Abstract
The inv(16) cytogenetic subtype of acute myeloid leukemia (AML) has a relatively good prognosis. Many patients achieve complete remission (CR). The prognostic uncertainty of negative qualitative reverse transcription–polymerase chain reaction (RT-PCR) assays suggests the need to identify prognostically significant critical thresholds by real-time RT-PCR. A reliable and sensitive (10−5) real-time RT-PCR assay was set up for the evaluation of relevant CBFβ-MYH11/ABL transcript ratios and was applied to the 21 patients with inv(16) AML routinely referred for cytogenetic and molecular monitoring in Seràgnoli Institute (Bologna, Italy) since 1990. Among the 18 patients who underwent ablative chemotherapy, all achieved CR with a 3-year disease-free survival probability of 63% (95% CI, 40%-87%) and no recorded events after 26 months. Five patients had relapses; 2 died of disease and 3 entered second CR. Analysis of the 125 bone marrow (or peripheral blood) samples studied by real-time RT-PCR showed that transcript ratios of samples taken during CR at any time before a relapse were always greater than 0.12%, whereas those of samples taken during first or second CR from patients who did not subsequently have relapses were always less than 0.25%. This suggests that transcript ratios greater than 0.25% may correspond to high risk for relapse, whereas ratios below 0.12% might indicate the patient is in a curable state. If confirmed, such thresholds could open the way to a new phase in post-CR therapeutic decision making for patients with inv(16) AML.
Introduction
Inversion of chromosome 16, inv(16)(p13q22), and its related translocation, t(16;16)(p13;q22), are recurrent rearrangements found in a subset of patients with acute myeloid leukemia (AML), particularly the M4Eo subtype.1-4 Inv(16) is associated with relatively good long-term, disease-free survival (DFS).5,6 It generates a chimeric mRNA transcript, CBFβ-MYH11, by fusing the CBFβ gene located on band q22 of chromosome 167,8 and the MYH11 gene, which encodes a heavy chain of the smooth-muscle myosin protein, located at the p13 breakpoint.9 10
Messenger RNA analysis by reverse transcription–polymerase chain reaction (RT-PCR) can help define leukemic subsets and provide potentially valuable diagnostic and prognostic information.11 Qualitative RT-PCR reveals the presence of detectable levels of minimal residual disease (MRD) in patients in complete clinical remission (CR) after induction chemotherapy.12 Introduction of real-time RT-PCR13 should allow routine quantitation of the transcript. The 5′-3′ nuclease activity of the Taq polymerase cleaves an internal fluorogenic probe specific for the target sequence, causing the emission of a fluorescent signal that can be detected during amplification. This allows rapid quantitation of specific RT-PCR products with a dynamic detection range of more than 5 orders of magnitude. Several groups have used real-time RT-PCR with TaqMan technology to quantitate MRD with leukemia-specific chromosome aberrations such as t(9;22), t(l5;17), and t(8;21).14-16
Most patients with inv(16) AML treated in our institute achieved long-term CR.17,18 However, several groups have reported persistence of the chimeric transcript even after allogeneic bone marrow transplantation (BMT).12,19-21 These considerations prompted us to investigate whether it is possible to predict relapse among patients who have achieved CR, and under what conditions patients can be considered in a curable state. To do this, we tested the prognostic potential of qualitative and quantitative RT-PCR analysis of the CBFβ-MYH11 transcript among the inv(16) AML patients routinely referred for cytogenetic and molecular diagnosis and follow-up in our institute. We designed the primers and probe necessary to apply TaqMan real-time RT-PCR quantitation to the analysis of the CBFβ-MYH11 transcript. We used this method alongside the recently defined BIOMED-1 Concerted Action qualitative RT-PCR protocol22 to conduct a retrospective analysis of the available samples of these patients. Our results suggest the existence of thresholds above and below which patients generally have relapses or remain in CR, respectively.
Patients and methods
Patients and therapy
This study concerns the 21 patients with inv(16) AML routinely referred for cytogenetic and molecular (qualitative RT-PCR) analysis at the Seràgnoli Institute of Hematology (Bologna, Italy) since June 1990. Of these, 19 were consecutive patients treated at the Seràgnoli Institute, whereas the remaining 2 were treated at the Institute of Hematology, University of Udine (patient 15) and the Institute of Hematology, University of Verona (patient 17). All patients provided written, informed consent to participate in the study. Diagnosis was made according to the French-American-British classification.23 Clinical and cytogenetic data are summarized in Table 1. Eighteen patients (patients 1-18) underwent molecular and cytogenetic analysis at diagnosis and during follow-up after ablative induction and consolidation chemotherapy, including transplantation procedures. Three patients (patients 19-21), who were older than 60, were referred for nonablative chemotherapy and underwent molecular and cytogenetic analysis only at diagnosis. Samples of bone marrow (n = 183) and peripheral blood (n = 5) were collected for routine patient care, according to institutional guidelines. Criteria for CR included morphologically normal bone marrow with less than 5% blasts, neutrophil count less than 1 × 109/L, platelet count less than 100 × 109/L, normalization of karyotype, and normal physical findings for more than 2 months.
Clinical and karyotype characteristics of patients with inv(16)(p13q22) AML
| Patient . | Sex/age, y . | Therapy . | Type of transcript . | Clinical outcome after CT/clinical status . | Karyotype . |
|---|---|---|---|---|---|
| 1 | F/47 | ICE/NOVIA/FLAG | A | CR/relapse, died | 46,XX(6)/46,XX,inv(16)(p13q22)(6)46,XX,imv(16) (p13q22)(14)/46,XX,t(2;17)(q31;p13)inv(16)(p13q22)(1)* |
| 2 | F/51 | ICE/NOVIA/FLAG/ABMT | A | CR/AW | 46,XX(3)/46,XX,inv(16)(p13q22)(7) |
| 3 | M/34 | ICE/NOVIA/ABMT | A | CR/AW | 46,XY(1)/46,XY,del(7)(q22),inv(16)(p13q22)(13) |
| 4 | M/38 | ICE/NOVIA/BMT | A | CR/AW | 46,XY(2)/46,XY,inv(16)(p13q22)(13) |
| 5 | F/51 | ICE/NOVIA/FLANG/FLANG/BMT | A | CR/breast cancer, died | 46,XX(1)/46,XX,inv(16)(p13q22)(21) |
| 6 | F/58 | ICE/NOVIA/ABMT-PBSC/FLANG/FLANG | A | 2nd CR/AW | 46,XX(2)/46,XX,inv(16)(p13q22)(28) |
| 7 | M/60 | ICE/NOVIA/FLANG/ABMT-PBSC | A | CR/AW | 46,XY(2)/46,XY,inv(16)(p13q22)(19) |
| 8 | M/37 | ICE/NOVIA/BMT | A | CR/AW | 46,XY(3)/46,XY,inv(16)(p13q22)(16) |
| 9 | M/57 | ICE/NOVIA/ABMT | A | CR/AW | 47,XY,inv(16)(p13q22),+22(12)/ 47,XY,t(9;19)(q22;q13),inv(16)(p13q22),+22(8) |
| 10 | M/57 | MEC6/MEC4/ABMT/FLANG/FLANG | A | 2nd CR/AW | 46,XY(1)/46,XY,inv(16)(p13q22)(14) |
| 11 | M/35 | ICE/FLANG/FLANG/ABMT-PBSC | A | 2nd CR/AW | 46,XY(1)/47,XY,inv(16)(p13q22),+9(14) |
| 12 | F/60 | ICE/FLANG/FLANG | D | CR/AW | 46,XX(1)/47,XX,inv(16)(p13q22),+8(30) |
| 13 | M/49 | ICE/FLANG/FLANG/ABMT | A | CR/AW | 46,XY(1)/47,XY,inv(16)(p13q22),+8(28) |
| 14 | M/49 | ICE/FLAN/FLAN/ABMT | A | CR/AW | 46,XY(2)/46,XY,inv(16)(p13q22)(23) |
| 15 | F/27 | FLAI/FLAN/FLAN/ABMT | A | CR/died in 2nd CR | 47,XX,inv(16)(p13q22),+X(25) |
| 16 | F/26 | ICE/FLAN/FLAN/ABMT | E | CR/AW | 46,XX,inv(16)(p13q22)(30) |
| 17 | M/17 | ICE/FLAN/FLAN | A | CR/AW | 46,XY(1)/46,XY,inv(16)(p13q22)(28) |
| 18 | M/16 | ICE/FLAN/FLAN | C | CR/AW | 46,XY,inv(16)(p13q22)(30) |
| 19 | M/63 | Nonablative | A | — | 46,XY(3)/46,XY,inv(16)(p13q22)(23) |
| 20 | M/61 | Nonablative | A | — | 46,XY(2)/46,XY,inv(16)(p13q22)(28) |
| 21 | F/65 | Nonablative | A | — | 46,XX(1)/46,XX,inv(16)(p13q22)(19) |
| Median (range, y) | 49 (16-65) |
| Patient . | Sex/age, y . | Therapy . | Type of transcript . | Clinical outcome after CT/clinical status . | Karyotype . |
|---|---|---|---|---|---|
| 1 | F/47 | ICE/NOVIA/FLAG | A | CR/relapse, died | 46,XX(6)/46,XX,inv(16)(p13q22)(6)46,XX,imv(16) (p13q22)(14)/46,XX,t(2;17)(q31;p13)inv(16)(p13q22)(1)* |
| 2 | F/51 | ICE/NOVIA/FLAG/ABMT | A | CR/AW | 46,XX(3)/46,XX,inv(16)(p13q22)(7) |
| 3 | M/34 | ICE/NOVIA/ABMT | A | CR/AW | 46,XY(1)/46,XY,del(7)(q22),inv(16)(p13q22)(13) |
| 4 | M/38 | ICE/NOVIA/BMT | A | CR/AW | 46,XY(2)/46,XY,inv(16)(p13q22)(13) |
| 5 | F/51 | ICE/NOVIA/FLANG/FLANG/BMT | A | CR/breast cancer, died | 46,XX(1)/46,XX,inv(16)(p13q22)(21) |
| 6 | F/58 | ICE/NOVIA/ABMT-PBSC/FLANG/FLANG | A | 2nd CR/AW | 46,XX(2)/46,XX,inv(16)(p13q22)(28) |
| 7 | M/60 | ICE/NOVIA/FLANG/ABMT-PBSC | A | CR/AW | 46,XY(2)/46,XY,inv(16)(p13q22)(19) |
| 8 | M/37 | ICE/NOVIA/BMT | A | CR/AW | 46,XY(3)/46,XY,inv(16)(p13q22)(16) |
| 9 | M/57 | ICE/NOVIA/ABMT | A | CR/AW | 47,XY,inv(16)(p13q22),+22(12)/ 47,XY,t(9;19)(q22;q13),inv(16)(p13q22),+22(8) |
| 10 | M/57 | MEC6/MEC4/ABMT/FLANG/FLANG | A | 2nd CR/AW | 46,XY(1)/46,XY,inv(16)(p13q22)(14) |
| 11 | M/35 | ICE/FLANG/FLANG/ABMT-PBSC | A | 2nd CR/AW | 46,XY(1)/47,XY,inv(16)(p13q22),+9(14) |
| 12 | F/60 | ICE/FLANG/FLANG | D | CR/AW | 46,XX(1)/47,XX,inv(16)(p13q22),+8(30) |
| 13 | M/49 | ICE/FLANG/FLANG/ABMT | A | CR/AW | 46,XY(1)/47,XY,inv(16)(p13q22),+8(28) |
| 14 | M/49 | ICE/FLAN/FLAN/ABMT | A | CR/AW | 46,XY(2)/46,XY,inv(16)(p13q22)(23) |
| 15 | F/27 | FLAI/FLAN/FLAN/ABMT | A | CR/died in 2nd CR | 47,XX,inv(16)(p13q22),+X(25) |
| 16 | F/26 | ICE/FLAN/FLAN/ABMT | E | CR/AW | 46,XX,inv(16)(p13q22)(30) |
| 17 | M/17 | ICE/FLAN/FLAN | A | CR/AW | 46,XY(1)/46,XY,inv(16)(p13q22)(28) |
| 18 | M/16 | ICE/FLAN/FLAN | C | CR/AW | 46,XY,inv(16)(p13q22)(30) |
| 19 | M/63 | Nonablative | A | — | 46,XY(3)/46,XY,inv(16)(p13q22)(23) |
| 20 | M/61 | Nonablative | A | — | 46,XY(2)/46,XY,inv(16)(p13q22)(28) |
| 21 | F/65 | Nonablative | A | — | 46,XX(1)/46,XX,inv(16)(p13q22)(19) |
| Median (range, y) | 49 (16-65) |
ICE (induction chemotherapy): idarubicin (endovenously infused, 10 mg/m2 per day) for days 1, 3, and 5, cytarabine (100 mg/m2 per day) continuously infused for 10 days, and etoposide (VePesid; 100 mg/m2 per day) for 5 days.29,30 NOVIA (consolidation chemotherapy: cytarabine (500 mg/m2 per day) for 6 days (edovenously infused twice a day for 4 hours) plus mithoxantrone (Novantrone; 12 mg/m2 per day) for 3 days (days 4, 5, 6).32FLAG: 5 days of treatment with a 30-minute infusion of fludarabine (30 mg/m2 per day) followed 4 hours later by a 4-hour infusion of cytarabine (2 g/m2 per day). G-CSF (300 μg/day) was administered subcutaneously before starting fludarabine, continued for 5 days and then given again 1 week after the end of therapy until complete neutrophil recovery.19-21 FLANG: 3 days of fludarabine (30 mg/m2 per day) as above, followed 4 hours later by a 2-hour infusion of cytarabine (1 g/m2 per day) and a 30-minute infusion of mithoxantrone (Novantrone; 10 mg/m2 per day). Twelve hours before fludarabine infusion, G-CSF (300 μg/day) was administered subcutaneously. This was continued for 3 days and then was restarted as in the FLAG regimen.33,34 MEC6 chemotherapy: 6 days of etoposide (VePesid; 100 mg/m2 per day), 30-minute infusion of mithoxantrone (Novantrone; 10 mg/m2 per day), and continuous infusion of cytarabine (100 mg/m2 per day), followed by consolidation chemotherapy (MEC4 protocol). MEC4: 4 days of etoposide (VePesid; 100 mg/m2 per day), 30-minute infusion of mithoxantrone (Novantrone; 10 mg/m2 per day),31 and continuous infusion of cytarabine (100 mg/m2 per day).
Karyotype at relapse. CR indicates complete remission; AW, alive and well.
Cytogenetic analysis
Cytogenetic analysis was routinely performed using a standard technique with Wright stain banding.24 At least 20 mitoses were analyzed for each sample.
Samples and RNA isolation
Mononuclear cells from samples were obtained by Ficoll-Hypaque density gradient centrifugation and were stored at −80°C in guanidinium thiocyanate. Total cellular RNA was extracted as previously described.18
Qualitative RT-PCR analysis
RT-PCR.
RT-PCR of ABL control gene.
To assess the quality and quantity of the amplifiable RNA isolated from samples, RT-PCR of ABL gene transcripts was performed as previously described.18
Criteria for qualitative RT-PCR negativity.
Stringent criteria for negativity were applied: no amplification of the CBFβ-MYH11 transcript in 3 independent assays at a sensitivity of at least 1:104 (see below), always accompanied by successful amplification of the ABL transcript. To determine the level of sensitivity of breakpoint sequence amplification, experiments using CBFβ-MYH11-positive RNA (types A, D, E, and C) were conducted by mixing serially diluted total RNA isolated from patients 7, 12, 16, and 18 with the HL60 cell line.
Reproducibility and accuracy of assays.
Positive and negative controls were included in all assays. In particular, RNA from the time of diagnosis was used for positive control. Negative controls included reactions with no RNA or no complementary DNA (cDNA) or HL60 cell line. Precautions taken to avoid contamination included the use of a specifically designed UV flow cabinet and PCR-designated pipettes with filtered tips. All tests were conducted twice to confirm the results.
Real-time RT-PCR
Primers and probe.
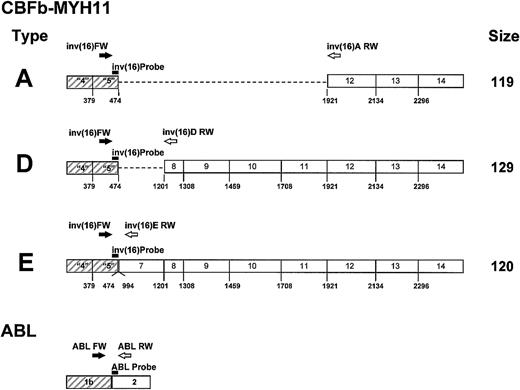
Figure 1 reports the primers and probes used for real-time RT-PCR quantification of the CBFβ-MYH11fusion gene and the ABL gene. Probes were labeled by a 5′ FAM reporter and a 3′ TAMRA quencher. Primers and TaqMan probes were all designed using Primer Express software (Perkin Elmer, Applied Biosystem, Foster City, CA).
Position of primers and probe for CBFβ-MYH11 transcript types A, D, and E, and for the
ABL gene. CBFβ-MYH11 transcript types A, D, and E: the probe (▬) inv(16) Probe (5′ TTC AAA TTC GCG TGT CCT TCT CCG A) and the sense primer (➡) inv(16) FW (5′ TTA GCA CAA CAG GCC TTT GAA) are positioned in exon 5 of the CBFβ gene. The reverse primers ( )inv(16)A RW (5′ CAG GGC CCG CTT GGA), inv(16)D RW(5′ CCT CGG CCT CGT TAA GCA T), and inv(16)E RW (5′ GCG TCT GCT TAT TCT TGT CTA GGT T) are positioned in exons 12, 8, and 7 of theMYH11 gene for transcript types A, D, and E, respectively.ABL gene: the probe (▬) ABL Probe (5′ CCA GTA GCA TCT GAC TTT GAG CCT CAG GG) and the reverse primer (
)inv(16)A RW (5′ CAG GGC CCG CTT GGA), inv(16)D RW(5′ CCT CGG CCT CGT TAA GCA T), and inv(16)E RW (5′ GCG TCT GCT TAT TCT TGT CTA GGT T) are positioned in exons 12, 8, and 7 of theMYH11 gene for transcript types A, D, and E, respectively.ABL gene: the probe (▬) ABL Probe (5′ CCA GTA GCA TCT GAC TTT GAG CCT CAG GG) and the reverse primer ( ) ABL RW (5′ TGG GTC CAG CGA GAA GGT T) are positioned in exon 2 of the ABL gene. The sense primer (➡)ABL FW (5′ TCC TCC AGC TGT TAT CTG GAA GA) is positioned in exon 1b of the ABL gene.
) ABL RW (5′ TGG GTC CAG CGA GAA GGT T) are positioned in exon 2 of the ABL gene. The sense primer (➡)ABL FW (5′ TCC TCC AGC TGT TAT CTG GAA GA) is positioned in exon 1b of the ABL gene.
Position of primers and probe for CBFβ-MYH11 transcript types A, D, and E, and for the
ABL gene. CBFβ-MYH11 transcript types A, D, and E: the probe (▬) inv(16) Probe (5′ TTC AAA TTC GCG TGT CCT TCT CCG A) and the sense primer (➡) inv(16) FW (5′ TTA GCA CAA CAG GCC TTT GAA) are positioned in exon 5 of the CBFβ gene. The reverse primers ( )inv(16)A RW (5′ CAG GGC CCG CTT GGA), inv(16)D RW(5′ CCT CGG CCT CGT TAA GCA T), and inv(16)E RW (5′ GCG TCT GCT TAT TCT TGT CTA GGT T) are positioned in exons 12, 8, and 7 of theMYH11 gene for transcript types A, D, and E, respectively.ABL gene: the probe (▬) ABL Probe (5′ CCA GTA GCA TCT GAC TTT GAG CCT CAG GG) and the reverse primer (
)inv(16)A RW (5′ CAG GGC CCG CTT GGA), inv(16)D RW(5′ CCT CGG CCT CGT TAA GCA T), and inv(16)E RW (5′ GCG TCT GCT TAT TCT TGT CTA GGT T) are positioned in exons 12, 8, and 7 of theMYH11 gene for transcript types A, D, and E, respectively.ABL gene: the probe (▬) ABL Probe (5′ CCA GTA GCA TCT GAC TTT GAG CCT CAG GG) and the reverse primer ( ) ABL RW (5′ TGG GTC CAG CGA GAA GGT T) are positioned in exon 2 of the ABL gene. The sense primer (➡)ABL FW (5′ TCC TCC AGC TGT TAT CTG GAA GA) is positioned in exon 1b of the ABL gene.
) ABL RW (5′ TGG GTC CAG CGA GAA GGT T) are positioned in exon 2 of the ABL gene. The sense primer (➡)ABL FW (5′ TCC TCC AGC TGT TAT CTG GAA GA) is positioned in exon 1b of the ABL gene.
PCR conditions.
Reaction mixtures of 25 μL contained 12.5 μL TaqMan buffer A with the ROX dye as the passive reference, 5 mM MgCl2, 200 μM dATP, dCTP, dGTP, 400 μM dUTP, 1.25 U AmpliTaq Gold DNA polymerase, 0.5 U AmpErase uracil N-glycosylase (UNG), 300 nM forward and reverse primers, 200 nM specific TaqMan probe, and 6 μL plasmid or cDNA (diluted 1:3). All reagents were from Perkin Elmer, Applied Biosystem.
All real-time RT-PCR experiments were performed at least in triplicate. Before determining the sensitivity of the target, the real-time RT-PCR set-up was optimized. In particular, the amounts of forward and reverse primer were determined so as to produce the highest ΔRn and the lowest threshold cycle (CT). In the primer–matrix experiment, 9 combinations of 50, 300, and 900 mM for each primer were tested in triplicate: 50/50, 50/300, 50/900; 300/50, 300/300, 300/900; 900/50, 900/300, 900/900. RT-PCR reactions were set in MicroAmp optical 96-well reaction plates closed with MicroAmp optical caps (Perkin Elmer/Applied Biosystem). After 2 minutes at 50°C to allow the destruction by UNG of potential contaminant RT-PCR products and 10 minutes at 95°C to denature UNG and activate AmpliTaq Gold, the amplification was carried out by 50 cycles at 95°C for 15 seconds and 65°C for 60 seconds in the ABI/Prism 7700 Sequence Detector System (Applied Biosystem, Perkin Elmer).
Construction of the standard curves forCBFβ-MYH11 fusion gene and ABL gene.
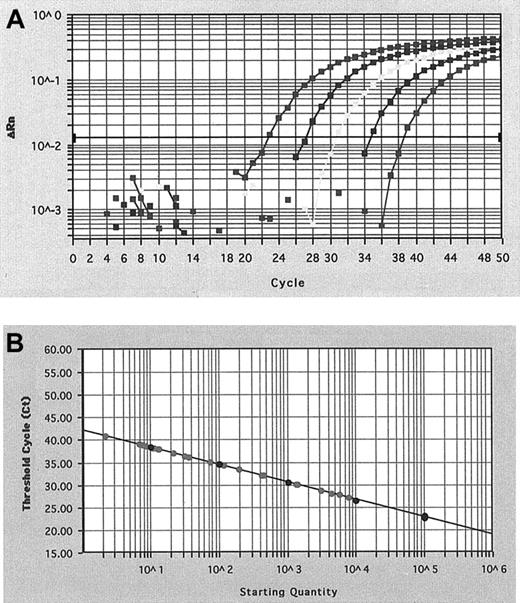
We cloned the RT-PCR products obtained by amplification with the primers listed in Figure 1, derived at diagnosis from patients 7, 12, and 16 and corresponding to CBFβ-MYH11 types A, D, and E, respectively, and the ABL amplification product. The 4 products were cloned to the pCR II-TOPO vector using the TOPO TA cloning kit (Invitrogen, Carlsbad, CA). We called the resultant plasmids pTA-A, pTA-D, pTA-E, and pTA-ABL, respectively. To construct the standard curve for the quantification of each type of CBFβ-MYH11 and ABL transcript, serial dilutions of the corresponding plasmid were used. For each assay, 10-fold dilutions, starting at 105plasmid copies, were analyzed in triplicate using Sequence Detector System software V1.6 (Perkin Elmer, Applied Biosystem) (Figure2A). A standard curve was established by plotting the CT and the known copy number on a logarithmic scale (Figure 2B).
CBFβ-MYHII amplification plot and standard curve.
(A) Amplification plot of a serial dilution of the CBFβ-MYH11 plasmid and (B) standard curve of CBFβ-MYH11 cDNA plasmid dilution for real-time RT-PCR. Amplification plot shows the increase of reporter fluorescence during amplification and CT. The CT value decrease was proportional to the increase of the target molecules. The standard curve shows the linear correlation between the CT value and the initial copy number of the CBFβ-MYH11 and can then be used to calculate the absolute CBFβ-MYH11 fusion transcript copy number in an unknown patient sample.
CBFβ-MYHII amplification plot and standard curve.
(A) Amplification plot of a serial dilution of the CBFβ-MYH11 plasmid and (B) standard curve of CBFβ-MYH11 cDNA plasmid dilution for real-time RT-PCR. Amplification plot shows the increase of reporter fluorescence during amplification and CT. The CT value decrease was proportional to the increase of the target molecules. The standard curve shows the linear correlation between the CT value and the initial copy number of the CBFβ-MYH11 and can then be used to calculate the absolute CBFβ-MYH11 fusion transcript copy number in an unknown patient sample.
Statistical analysis
Overall survival (OS) and DFS probability were calculated from the date of diagnosis to the last contact or death, and to death or relapse, respectively, according to the Kaplan-Meier method.25 Comparisons of the CBFβ-MYH11/ABL ratios at diagnosis and relapse and during treatment and CR were performed by the Kruskal-Wallis test. χ2 analysis was used for binary variables. All analyses were performed using the SPSS software package (SPSS, Chicago, IL).
Results
Clinical follow-up, survival, and status
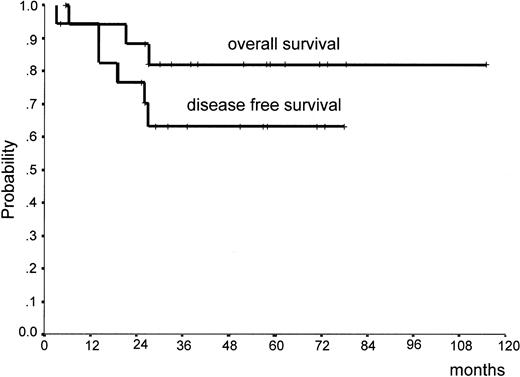
Figure 3 reports the clinical outcomes and OS and DFS of the 18 patients who underwent ablative chemotherapy. At a median follow-up of 51 months (range, 4-115 months), 13 (72%) patients are in first CR and 3 (17%) are in second CR. Thirteen (72%) patients achieved qualitative RT-PCR negativity after induction chemotherapy, and 3 (17%) achieved it after consolidation chemotherapy. Figure 3 illustrates that the OS and DSF probabilities at 3 years were 82% (95% confidence interval [CI], 63%-100%) and 63% (95% CI, 40%-87%), respectively. In either case, no events were recorded after the 26th month.
Overall survival and disease-free survival of 18 patients with inv(16) AML.
Cytogenetic analysis
Cytogenetic results are summarized in Figure4A. All 21 patients showed inv(16)(p13q22) at diagnosis, and 5 also had additional karyotypic abnormalities (Table 1). In one patient (patient 1), an additional translocation was detected at relapse [46,XX, inv(16)(p13q22), t(2;17) (q31;p13)].
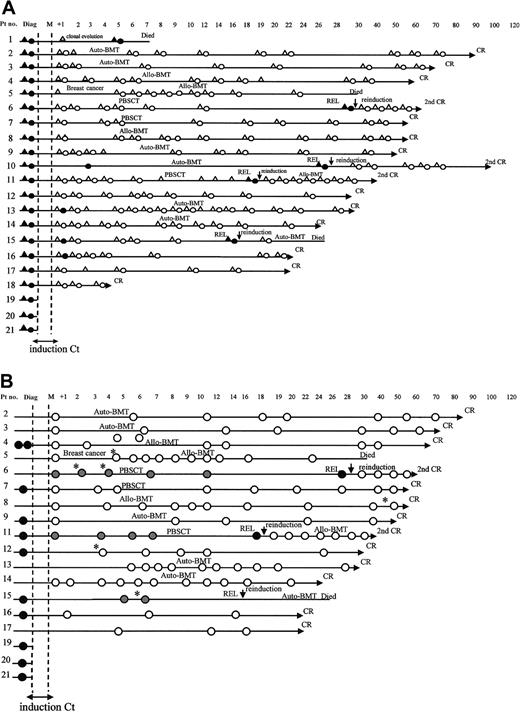
Schematic representations of karyotypic and qualitative RT-PCR and real-time RT-PCR follow-up of patients with inv(16) AML.
(A) Karyotypic and qualitative RT-PCR follow-up of 21 patients: karyotypic analysis negative (▵) or positive (▴) for inv(16); molecular analysis negative (○) or positive (●) for CBFβ-MYH11. (B) Real-time RT-PCR of 18 patients (patients 1, 10, 18 had insufficient material for analysis): samples taken at diagnosis or at the moment or relapse (●); samples taken at any time during or after treatment before relapse ( ); samples taken during follow-up when no subsequent relapse was recorded (○). *Samples that fell in the gray zone with CBFβ-MYH11/ABL ratios between 0.12% and 0.25%. Auto-BMT indicates autologous bone marrow transplantation; allo-BMT, allogeneic bone marrow transplantation; and PBSCT, autologous transplantation with peripheral blood stem cells.
); samples taken during follow-up when no subsequent relapse was recorded (○). *Samples that fell in the gray zone with CBFβ-MYH11/ABL ratios between 0.12% and 0.25%. Auto-BMT indicates autologous bone marrow transplantation; allo-BMT, allogeneic bone marrow transplantation; and PBSCT, autologous transplantation with peripheral blood stem cells.
Schematic representations of karyotypic and qualitative RT-PCR and real-time RT-PCR follow-up of patients with inv(16) AML.
(A) Karyotypic and qualitative RT-PCR follow-up of 21 patients: karyotypic analysis negative (▵) or positive (▴) for inv(16); molecular analysis negative (○) or positive (●) for CBFβ-MYH11. (B) Real-time RT-PCR of 18 patients (patients 1, 10, 18 had insufficient material for analysis): samples taken at diagnosis or at the moment or relapse (●); samples taken at any time during or after treatment before relapse ( ); samples taken during follow-up when no subsequent relapse was recorded (○). *Samples that fell in the gray zone with CBFβ-MYH11/ABL ratios between 0.12% and 0.25%. Auto-BMT indicates autologous bone marrow transplantation; allo-BMT, allogeneic bone marrow transplantation; and PBSCT, autologous transplantation with peripheral blood stem cells.
); samples taken during follow-up when no subsequent relapse was recorded (○). *Samples that fell in the gray zone with CBFβ-MYH11/ABL ratios between 0.12% and 0.25%. Auto-BMT indicates autologous bone marrow transplantation; allo-BMT, allogeneic bone marrow transplantation; and PBSCT, autologous transplantation with peripheral blood stem cells.
Qualitative RT-PCR analysis
Qualitative RT-PCR results are summarized in Figure 4A. Assays of all 188 samples from the 21 patients were routinely studied. Of these, 172 samples were also fully studied according to the BIOMED-1 Concerted Action protocol22; the 16 samples that turned out to be ABL-negative were excluded from subsequent analysis. Log sensitivities of the transcript primer sets were 10−4 for types A, C, D, and E.
At diagnosis, 18 patients displayed type A transcript, one had type D, one had type E, and one had a type C transcript (GenBank accession numbers AF249898, AF249897, AF251768, respectively).26 At follow-up, 9 of 149 samples taken from the 18 patients who received ablative chemotherapy were positive for CBFβ-MYH11 according to the BIOMED-1 Concerted Action protocol. Fifteen of 18 patients became negative after induction chemotherapy. The remaining 3 patients (patients 13, 15, 16) became negative after the first consolidation course. One patient (patient 10), who was found to be positive 3 months after completing consolidation chemotherapy, had insufficient available samples for molecular follow-up before relapse.
Application of real-time RT-PCR for CBFβ-MYH11 analysis
For 3 transcript types (A, D, E) and the ABL gene, it was possible to develop primer–probe combinations that permitted amplification and real-time RT-PCR analysis. In primer–matrix experiments, different ΔRn values were found for each primer combination. CT values were comparable. The 300/300 mM primer combination was used in all experiments. As can be seen from Figure 1, for the 3 types of transcript, the TaqMan probe and the forward primer were located in exon 5 of the CBFβ gene. Reverse primers were located in exons 12, 8, and 7 of theMYH11 gene for transcript types A, D, and E, respectively. The number of target molecules of transcript in each sample was expressed as a percentage ratio between CBFβ-MYH11 and ABL in 6 μL cDNA. Three patients (patients 19-21) were tested at diagnosis only. For 16 patients, transcript levels were quantified in sequential bone marrow or peripheral blood samples, but for 2 patients (patients 1 and 10), insufficient amounts of material precluded transcript quantification at diagnosis and during follow-up.
Sensitivity of real-time RT-PCR for CBFβ-MYH11.
Sensitivity was tested at the cellular level in HL60 with serial dilutions of diagnostic samples from patients 7, 12, and 16. This method allowed quantify the amount of CBFβ-MYH11 transcript up to a 1:105 dilution.
Reliability of real-time RT-PCR for CBFβ-MYH11 and ABL.
Analysis of 20 identical samples of type A CBFβ-MYH11 in a single run (intra-assay comparison) revealed a coefficient of variation (CV; calculated for the determined concentrations) of 0.07. Respective cycle threshold crossing points gave a CV of 0.004. Analysis of 2 identical samples of 105 molecules of pTA-ABL plasmid and pTA-A plasmid, respectively, in 10 runs on 10 different days (interassay comparison, day-to-day variation with new mixtures of reagents) resulted in CVs of 0.06 and 0.08, respectively, and cycle threshold crossing points with CVs of 0.038 and 0.097, respectively.
Real-time RT-PCR quantification of CBFβ-MYH11 and ABL.
One hundred twenty-five samples were retrospectively studied by real-time RT-PCR (the other 47 were insufficient for study). ABL quantification was possible in all samples, with amounts of transcript ranging from 38 to 93 544 (median, 4217). Regarding CBFβ-MYH11, 64 samples appeared positive, with amounts of transcript ranging from 10 to 9309 (median, 20). The CBFβ-MYH11/ABL ratio ranged from 0% to 60% (median, 0.01%). Forty-eight of the samples positive at real-time RT-PCR had been negative at qualitative RT-PCR using the BIOMED-1 Concerted Action protocol. χ2 analysis revealed that real-time RT-PCR was significantly more sensitive (P < .001).
Significance of CBFβ-MYH11/ABL ratios evaluated by real-time RT-PCR
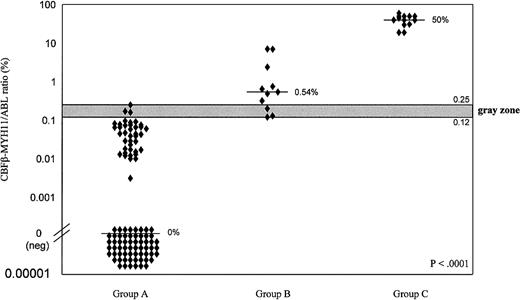
We retrospectively analyzed CBFβ-MYH11/ABL ratios in terms of samples obtained at any time during or after treatment in the absence of subsequent relapse (group A), those obtained at any time during follow-up before relapse (group B), and those obtained at the time of diagnosis or relapse (group C). Results are summarized in Figures 4B and 5. The Kruskal-Wallis test showed that the differences among the 3 groups were all highly significant (P < .0001). At diagnosis or relapse (group C), the CBFβ-MYH11/ABL ratio ranged from 19% to 60%. Ratios (n = 101) obtained during or after treatment from patients who did not have relapses (group A) were always 0.25% or less (minimum, 0%). By contrast, ratios obtained during or after treatment from patients who did have relapses (group B) were always 0.12% or more (maximum, 7.1%). Figure 5 illustrates these 2 thresholds and the overlapping gray zone. Among the 6 assays that fell within the gray zone, 3 refer to 2 patients (patients 6, 15) who had relapses and 3 to 3 patients (patients 5, 8, 12) who remained in CR (Figure 4B).
CBFβ-MYH11/ABL ratios, as evaluated by real-time RT-PCR.
One hundred twenty-five samples were taken from 21 patients with inv(16) AML during or after treatment in the absence of subsequent relapse (Group A), at any time during follow-up before relapse (Group B), or at the time of diagnosis or relapse (Group C). Differences among the 3 groups were all highly significant (P < .0001), as determined by the Kruskal-Wallis test. When values were lower than 0.12%, no subsequent relapse was recorded, but when values were greater than 0.25%, relapse always occurred. Values fell within the intermediate gray zone for 6 samples. Median values of each group (—).
CBFβ-MYH11/ABL ratios, as evaluated by real-time RT-PCR.
One hundred twenty-five samples were taken from 21 patients with inv(16) AML during or after treatment in the absence of subsequent relapse (Group A), at any time during follow-up before relapse (Group B), or at the time of diagnosis or relapse (Group C). Differences among the 3 groups were all highly significant (P < .0001), as determined by the Kruskal-Wallis test. When values were lower than 0.12%, no subsequent relapse was recorded, but when values were greater than 0.25%, relapse always occurred. Values fell within the intermediate gray zone for 6 samples. Median values of each group (—).
Discussion
Among the distinct subset of AML patients carrying inv(16)(p13q22), a high percentage achieves CR.12,17 18 It is thought that some of these patients may be considered cured. This report on a large series of patients with inv(16) AML examines what, to our knowledge, is the longest clinical, cytogenetic, and molecular follow-up thus far published. Our results suggest that the high proportion of patients who remain in CR for as long as 2 years (89% in our series) seems to have a very good prognosis because none, at the time of writing, have had relapses. To search for prognostic indications at a molecular level, we developed primer–probe combinations that permitted effective real-time RT-PCR quantification of CBFβ-MHY11 transcripts. Our results provide the first evidence that cutoff levels of transcripts probably do exist, below which or above which cure or relapse, respectively, is likely.
The clinical results from our series are fully in line with the concept that long-term CR can be achieved in a high proportion of patients with inv(16) AML. At a median follow-up of 51 months (range, 4-115 months), 16 of 18 (89%) of the patients are stable in either first (13 patients) or second (3 patients) CR, with a DFS probability of 63% at 3 years. No relapses or deaths have been observed after the 26th month. These encouraging data highlight the need to assess whether any or all patients who achieve durable CR can be considered cured.
At qualitative RT-PCR, most of our patients achieved molecular remission, defined as undetectability of the neoplastic transcript at the sensitivity threshold level (1:104) permitted by the BIOMED-1 Concerted Action protocol.22 Probably because of different sensitivity methods applied,12,19-21 some authors reported that most patients with AML inv(16) show persistence of minimum residual disease after CR. Using BIOMED-1 criteria, 72% of our patients achieved qualitative molecular remission after induction chemotherapy, and 17% achieved it after consolidation chemotherapy. The ultimate prognostic impact of qualitative RT-PCR negativity is uncertain.12 Our series confirmed that patients who achieve this status are still susceptible to eventual relapse. Indeed, all 3 of our patients who had relapses after receiving ablative chemotherapy had had at least one negative qualitative RT-PCR assay. Like other authors,27 we found that qualitative RT-PCR did not allow cutoff levels to be identified for prognostic purposes.
Using TaqMan technology, we therefore set up a quantitative real-time RT-PCR assay. To our knowledge, this is the first time that real-time RT-PCR has been applied to the analysis of inv16. Our data demonstrate that the method used is reliable and consistently more sensitive than qualitative RT-PCR (1:105 vs 1:104). Indeed, a high proportion of the samples in which the CBFβ-MYH11 transcript was detectable at real-time RT-PCR were negative at qualitative RT-PCR (P < .0001). Inv(16) could be a particularly promising testing ground for the clinical possibilities of real-time RT-PCR because it has a better prognosis than other subtypes of AML, and identification of patients at different risk levels for relapse could be of major clinical relevance.
When we analyzed the CBFβ-MYH11/ABL ratios among the 125 samples on which real-time RT-PCR was performed, we found that samples taken at diagnosis and relapse gave similar values (median, 50%). Furthermore, the samples taken during CR at any time before relapse always had transcript ratios greater than 0.12%. On the other hand, samples taken from patients in first or second CR who did not have subsequent relapses always had transcript ratios less than 0.25%. This suggests the existence of a cutoff point at approximately 0.12%, beneath which durable remission is likely, and of another at 0.25%, above which relapse is probable. Only 6 assays fell within the intermediate gray zone (Figure 4B). Longer follow-up should help further define the long-term predictive value of the cutoff points. If, as seems probable, peripheral blood samples are thought to be adequate for routine real-time RT-PCR analysis,28 regular study at set time points would not place a major burden on patients.
We also found that some samples were negative at qualitative and quantitative RT-PCR. This finding may shed some light12 on the prognostic significance of an undetectable CBFβ-MYH11 transcript in inv(16) AML patients in CR. Considering that none of the patients who reached this status had subsequent relapses, the achievement of real-time RT-PCR negativity may be predictive of eventual cure.
In conclusion, our series suggests that most patients with inv(16) AML can achieve durable CR after ablative chemotherapy and that many achieve negative assays at qualitative RT-PCR. The real-time RT-PCR assay set up by us can be recommended as a reliable and more sensitive method for routine monitoring of minimum residual disease in such patients. Our data suggest that samples with CBFβ-MYH11/ABL ratios above 0.25% correspond to a high risk for relapse. In contrast, in the presence of ratios lower than 0.12%, relapse seems more unlikely and patients might be considered to be in a curable state. It is possible that complete negativity at real-time RT-PCR could be associated with a cure from this AML subtype. More generally, our findings suggest that in inv(16) AML, real-time RT-PCR could provide indications regarding the patient's clinical state similar to those that can apparently be obtained (through CBFα) in t(8;21) AML, where high- and low-risk thresholds have already been reported.28
We thank SANCO (European Concerted Action Project), Dr Clara Bloomfield, and A. K. Burnett for their valuable suggestions. We thank Dr Achille Ambrosetti and Dr Domenico Russo of the Institutes of Hematology of the Universities of Verona and Udine, respectively, for providing the material for the molecular analysis of 2 patients. We also thank Barbara Giannini, Simona Soverini, and Maria Stella Zagarella for expert technical assistance, and Robin M. T. Cooke for helping work up the manuscript.
Supported by Associazione Italiana per la Ricerca sul Cancro, MURST 40% (Sante Tura) (AML), Associazione italiana contro le leucemie, and the Italian Consiglio nazionale delle ricerche target.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Giovanni Martinelli, Institute of Hematology and Medical Oncology L. A. Seràgnoli, University of Bologna, H. S. Orsola-Malpighi, Via Massarenti 9-40138 Bologna, Italy; e-mail: gmartino@kaiser.alma.unibo.it.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal