germany). Cell lysates were clarified by centrifugation at 16 000g for 5 minutes and subjected to immunoprecipitation using agarose-conjugated anti-Flag M2 antibodies (Sigma, St Louis, MO). Immunocomplexes were fractionated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes. The resulting blots were incubated with anti-Myc (1:100 vol/vol, Santa Cruz Biotechnology, Santa Cruz, CA) followed by horseradish peroxidase–conjugated secondary antibodies and detection by an enhanced chemiluminescence method (ECL; Amersham Pharmacia Biotech, Buckinghamshire, United Kingdom). Alternatively, lysates were analyzed directly by immunoblotting after normalization for total protein content.
Apoptosis assay
The 293 cells (106) were plated per well in a 2-chamber slide and transfected by the Superfect transfection reagent (Qiagen) with various combinations of expression plasmids (total 1.3 μg) and 0.2 μg green fluorescence protein (GFP) marker plasmid pEGFP (Clontech, Palo Alto, CA). Twenty-four hours after the transfection, the percentage of GFP+ cells with nuclear apoptotic morphology was determined by fixing in 10% methanol for 15 minutes and staining with 1 μg/mL 4′-6-diamidino-2-phenylindole (DAPI) for 15 minutes (mean ± SD, n = 4).
Results
CASP10 gene mutations
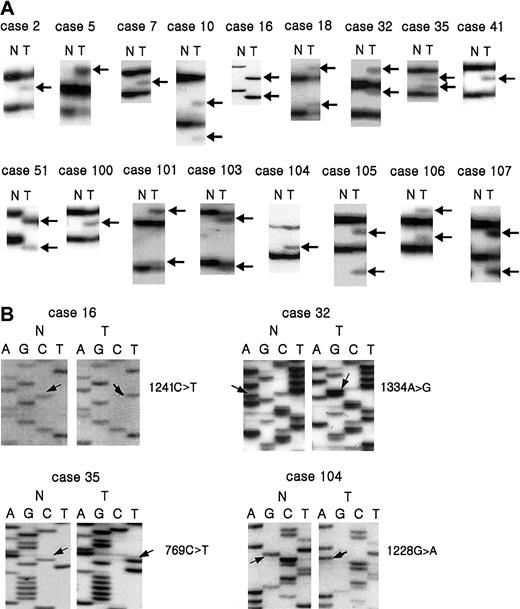
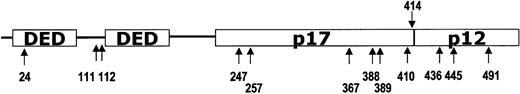
Through the microdissection technique, we selectively procured tumor cells from histologic sections of 117 NHLs as shown previously.20 Genomic DNA was isolated and analyzed for potential mutations in all 10 exons in the coding region of theCASP10 gene by PCR-SSCP analysis. Enrichment and direct sequence analysis of aberrantly migrating bands led to the identification of mutations in 17 of the samples (14.5%; Table2 and Figures1 and2). The mutations consisted of 14 missense, 1 nonsense, and 2 frame shift mutations (Table 2). The mutations were identified in exon 2 (n = 3), exon 7 (n = 2), exon 9 (n = 11), and exon 10 (n = 1) of the CASP10 gene. Five of the mutations in exon 9 showed an identical missense mutation. The mutation resulted in the substitution of Ala by Val at amino acid residue 414, which is next to the cleavage site between the p17 large subunit and the p12 small subunit (Table 2 and Figure 1). As for the relationship between the histologic types and the CASP10gene mutations, the tumors with the mutations consisted of 8 diffuse large B-cell lymphomas, 1 B-cell small lymphocytic lymphoma, 2 MALT lymphomas, 2 angiocentric lymphomas, and 2 peripheral T-cell lymphomas, unclassified (Table 2). None of the corresponding normal samples showed evidence of mutations by SSCP (Figure 1), indicating the mutations detected in the NHL specimens had risen somatically. We repeated the experiments 4 times, including tissue microdissection, PCR, SSCP, and sequencing analysis to ensure the specificity of the results, and found that the data were consistent (data not shown).
Summary of CASP10 mutations identified in NHL
| Case no. . | Histologic type . | LOH* analysis . | SSCP patterns at mutation sites† . | Mutation site . | Mutation domain‡ . | Nucleotide change2-153 (predicted amino acid change) . | |
|---|---|---|---|---|---|---|---|
| 117 . | 1337 . | ||||||
| 2 | Peripheral T-cell lymphoma | NI | NI | M + W | Exon 9 | p17 | 1241C>T (Ala414Val) |
| 5 | B-cell small lymphocytic lymphoma | NI | NI | M + W | Exon 2 | DED | 70C>A (Leu24Ile) |
| 7 | Angiocentric lymphoma | NI | NI | M + W | Exon 7 | p17 | 740G>A (Gly247Asp) |
| 10 | Diffuse large B-cell lymphoma | NI | NI | M + W | Exon 2 | Between DED/DED | 334G>A (Val112Ile) |
| 16 | Angiocentric lymphoma | HET | NI | M | Exon 9 | p17 | 1241C>T (Ala414Val) |
| 18 | Follicular center cell lymphoma | NI | NI | M + W | Exon 10 | p12 | 1471G>T (Val491Leu) |
| 32 | Diffuse large B-cell lymphoma | NI | NI | M + W | Exon 9 | p12 | 1334G>A (Gly445Asp) |
| 35 | Diffuse large B-cell lymphoma | NI | NI | M + W | Exon 7 | p17 | 769C>T (Gln257Stop) |
| 41 | Peripheral T-cell lymphoma | NI | NI | M + W | Exon 9 | p17 | 1241C>T (Ala414Val) |
| 51 | B-cell small lymphocytic lymphoma | NI | NI | M | Exon 9 | p17 | 1241C>T (Ala414Val) |
| 100 | Diffuse large B-cell lymphoma | NI | NI | M + W | Exon 9 | p17 | 1241C>T (Ala414Val) |
| 101 | MALT lymphoma | NI | NI | M + W | Exon 2 | Between DED/DED | 332G>C (Arg111Thr) |
| 103 | MALT lymphoma | NI | NI | M + W | Exon 9 | p17 | 1042insA (frameshift termination at position 367) |
| 104 | Diffuse large B-cell lymphoma | NI | NI | M + W | Exon 9 | p17 | 1228G>A (Val410Ile) |
| 105 | Diffuse large B-cell lymphoma | NI | NI | M + W | Exon 9 | p12 | 1315G>T (Gly436Cys) |
| 106 | Diffuse large B-cell lymphoma | NI | NI | M + W | Exon 9 | p17 | 1164insG (frameshift termination at position 389) |
| 107 | Diffuse large B-cell lymphoma | NI | NI | M + W | Exon 9 | p17 | 1163G>T (Arg388Ile) |
| Case no. . | Histologic type . | LOH* analysis . | SSCP patterns at mutation sites† . | Mutation site . | Mutation domain‡ . | Nucleotide change2-153 (predicted amino acid change) . | |
|---|---|---|---|---|---|---|---|
| 117 . | 1337 . | ||||||
| 2 | Peripheral T-cell lymphoma | NI | NI | M + W | Exon 9 | p17 | 1241C>T (Ala414Val) |
| 5 | B-cell small lymphocytic lymphoma | NI | NI | M + W | Exon 2 | DED | 70C>A (Leu24Ile) |
| 7 | Angiocentric lymphoma | NI | NI | M + W | Exon 7 | p17 | 740G>A (Gly247Asp) |
| 10 | Diffuse large B-cell lymphoma | NI | NI | M + W | Exon 2 | Between DED/DED | 334G>A (Val112Ile) |
| 16 | Angiocentric lymphoma | HET | NI | M | Exon 9 | p17 | 1241C>T (Ala414Val) |
| 18 | Follicular center cell lymphoma | NI | NI | M + W | Exon 10 | p12 | 1471G>T (Val491Leu) |
| 32 | Diffuse large B-cell lymphoma | NI | NI | M + W | Exon 9 | p12 | 1334G>A (Gly445Asp) |
| 35 | Diffuse large B-cell lymphoma | NI | NI | M + W | Exon 7 | p17 | 769C>T (Gln257Stop) |
| 41 | Peripheral T-cell lymphoma | NI | NI | M + W | Exon 9 | p17 | 1241C>T (Ala414Val) |
| 51 | B-cell small lymphocytic lymphoma | NI | NI | M | Exon 9 | p17 | 1241C>T (Ala414Val) |
| 100 | Diffuse large B-cell lymphoma | NI | NI | M + W | Exon 9 | p17 | 1241C>T (Ala414Val) |
| 101 | MALT lymphoma | NI | NI | M + W | Exon 2 | Between DED/DED | 332G>C (Arg111Thr) |
| 103 | MALT lymphoma | NI | NI | M + W | Exon 9 | p17 | 1042insA (frameshift termination at position 367) |
| 104 | Diffuse large B-cell lymphoma | NI | NI | M + W | Exon 9 | p17 | 1228G>A (Val410Ile) |
| 105 | Diffuse large B-cell lymphoma | NI | NI | M + W | Exon 9 | p12 | 1315G>T (Gly436Cys) |
| 106 | Diffuse large B-cell lymphoma | NI | NI | M + W | Exon 9 | p17 | 1164insG (frameshift termination at position 389) |
| 107 | Diffuse large B-cell lymphoma | NI | NI | M + W | Exon 9 | p17 | 1163G>T (Arg388Ile) |
NI indicates not informative (homozygosity); HET, retention heterozygosity.
177 indicates the polymorphism at nucleotide 177 ofCASP10; 1337, the polymorphism at nucleotide at 1337 ofCASP10.
M is aberrant bands of mutant allele without those of wild-type allele; M + W, aberrant bands of mutant allele with those of wild-type allele.
Cleavage site (p17/p12) is cleavage site between p17 large subunit and p12 small subunit; p17, p17 large subunit; p12, p12 small subunit.
Numbering of cDNA of CASP10 was done with respect to the ATG start codon according to Ng et al.21
Mutations and deletions of
CASP10 gene in NHL. SSCP (A) and sequencing analysis (B) of caspase 10 DNA from tumors (lane T) and normal tissues (lane N). (A) Arrows (lane T) indicate aberrant bands compared to SSCP from normal tissue (N). (B) Arrows indicate nucleotide substitutions in tumor tissue compared to normal tissue.
Mutations and deletions of
CASP10 gene in NHL. SSCP (A) and sequencing analysis (B) of caspase 10 DNA from tumors (lane T) and normal tissues (lane N). (A) Arrows (lane T) indicate aberrant bands compared to SSCP from normal tissue (N). (B) Arrows indicate nucleotide substitutions in tumor tissue compared to normal tissue.
Schematic organization of caspase 10 cDNA.
The positions of the predicted amino acid changes of the detectedCASP1010 mutations are indicated by arrows, and their amino acid numbers are presented under the corresponding arrows.
Schematic organization of caspase 10 cDNA.
The positions of the predicted amino acid changes of the detectedCASP1010 mutations are indicated by arrows, and their amino acid numbers are presented under the corresponding arrows.
Allelic status
Because one of the 2 CASP10 gene mutations detected in the ALPS patients has been suggested to affect apoptosis function in a dominant-negative fashion,4 we examined the allelic status of the CASP10 gene in the tumors carrying the mutations. In addition to the previously described CASP10 gene polymorphism in exon 9,4 we also detected a novel polymorphism in exon 2 (nucleotide 177 A/G). The heterozygosity rate was 18% (18 of 100 normal healthy persons). We also analyzed the frequency of this polymorphism in the normal tissues of the NHLs. We could get normal tissues from 82 of the 117 NHLs. We found that the normal tissues from 22 cases showed this polymorphism (27%). However, the difference of these frequencies between the normal tissues of healthy population and NHLs were not statistically significant (Fisher exact test; 2-tailed, P > .05). Unfortunately, all cases with CASP10 mutations are noninformative or showed retention of heterozygosity with these 2 polymorphisms (Table 2).
Instead, SSCP patterns in 2 of 17 mutations (cases 16 and 51) showed only aberrant bands of the mutant allele without those of the wild-type allele (Figure 1), and sequencing analysis also revealed only mutated sequences without wild-type ones, indicating either a homozygous mutation or a hemizygous mutation with allelic loss (Figure 1). In contrast, the other 15 cases with caspase 10 mutation showed that bands of the wild-type allele were present together with aberrant bands of the mutant allele (Figure 1).
Abrogation of apoptosis activities by CASP10mutations
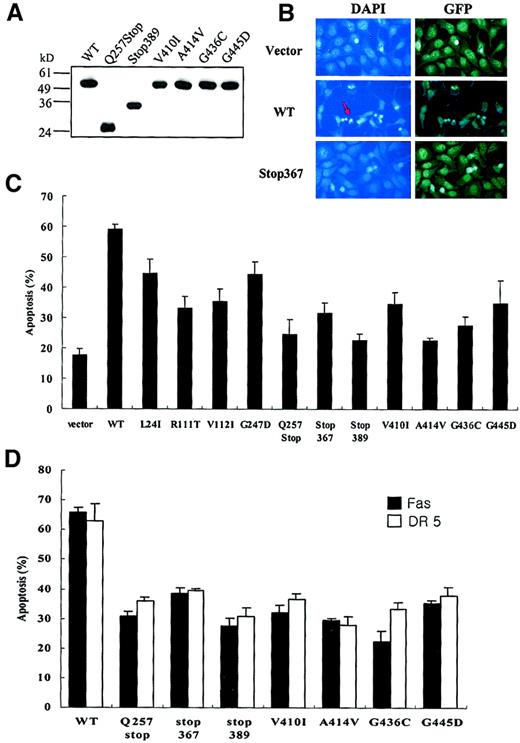
To determine whether the mutant forms of caspase 10 are functionally defective, we generated caspase 10 mammalian expression constructs containing the mutations found in this study by using site-directed mutagenesis. On transfection into 293 cells, we found that all mutant types showed a significant defect in apoptosis function (Figure 3B-C).
Defective apoptotic activities of tumor-derived
CASP10 mutants. (A) Myc-tagged expression constructs of wild-type (WT) CASP10 or tumor-derived CASP10mutants were transfected into 293 cells. Cell lysates were immunoblotted with anti-Myc antibody. (B) The 293 cells were transfected with 1.3 μg wild-type (WT) caspase 10, Stop367, or vector only, together with 0.2 μg of pEGEF. Twenty-four hours after transfection, cell were fixed in 10% methanol for 15 minutes, and stained with 1 μg/mL DAPI for 15 minutes, and the nuclei were examined by fluorescence microscopy. Arrow indicates apoptotic cells with condensed nuclei. Original magnification × 400. (C) The percentage of apoptosis was measured 48 hours later by DAPI staining (mean ± SD; n = 4). The 293 cells were cotransfected with Fas and TRAIL receptor 2 constructs and a 4-fold excess of each mutant caspase 10. Cell death was analyzed as described in panel B.
Defective apoptotic activities of tumor-derived
CASP10 mutants. (A) Myc-tagged expression constructs of wild-type (WT) CASP10 or tumor-derived CASP10mutants were transfected into 293 cells. Cell lysates were immunoblotted with anti-Myc antibody. (B) The 293 cells were transfected with 1.3 μg wild-type (WT) caspase 10, Stop367, or vector only, together with 0.2 μg of pEGEF. Twenty-four hours after transfection, cell were fixed in 10% methanol for 15 minutes, and stained with 1 μg/mL DAPI for 15 minutes, and the nuclei were examined by fluorescence microscopy. Arrow indicates apoptotic cells with condensed nuclei. Original magnification × 400. (C) The percentage of apoptosis was measured 48 hours later by DAPI staining (mean ± SD; n = 4). The 293 cells were cotransfected with Fas and TRAIL receptor 2 constructs and a 4-fold excess of each mutant caspase 10. Cell death was analyzed as described in panel B.
To explore how the mutants of caspase 10 impair apoptosis, we coexpressed some of the caspase 10 mutants either with Fas or TRAIL receptor 2 in 293 cells, and found that these mutants interfered with death induction by Fas or TRAIL receptor overexpression, indicating these mutants have dominant-negative inhibition of the death receptor–induced apoptosis (Figure 3D).
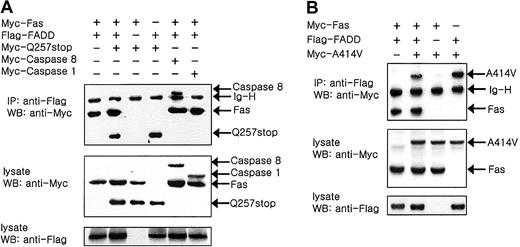
CASP10 mutants are recruited in DISC
The interference in death pathways caused by the caspase mutants (Figure 3) led us to hypothesize that the caspase mutants might be recruited into the DISC. To confirm this, we coexpressed Fas and FADD either with the Gln 257 stop mutant or the Ala414Val mutant. The coimmunoprecipitation analysis shows that FADD coimmunoprecipitates both with the Gln 257 stop mutant and FADD indicating this mutant is recruited in the DISC (Figure 4A). As a positive control, Figure 4A also shows that FADD coimmunoprecipitates both with Fas and caspase 8. In contrast, FADD did not coimmunoprecipitate with caspase 1, thus demonstrating the specificity of these results (Figure 4A). Figure 4B also shows that FADD coimmunoprecipitates both with Fas and Ala414Val mutant.
Recruitment of
CASP10 mutants to DISC. The 293 cells were cotransfected with expression constructs for human Myc-Fas and Flag-FADD either with the Myc-Q257stop mutant (A) or the Myc-A414V mutant (B) as indicated. The transfected cells were harvested and lysed 2 days later and immunoprecipitated with anti-Flag antibodies and sequentially immunoblotted with anti-Myc antibodies as indicated. Aliquots of the same lysates (normalized for total protein content) were also analyzed directly by SDS-PAGE/immunoblotting as indicated. IP indicates immunoprecipitation; WB, Western blotting.
Recruitment of
CASP10 mutants to DISC. The 293 cells were cotransfected with expression constructs for human Myc-Fas and Flag-FADD either with the Myc-Q257stop mutant (A) or the Myc-A414V mutant (B) as indicated. The transfected cells were harvested and lysed 2 days later and immunoprecipitated with anti-Flag antibodies and sequentially immunoblotted with anti-Myc antibodies as indicated. Aliquots of the same lysates (normalized for total protein content) were also analyzed directly by SDS-PAGE/immunoblotting as indicated. IP indicates immunoprecipitation; WB, Western blotting.
Discussion
The germ line mutations of the Fas gene result in ALPS type Ia12,13 and NHL has been reported to have somatic mutations of the Fas gene.20 Inherited mutations in Fas are currently the best known genetic link between apoptosis and the regulation of lymphocyte homeostasis and immune tolerance. Moreover, caspase 10, one of the initiator caspases, is known to be essential for maintaining the equilibrium in the cells of the immune system as shown in ALPS type II.4 In addition, NHL is one of the human tumors with LOH at chromosome 2q33-34 where the CASP10 gene resides.22 Thus, we hypothesized that the neoplastic cells of NHL have the somatic mutations of the CASP10 gene and detected the somatic mutations of the CASP10 gene in 17 of 117 NHLs (14.5%). The transfection study revealed that the NHL-derived caspase 10 mutants showed a loss of apoptotic function. Also, transfection of the NHL-derived caspase 10 mutants curtailed apoptosis induction both by Fas and TRAIL receptor 2, suggesting that these caspase 10 mutants affect a signal integration point for the death receptor. Furthermore, coimmunoprecipitation revealed that the representative cases of the mutant caspase 10 are recruited into DISC together with Fas and FADD, suggesting that the DISCs formed with these mutants are not functional and block the apoptosis signaling. This is the first report on theCASP10 gene mutations in human cancers, and our data suggest that the inactivating mutations of the CASP10 gene might lead to the loss of its apoptotic signaling function and contribute to the pathogenesis of some NHLs.
Caspase 10, like caspase 8, is present as an inactive proenzyme comprising a prodomain that contains 2 death effector domains (DEDs) to allow caspase 10 to interact with the DED of FADD and a catalytic protease domain that can be further processed to give a large and a small subunit.3 It has been reported that caspase 10 lacking the DEDs did not bind FADD,3 suggesting that the DED is necessary for the transduction of an apoptotic signal. Among the mutations of CASP10 found in this study, 3 mutations were identified in the coding regions of the prodomain. Interestingly, the point mutations at codons 111 and 112, which encode the region between the 2 DEDs, showed impairment of apoptosis in transfected cells (Figure 3C). These amino acid changes might lead to structural change of caspase 10 to prevent DED-DED interactions between FADD and caspase 10.
A high proportion of the mutations of the CASP10 gene in the study were identified in the coding region of the p17 large subunit. The 2 inherited mutations of CASP10 gene reported in ALPS were found in the p17 large subunit and both mutants showed impaired caspase function,4 suggesting that the p17 large subunit is necessary for the enzymatic activity of caspase 10. Among the 11 tumors with the CASP10 gene mutation detected in the p17 large subunit, one nonsense mutation and 2 frame shift mutations were identified. These mutations are predicted to cause premature termination of protein synthesis, and hence resemble typical loss-of-function mutations. In case 35, C-to-T transition at bp 769 leads to a termination at codon 257, resulting in a protein that is truncated shortly after the DED-containing prodomain. This truncated mutant may be similar to FLICE-inhibitory protein (FLIP),23 which retains the ability to bind FADD but lacks the catalytic activity. Overexpression of this mutant confirmed that this mutant has a defect in induction of apoptosis (Figure 3C). We also found one mutation (case 104) in the p17 large protease domain, which is the same mutation as detected in the ALPS type II patient,4 suggesting that this mutation is functionally defective.
Caspase 10 is cleaved into the p17 large protease subunit and the p12 small protease subunit at highly conserved residue (Asp 415), and this cleavage is very important for the activation of this protein.17 We identified a missense mutation that would result in an amino acid change at residue 414 just next to the cleavage site. Apoptosis by this mutant was significantly lower than that by the wild-type caspase 10 (Figure 3C), but the cleavage of the mutant caspase 10 was not different from the cleavage of wild-type caspase 10 (data not shown), indicating that apoptotic defect of this mutant dose not result from the cleavage itself.
Of the 2 ALPS type II patients reported, one carried a heterozygous mutation of the CASP10 gene with dominant interference of death receptor–induced apoptosis.4 This mutant caspase 10 bound with caspase 8 and was corecruited with caspase 8 into the DISC.4 In the NHLs, 13 mutations of the CASP10gene showed evidence of retention of the wild-type allele (Table 2). Therefore, in these cases, it is possible that the heterozygously mutated caspase 10, which inefficiently autoprocesses, and normal caspase 10 are corecruited into the DISC together with caspase 8 and could therefore block the caspase activation in a dominant-negative fashion. In contrast, the other ALPS type II patient had inherited 2 identical defective alleles from unaffected heterozygous parents in a manner of a classic mendelian recessive trait.4 The 2CASP10 mutations in the NHL (cases 16 and 51) showed evidence of alterations of both alleles (Table 2). These biallelic inactivations of the CASP10 gene (a LOH and a mutation) may also lead to the production of mutant caspase 10 without any normal caspase 10, construct a nonfunctional DISC, and block the apoptosis signaling.
Regarding 1228G>A substitution of CASP10 gene, Wang and colleagues described that none of this substitution had been found on 440 normal alleles.4 However, Grønbaek and coworkers reported that in a normal Danish population they had found the 1228G>A substitution in the heterozygous constellation (6.8%).24In the current study, one NHL (case 106) was shown to have the 1228G>A substitution. The SSCP from the tumor DNA (case 106) showed mobility shift, whereas the SSCP from the corresponding normal DNA from the same slide section did not show any mobility shift (Figure 1A). Therefore, we concluded that the 1228G>A substitution in this study is not a polymorphism, but a mutation.
There is a DED-caspase gene cluster on human chromosome 2q33-34, which contains the genes for FLIP, CASP8, andCASP10, suggesting that all of these genes have arisen by tandem duplication.17,23 Such duplications may reflect the relative instability of the chromosomal region.25 The occurrences of the CASP10 gene mutations in the NHL in the present study and the caspase 8 mutation in the head and neck cancer by another group26 indicate that CASP8 andCASP10 may be candidate tumor suppressor genes at chromosome 2q33-34. In our preliminary study, we failed to find any mutation ofCASP8 gene in NHL (data not shown), suggesting thatCASP10, not CASP8, may be a tumor suppressor gene in the locus.
Several lines of evidence suggest that the loss of apoptosis function can enhance lymphoid tumor development. For example, lymphomatogenesis driven by the Eμ-myc transgene was shown to be markedly accelerated in lpr mice compared to wild-type mice, confirming a causal, rather than correlative, role for apoptosis loss in tumor development.27 This is well correlated with the fact that patients with ALPS type I and II have shown phenotypical abnormalities mainly in the lymphoid system.4,14,15,19 20In this study, we have shown another suggestion, namely, that the possible apoptosis dysregulation due to the CASP10 mutation might be involved in lymphomatogenesis.
Supported by grant no. R01-1999-00093 from the Korea Science and Engineering Foundation.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Sug Hyung Lee, Department of Pathology, College of Medicine, The Catholic University of Korea, 505 Banpo-dong, Socho-gu, Seoul 137-701, Korea; e-mail: suhulee@cmc.cuk.ac.kr.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal