Abstract
Angioimmunoblastic T-cell lymphoma (AITL) is a systemic disease involving lymph nodes, spleen, and bone marrow. Although the histologic features have been well described, the diagnosis is often challenging, as there are no specific phenotypic or molecular markers available. This study shows that the neoplastic cells of AITL can be identified by aberrant CD10 expression. Archival material from 30 cases of AITL, 10 cases of peripheral T-cell lymphoma unspecified (PTL), and 10 cases of reactive lymphoid hyperplasia were reviewed. Single and double immunostaining for CD3, CD4, CD8, CD20, CD21, CD10, BCL6, Ki67, and LMP-1 in situ hybridization for Epstein-Barr early region and polymerase chain reaction (PCR) for T-cell receptor gamma chain gene and immunoglobulin heavy chain gene were performed. Three overlapping histologic patterns with hyperplastic follicles, depleted follicles, or without follicles were identified in AITL. Of the 30 cases of AITL, 27 contained CD10+ T cells. No CD10+ T cells were present in the cases of PTL or reactive hyperplasia. PCR confirmed a monoclonal or oligoclonal T-cell population in 29 of 30 cases of AITL and a monoclonal B-cell population in 6 cases. Analysis of microdissected CD10+ single cells showed that they belonged to the neoplastic clone. In conclusion CD10 is a phenotypic marker that specifically identifies the tumor cells in 90% of AITL, including the early cases. The presence of these cells distinguishes AITL from other PTLs. This finding provides an objective criterion for accurate and early diagnosis of AITL.
Introduction
Angioimmunoblastic T-cell lymphoma (AITL) is a systemic disease involving lymph nodes, spleen, and bone marrow.1,2 Histologically, the lymph nodes show partial or total obliteration of the normal architecture by a polymorphic infiltrate of lymphocytes, plasma cells, and eosinophils and proliferation of venules and follicular dendritic cells (FDCs).3-5 In many instances cytologic features of malignancy are not readily identifiable. For this reason the disease was first described as an atypical reactive process, angioimmunoblastic lymphadenopathy.1 However, cytogenetic and molecular studies have consistently shown clonal chromosomal abnormalities6 and monoclonal or oligoclonal T-cell populations in most cases, which strongly supports a T-cell neoplasm.7,8 Some cases also contain oligoclonal or monoclonal B-cell populations7,8 that are attributed to the expansion of B cells infected with Epstein-Barr virus (EBV),9 possibly secondary to the immunodeficiency associated with the T-cell lymphoma. Whether AITL is a neoplastic process from the inception or whether it is preceded by a polyclonal or oligoclonal reactive state, angioimmunoblastic lymphadenopathy is a matter of debate.4,10 11
The outcome for patients with AITL remains dismal with a median survival of less than 3 years.12-14 There are 2 major reasons for difficulties in dealing with this disease. First, although the histologic features of AITL are well described, there is considerable morphologic overlap with reactive lymphoid proliferations and other lymphomas. Unlike many other lymphomas, no specific phenotypic or molecular markers have been identified so far to assist the diagnosis that often leads to delayed or wrong histologic diagnosis, denying the patient the chance of early treatment. Second, very little is known about the biology of the disease. This is partly due to the lack of objective diagnostic criteria, but, more critically, in most cases the tumor cell population cannot be specifically identified within the polymorphic reactive and inflammatory cells. In a subset of the cases it is thought that perivascular collections of CD3+ cells with clear cytoplasm represent the neoplastic population.15
In this paper we show that the neoplastic cells in most cases of AITL can be identified by aberrant expression of CD10. The presence of such a marker has provided the opportunity to establish objective criteria for the diagnosis of this disease, even in its early stage of evolution, as well as the ability to investigate the biologic characteristics of the neoplastic T cells.
Materials and methods
Tissues
Thirty-two formalin-fixed, paraffin-embedded, lymph node biopsies from 30 cases of AITL, 10 blocks from 10 cases of nodal peripheral T-cell lymphoma unspecified (PTL), and 10 blocks from 10 cases of reactive lymph node hyperplasia were retrieved from the archives of the Department of Histopathology, University College London Hospital. The histology was reviewed and the diagnosis was confirmed in each case. Of the cases of AITL, 4 were local cases, whereas the remaining 26 cases had been referred from other centers for a second opinion. The original diagnosis of the referring center was obtained where possible.
Immunohistochemistry
Paraffin sections (3 μm) were immunostained by the streptavidin immunoperoxidase method (ChemMate Streptavidin Peroxidase kit [Dako, Cambridge, United Kingdom] and DAB chromogen [Sigma, Poole, United Kingdom]) after heat-mediated antigen retrieval as previously described.16 Primary antibodies included CD3 (polyclonal anti-CD3+; Dako), CD20 (L26; Dako), CD4 (1F6; Vector Labs, Peterborough, United Kingdom), CD8 (C8/144B; Dako), CD20, CD79a (JCB117; Dako), CD21 (1F8; Dako), Ki67 (MIB-1; Dako), LMP-1 (CS1-4; Dako), and CD10 (56C6; Novacastra Labs, Newcastle, United Kingdom).
Sequential double-staining was performed by using CD20/CD10, CD3/CD10, CD4/CD10, and Ki67/CD10. The first antibody was revealed with peroxidase and the second with alkaline phosphatase with a fast blue chromogen (Fast Blue/Naphthol AS-BI; Sigma) in each case.17 No counterstain was used. The approximate ratio of CD10+ T cells to all T cells was assessed from serial sections immunostained for CD3 and CD20/CD10. The proliferation fraction was calculated by counting 500 CD10+ cells in 5 to 7 different areas of the lymph node on sections double immunostained for Ki67/CD10.
In situ hybridization for EBV
In situ hybridization was carried out with a polymerase chain reaction (PCR)–generated EBV DNA probe labeled with digoxigenin, followed by incubation with antidigoxigenin-AP (Boehringer Mannheim, Germany) and visualization with 5-bromo-4-chloro-3-indolyl phosphate and nitroblue tetrazolium as previously described.18
PCR for T-cell receptor γ chain gene and immunoglobulin heavy chain gene
PCR was performed to analyze clonal expansion of T cells and B cells. DNA was extracted from paraffin sections by using proteinase K digestion without subsequent organic extraction as previously described.17 T-cell clonal expansion was detected by analysis of T-cell receptor γ (TCR-γ) chain gene rearrangement. Duplicate aliquots of each sample were analyzed for rearrangement by using 2 sets of primers.17 Set 1 consisted of primers directed to the Vγ-I, Vγ-II, Vγ-III-IV, and Jγ-1/2 segments and set 2 of primers targeted Vγ-I, Vγ-II, Vγ-III-IV, and JPγ-1/2. Forty cycles of PCR of 30 seconds at 93°C, 30 seconds at 55°C, and 1 minute at 73°C were performed after hotstart by addition of Taq polymerase at 55°C after 5 minutes at 95°C. The final extension time was extended to 6 minutes. DNA extracted from a paraffin block of a T-cell lymphoma was used as a positive control and a reaction without template DNA was run as a negative control in all experiments.
B-cell clonal expansion was detected by analysis of immunoglobulin heavy (IgH) chain gene rearrangement. Samples were subjected to amplification of the IgH chain gene by using a seminested protocol with primers directed to the framework 3 and joining regions of the gene, as described.19 Test samples were studied in parallel with appropriate positive and negative controls. PCR products were analyzed on 10% polyacrylamide minigels, stained with ethidium bromide, and viewed under UV light.
Microdissection of CD10+ cells with laser capture microscopy
Microdissection of CD10+ cells and CD10− areas was performed by using PixCell II Laser Capture Microdissection system (Arcturus, Mountain View, CA), using 5-μm sections immunostained for CD10.20 A laser beam diameter of 7.5 μm was used to achieve single cell microdissection. Between 30 and 50 single cells were microdissected onto Capsure Transfer film (Arcturus). The DNA was extracted and PCR for TCR-γ chain gene was performed as described above. Each PCR reaction was estimated to contain DNA from 6 CD10+ cells.
Cloning and sequencing
To confirm the monoclonal nature of dominant bands and the clonal identity of PCR products of the same size from whole sections and microdissected CD10+ cells, the individual bands were isolated, cloned, and sequenced as previously described.18PCR products were purified by using QIA Quick Gel Extraction Kit (QIAGEN, West Sussex, United Kingdom), then ligated into the pGEM-T vector and transformed into JM109 competent cells (Promega, Southampton, United Kingdom). The transformed cells were selected on LB-ampicillin agar plates containing X-gal and isopropyl β-D-thiogalactoside. White colonies were screened by using PCR with vector primers (Sp6 and T7). The PCR products showing the expected insert size were sequenced in both directions by using an ABI sequencer with dRhodamine terminators (Perkin-Elmer, Foster City, CA). Five to 10 clones from each PCR band were sequenced.
Results
Clinical features
The clinical features are summarized in Table1. There were 17 men and 13 women between the ages of 37 and 84 years. Most patients presented with generalized lymphadenopathy. Other clinical features included weight loss, fever, skin rash, anemia, and hepatosplenomegaly. The initial histologic diagnosis was AITL in only 13 cases. In the remaining 17 cases the diagnoses included reactive lymphoid hyperplasia (4 cases), T-cell rich B-cell lymphoma (4 cases), and other lymphomas (8 cases).
Clinical features of AITL cases
| Case no. . | Age/sex . | Clinical features . | Site . | Initial diagnosis . |
|---|---|---|---|---|
| 1 | 65/M | Fever, generalized lymphadenopathy, hemolytic anemia | Lymph node | AITL |
| 2 | 62/M | Generalized lymphadenopathy | Lymph node | PTL |
| 3 | 59/M | Fever, cervical lymphadenopathy, abnormal liver function tests | Lymph node | AIL |
| 4 | 37/M | Generalized lymphadenopathy | Inguinal lymph node | Reactive |
| 5 | 82/M | Generalized lymphadenopathy, splenomegaly, positive Coombs test, night sweats | Lymph node | Reactive |
| 6 | 59/F | Cervical lymphadenopathy, pleural effusion | Lymph node | DLBL |
| 7 | 62/F | Lymphadenopathy, polyclonal gammopathy, hepatosplenomegaly | Lymph node | AIL |
| 8 | 70/M | Generalized lymphadenopathy, weight loss, rash | Axillary lymph node | HL, TRBL |
| 9 | 48/M | Not available | Lymph node | AITL |
| 10 | 64/F | Generalized lymphadenopathy, fever, pruritus, night sweats | Axillary lymph node | AIL?, reactive? |
| 11 | 76/F | Generalized lymphadenopathy, hepatosplenomegaly | Lymph node | AIL?, CD?, reactive? |
| 12 | 79/F | Not available | Lymph node | AITL?, PTL? |
| 13 | 84/F | Lymphadenopathy, rash | Inguinal lymph node | PTL?, TRBL? |
| 14 | 78/M | Fever, generalized lymphadenopathy, splenomegaly, skin rash | Lymph node | NHL |
| 15A | 42/M | Generalized lymphadenopathy, fever, hepatosplenomegaly, hemolytic anemia, weight loss | Lymph node February 1998 | Reactive?, CD? |
| 15B | Lymph node March 2000 | |||
| 16A | 60/F | Fever, bilateral inguinal lymphadenopathy, splenomegaly | Inguinal lymph node 2000 | NHL |
| 16B | Lymph node 2001 | DLBL | ||
| 17 | 58/F | Generalized lymphadenopathy, “B” symptoms | Lymph node | DLBL |
| 18 | 78/M | Generalized lymphadenopathy, positive Coombs test, hemolytic anemia, hypergammaglobulinemia, | Cervical lymph node | AIL |
| 19 | 55/M | Generalized lymphadenopathy, fever, weight loss | Inguinal lymph node 2000 | Reactive |
| 20 | 71/M | Inguinal lymphadenopathy, weight loss | Inguinal lymph node | DLBL |
| 21 | 39/F | Generalized lymphadenopathy, night sweats | Lymph node | AITL |
| 22 | 77/F | Generalized lymphadenopathy, splenomegaly, “B” symptoms | Axillary lymph node | PTL?, HL? |
| 23 | 52/F | Lymphadenopathy, high ESR | Cervical lymph node | DLBL?, HL? |
| 24 | 61/M | Axillary, inguinal lymphadenopathy, hepatomegaly, pneumonia | Axillary lymph node | AIL |
| 25 | 61/M | Not available | Lymph node | AITL |
| 26 | 74/F | Generalized lymphadenopathy, pruritus | Cervical lymph node | AIL |
| 27 | 78/F | Lymphadenopathy, “B” symptoms | Inguinal lymph node | TRBL |
| 28 | 78/M | Generalized lymphadenopathy, rash, weight loss | Inguinal lymph node | TRBL?, PTL? |
| 29 | 52/M | Generalized lymphadenopathy, hepatosplenomegaly, anemia | Lymph node | FDC tumor |
| 30 | 59/M | Generalized lymphadenopathy | Lymph node | AIL |
| Case no. . | Age/sex . | Clinical features . | Site . | Initial diagnosis . |
|---|---|---|---|---|
| 1 | 65/M | Fever, generalized lymphadenopathy, hemolytic anemia | Lymph node | AITL |
| 2 | 62/M | Generalized lymphadenopathy | Lymph node | PTL |
| 3 | 59/M | Fever, cervical lymphadenopathy, abnormal liver function tests | Lymph node | AIL |
| 4 | 37/M | Generalized lymphadenopathy | Inguinal lymph node | Reactive |
| 5 | 82/M | Generalized lymphadenopathy, splenomegaly, positive Coombs test, night sweats | Lymph node | Reactive |
| 6 | 59/F | Cervical lymphadenopathy, pleural effusion | Lymph node | DLBL |
| 7 | 62/F | Lymphadenopathy, polyclonal gammopathy, hepatosplenomegaly | Lymph node | AIL |
| 8 | 70/M | Generalized lymphadenopathy, weight loss, rash | Axillary lymph node | HL, TRBL |
| 9 | 48/M | Not available | Lymph node | AITL |
| 10 | 64/F | Generalized lymphadenopathy, fever, pruritus, night sweats | Axillary lymph node | AIL?, reactive? |
| 11 | 76/F | Generalized lymphadenopathy, hepatosplenomegaly | Lymph node | AIL?, CD?, reactive? |
| 12 | 79/F | Not available | Lymph node | AITL?, PTL? |
| 13 | 84/F | Lymphadenopathy, rash | Inguinal lymph node | PTL?, TRBL? |
| 14 | 78/M | Fever, generalized lymphadenopathy, splenomegaly, skin rash | Lymph node | NHL |
| 15A | 42/M | Generalized lymphadenopathy, fever, hepatosplenomegaly, hemolytic anemia, weight loss | Lymph node February 1998 | Reactive?, CD? |
| 15B | Lymph node March 2000 | |||
| 16A | 60/F | Fever, bilateral inguinal lymphadenopathy, splenomegaly | Inguinal lymph node 2000 | NHL |
| 16B | Lymph node 2001 | DLBL | ||
| 17 | 58/F | Generalized lymphadenopathy, “B” symptoms | Lymph node | DLBL |
| 18 | 78/M | Generalized lymphadenopathy, positive Coombs test, hemolytic anemia, hypergammaglobulinemia, | Cervical lymph node | AIL |
| 19 | 55/M | Generalized lymphadenopathy, fever, weight loss | Inguinal lymph node 2000 | Reactive |
| 20 | 71/M | Inguinal lymphadenopathy, weight loss | Inguinal lymph node | DLBL |
| 21 | 39/F | Generalized lymphadenopathy, night sweats | Lymph node | AITL |
| 22 | 77/F | Generalized lymphadenopathy, splenomegaly, “B” symptoms | Axillary lymph node | PTL?, HL? |
| 23 | 52/F | Lymphadenopathy, high ESR | Cervical lymph node | DLBL?, HL? |
| 24 | 61/M | Axillary, inguinal lymphadenopathy, hepatomegaly, pneumonia | Axillary lymph node | AIL |
| 25 | 61/M | Not available | Lymph node | AITL |
| 26 | 74/F | Generalized lymphadenopathy, pruritus | Cervical lymph node | AIL |
| 27 | 78/F | Lymphadenopathy, “B” symptoms | Inguinal lymph node | TRBL |
| 28 | 78/M | Generalized lymphadenopathy, rash, weight loss | Inguinal lymph node | TRBL?, PTL? |
| 29 | 52/M | Generalized lymphadenopathy, hepatosplenomegaly, anemia | Lymph node | FDC tumor |
| 30 | 59/M | Generalized lymphadenopathy | Lymph node | AIL |
AITL indicates angioimmunoblastic T-cell lymphoma; PTL, peripheral T-cell lymphoma; AIL, angioimmunoblastic lymphadenopathy; DLBL, diffuse large B-cell lymphoma; HL, Hodgkin lymphoma; TRBL, T-cell–rich B-cell lymphoma; NHL, non-Hodgkin lymphoma; CD, Castleman disease; FDC, follicular dendritic cell.
Histology
Angioimmunoblastic T-cell lymphoma.
The histologic features are summarized in Table2. At the time of initial diagnosis, the cases could be separated into 3 overlapping patterns. In pattern I (6 cases) there was partial preservation of the lymph node architecture. Hyperplastic B-cell follicles with poorly developed mantle zones and ill-defined borders were easily identifiable in the cortex of the lymph node. These structures merged into the expanded paracortex containing a polymorphic infiltrate of lymphocytes and transformed large lymphoid blasts, plasma cells, macrophages, and eosinophils within a prominent vascular network. Increase in FDCs was not evident (Figure1A).
Summary of histologic features, immunophenotype, and molecular analysis of AITL cases
| Case no. . | Pattern* . | CD10/ CD3†(%) . | Clear cells‡ . | CD10/ Ki672-153(%) . | TCR PCR . | IgH PCR . |
|---|---|---|---|---|---|---|
| 1 | I | 5 | Absent | O | P | |
| 2 | I | 5 | Absent | 16 | M | P |
| 3 | I | 10 | Present | O | P | |
| 4 | I | 10 | Absent | M | P | |
| 5 | I | 20 | Present | 8 | M | P |
| 6 | I | 0 | Absent | M | P | |
| 7 | II | 5 | Absent | P | P | |
| 8 | II | 5 | Absent | M | P | |
| 9 | II | 5 | Absent | 11 | M | P |
| 10 | II | 10 | Present | 19 | O | P |
| 11 | II | 10 | Present | M | M | |
| 12 | II | 15 | Present | M | P | |
| 13 | II | 20 | Absent | M | M | |
| 14 | II | 20 | Absent | O | M | |
| 15A | II | 5 | Absent | M | P | |
| 15B | III | 5 | Present | M | P | |
| 16A | II | 0 | Absent | M | P | |
| 16B | III | 0 | Absent | M | M | |
| 17 | III | 5 | Present | M | P | |
| 18 | III | 5 | Present | M | P | |
| 19 | III | 5 | Present | M | P | |
| 20 | III | 5 | Present | O | M | |
| 21 | III | 10 | Absent | M | P | |
| 22 | III | 10 | Absent | M | P | |
| 23 | III | 20 | Present | M | P | |
| 24 | III | 20 | Present | M | NA | |
| 25 | III | 20 | Present | 10 | M | P |
| 26 | III | 30 | Absent | 11 | O | P |
| 27 | III | 30 | Present | 19 | M | M |
| 28 | III | 30 | Present | M | P | |
| 29 | III | 30 | Present | O | P | |
| 30 | III | 0 | Present | M | P |
| Case no. . | Pattern* . | CD10/ CD3†(%) . | Clear cells‡ . | CD10/ Ki672-153(%) . | TCR PCR . | IgH PCR . |
|---|---|---|---|---|---|---|
| 1 | I | 5 | Absent | O | P | |
| 2 | I | 5 | Absent | 16 | M | P |
| 3 | I | 10 | Present | O | P | |
| 4 | I | 10 | Absent | M | P | |
| 5 | I | 20 | Present | 8 | M | P |
| 6 | I | 0 | Absent | M | P | |
| 7 | II | 5 | Absent | P | P | |
| 8 | II | 5 | Absent | M | P | |
| 9 | II | 5 | Absent | 11 | M | P |
| 10 | II | 10 | Present | 19 | O | P |
| 11 | II | 10 | Present | M | M | |
| 12 | II | 15 | Present | M | P | |
| 13 | II | 20 | Absent | M | M | |
| 14 | II | 20 | Absent | O | M | |
| 15A | II | 5 | Absent | M | P | |
| 15B | III | 5 | Present | M | P | |
| 16A | II | 0 | Absent | M | P | |
| 16B | III | 0 | Absent | M | M | |
| 17 | III | 5 | Present | M | P | |
| 18 | III | 5 | Present | M | P | |
| 19 | III | 5 | Present | M | P | |
| 20 | III | 5 | Present | O | M | |
| 21 | III | 10 | Absent | M | P | |
| 22 | III | 10 | Absent | M | P | |
| 23 | III | 20 | Present | M | P | |
| 24 | III | 20 | Present | M | NA | |
| 25 | III | 20 | Present | 10 | M | P |
| 26 | III | 30 | Absent | 11 | O | P |
| 27 | III | 30 | Present | 19 | M | M |
| 28 | III | 30 | Present | M | P | |
| 29 | III | 30 | Present | O | P | |
| 30 | III | 0 | Present | M | P |
TCR indicates T-cell receptor-γ chain gene; PCR, polymerase chain reaction; IgH, immunoglobulin heavy chain gene; M, monoclonal; O, oligoclonal; P, polyclonal; NA, no amplification.
The histologic pattern as described in “Results.”
Percentage of CD10+ and CD3+ T cells.
Cases containing aggregates of atypical lymphoid cells with clear cytoplasm.
Percentage of CD10+ cells expressing Ki67.
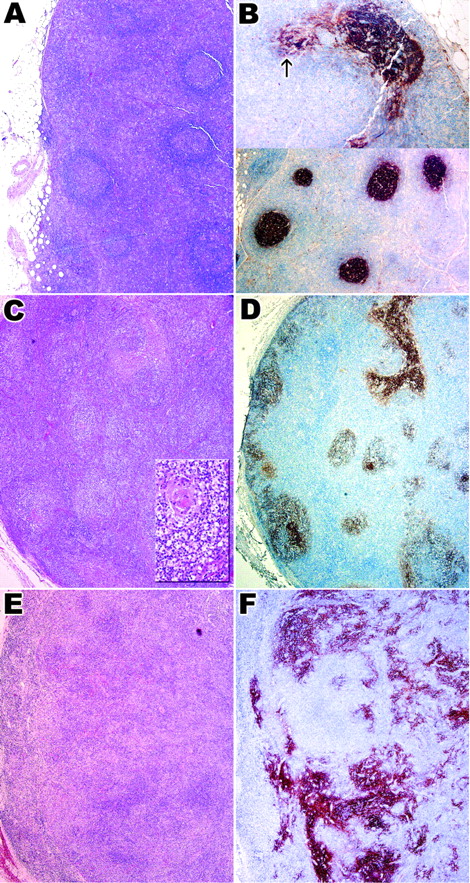
Histologic features and CD21 expression in 3 histologic patterns of AITL.
Panels A and B show case 2 with histologic pattern I, panels C and D show case 10 with histologic pattern II, and panels E and F show case 28 with histologic pattern III. (A,C,E) Hematoxylin and eosin–stained sections showing 3 different histologic patterns. Inset in panel C highlights a depleted follicle. Follicular architecture is retained in pattern I (A) and II (C) but is completely lost in pattern III (E). (B,D,F) Immunostaining for CD21 in 3 histologic patterns. There is only focal proliferation of FDCs in pattern I (B, upper panel; arrow); in patterns II and III (panels D and F), there is increasing FDC proliferation. Original magnification A-F, × 40; C inset, × 200.
Histologic features and CD21 expression in 3 histologic patterns of AITL.
Panels A and B show case 2 with histologic pattern I, panels C and D show case 10 with histologic pattern II, and panels E and F show case 28 with histologic pattern III. (A,C,E) Hematoxylin and eosin–stained sections showing 3 different histologic patterns. Inset in panel C highlights a depleted follicle. Follicular architecture is retained in pattern I (A) and II (C) but is completely lost in pattern III (E). (B,D,F) Immunostaining for CD21 in 3 histologic patterns. There is only focal proliferation of FDCs in pattern I (B, upper panel; arrow); in patterns II and III (panels D and F), there is increasing FDC proliferation. Original magnification A-F, × 40; C inset, × 200.
Pattern II (10 cases) was characterized by loss of normal architecture except for the presence of occasional depleted follicles with concentrically arranged FDCs. In some cases FDC proliferation extending beyond the follicles could be identified. The rest of the node showed a polymorphic infiltrate with increased numbers of transformed lymphoid blasts and vascular proliferation similar to that described for pattern I (Figure 1C).
In pattern III (14 cases) the normal architecture was completely effaced and no B-cell follicles could be identified. Prominent irregular proliferation of FDCs could be seen in hematoxylin and eosin–stained sections in most cases, and this situation was accompanied by extensive vascular proliferation and a polymorphic infiltrate similar to that seen in patterns I and II (Figure 1E).
In 16 cases perivascular collections of cells with clear cytoplasm were evident. In 2 cases in which consecutive biopsies from the same patient were available, the earlier biopsies showed pattern II and the later pattern III.
Peripheral T-cell lymphoma.
All cases of PTL showed effacement of nodal architecture with a relatively monomorphic infiltrate of medium to large atypical lymphoid cells. A moderate degree of vascular proliferation was present without the prominent arborizing networks seen in AITL. FDC proliferation was not a feature.
Reactive lymph node hyperplasia.
All reactive lymph nodes showed preservation of the architecture and hyperplastic lymphoid follicles with moderate paracortical expansion.
Immunohistochemistry
Angioimmunoblastic T-cell lymphoma.
In pattern I sprouts of CD21+ FDCs extended beyond the confines of the CD21+ germinal centers and occasionally enveloped small blood vessels (Figure 1B). Paracortical T cells, including some transformed blasts, expressed CD3 and predominantly CD4. A smaller CD8+ population was present. Preserved germinal centers expressed CD20 and CD10. In addition to CD10+germinal center cells there was a population of smaller strongly staining lymphoid cells at the outer rim of the germinal center that extended into the paracortex (Figure2A-B).
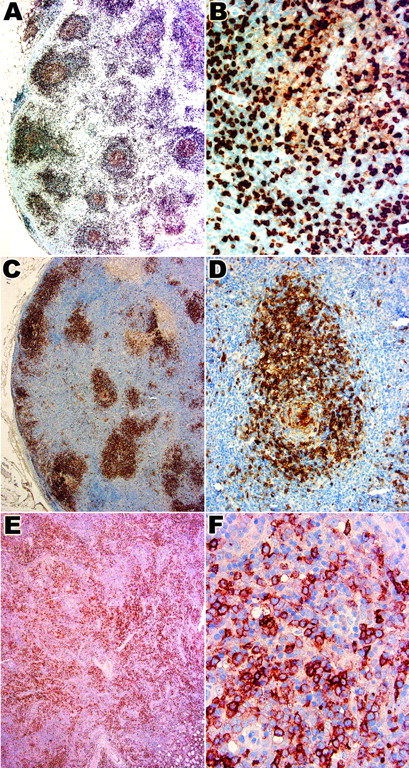
CD10 expression in the 3 histologic patterns of AITL.
Panels A and B show case 2 with histologic pattern I, panels C and D show case 10 with histologic pattern II, and panels E and F show case 28 with histologic pattern III. Numerous medium-sized lymphoid cells in and around the follicles in pattern I (A,B) and II (C,D) are CD10+. Panel B shows that the intensity of staining is much higher in these cells compared with weakly staining follicle center B cells. The follicular distribution is lost in pattern III (E) in which a diffuse infiltrate of CD10+ cells is seen. Panel F shows that some of these are large atypical lymphoid cells. Original magnification A, C, E, × 40; B, F, × 400; D, × 200.
CD10 expression in the 3 histologic patterns of AITL.
Panels A and B show case 2 with histologic pattern I, panels C and D show case 10 with histologic pattern II, and panels E and F show case 28 with histologic pattern III. Numerous medium-sized lymphoid cells in and around the follicles in pattern I (A,B) and II (C,D) are CD10+. Panel B shows that the intensity of staining is much higher in these cells compared with weakly staining follicle center B cells. The follicular distribution is lost in pattern III (E) in which a diffuse infiltrate of CD10+ cells is seen. Panel F shows that some of these are large atypical lymphoid cells. Original magnification A, C, E, × 40; B, F, × 400; D, × 200.
In pattern II, CD21+ FDCs extended beyond the follicles into the paracortex, often surrounding proliferating small vessels, whereas in pattern III they showed a more haphazard arrangement, surrounding the arborizing small vessels (Figure 1D,F). In both patterns II and III, CD20, IgM, and IgD+ but CD10− clusters of small B cells, mostly not associated with germinal centers, could be identified within meshworks of CD21+ FDCs. In all but 3 cases (4 lymph node biopsies) there were concentrations of CD10+ T cells around the CD20+ B-cell clusters as described for pattern I (Figure 2C-F). In addition isolated CD10+ T cells were scattered throughout the entire lymph node and formed perivascular aggregates, corresponding to the clear cells noted in hematoxylin and eosin–stained sections (Figure 2F). CD10 was also expressed by granulocytes most of which were within blood vessels. Their characteristic cytologic features easily distinguished them from the CD10+ lymphoid cells. In biopsies showing patterns II and III isolated transformed CD20+ blasts were identified within the predominantly CD3+ infiltrate.
PTL and reactive lymph node hyperplasia.
No CD10+ lymphoid cells were present in any of the cases of PTL, whereas in reactive lymph nodes CD10+ cells were confined to the germinal centres.
Sequential double staining
Double staining with CD20 followed by CD10 was performed on all lymph node biopsies to show that the lymphoid cells expressing CD10 were not germinal center B cells. In 28 of 32 biopsies (27 of 30 cases) there were varying numbers of CD20−, CD10+lymphoid cells, strongly suggesting that these were T cells (Figure3A-F). Double staining with CD79a and CD10 gave identical results. The T-cell phenotype of the CD20−, CD10+ lymphoid cells was also confirmed by CD3/CD10 double staining that showed a population of “purple” cells coexpressing CD3 and CD10. The ratio of CD10+ T cells to all T cells for each case is shown in Table 2.
CD20/CD10 double immunostaining in the 3 histologic patterns of AITL.
Panels A and B show case 2 with histologic pattern I, panels C and D show case 10 with histologic pattern II, and panels E and F show case 28 with histologic pattern III. (A-F) Double immunostaining for CD20 in brown (DAB) and CD10 in blue (fast blue). Numerous single blue cells expressing CD10 but not CD20 are seen around the follicles and in the paracortical areas. Panels B, D, and F show high-power view of double immunostaining for CD20 and CD10. Original magnification A, C, E, × 40; B, D, F, × 400.
CD20/CD10 double immunostaining in the 3 histologic patterns of AITL.
Panels A and B show case 2 with histologic pattern I, panels C and D show case 10 with histologic pattern II, and panels E and F show case 28 with histologic pattern III. (A-F) Double immunostaining for CD20 in brown (DAB) and CD10 in blue (fast blue). Numerous single blue cells expressing CD10 but not CD20 are seen around the follicles and in the paracortical areas. Panels B, D, and F show high-power view of double immunostaining for CD20 and CD10. Original magnification A, C, E, × 40; B, D, F, × 400.
Further double-staining experiments showed that the CD10+ T cells expressed CD4 but not CD8, whereas a subset of the CD10+ T cells showed nuclear positivity for BCL6 in all cases with CD10+ T cells. Double staining of the CD10+ cells with Ki-67 showed that these phenotypically aberrant T cells had a low proliferation fraction (Table 2).
In situ hybridization for EBV–Epstein-Barr early region
All cases of AITL showed hybridization for EBV–Epstein-Barr early region (EBER) in a subset of the cells. The number of cells expressing EBV-EBER varied markedly from case to case; in some only scattered cells were positive, and in others numerous large blasts were labeled. The distribution of EBER+ cells was similar to the distribution of large B blasts and did not correspond to the clusters of CD10+ T cells.
PCR for TCR-γ and IgH gene rearrangement
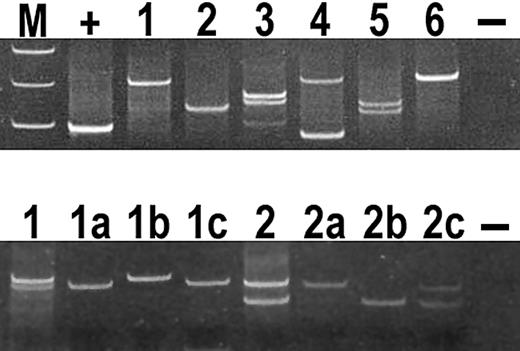
The results of PCR for TCR-γ gene and IgH chain gene are shown in Table 2 and Figure 4. There was a monoclonal (1-2 dominant bands) or oligoclonal (3-4 dominant bands) T-cell population in all cases except one. Six of the cases also showed evidence of a monoclonal B-cell population. In all of these cases numerous EBV-infected transformed lymphocytes were present.
Analysis of PCR products for TCR-γ chain gene on polyacrylamide gels in AITL.
(Top panel) All cases demonstrated in the figure show 1 or 2 dominant bands consistent with a monoclonal T-cell population. Lane M, molecular weight markers; lane +, positive control; lane 1, case 21; lane 2, case 23; lane 3, case 26; lane 4, case 22; lane 5, case 3; lane 6, case 4; and lane −, negative control. (Bottom panel) Microdissection of CD10+ cells in cases 26 and 13 gave PCR products identical in size to products obtained from whole-section PCR. Lane 1, whole-section PCR products of case 26; lanes 1a,b,c, microdissected PCR products of case 26; lane 2, whole section PCR products of case 13; lanes 2a,b,c, microdissected PCR products of case 13; and lane −, negative control.
Analysis of PCR products for TCR-γ chain gene on polyacrylamide gels in AITL.
(Top panel) All cases demonstrated in the figure show 1 or 2 dominant bands consistent with a monoclonal T-cell population. Lane M, molecular weight markers; lane +, positive control; lane 1, case 21; lane 2, case 23; lane 3, case 26; lane 4, case 22; lane 5, case 3; lane 6, case 4; and lane −, negative control. (Bottom panel) Microdissection of CD10+ cells in cases 26 and 13 gave PCR products identical in size to products obtained from whole-section PCR. Lane 1, whole-section PCR products of case 26; lanes 1a,b,c, microdissected PCR products of case 26; lane 2, whole section PCR products of case 13; lanes 2a,b,c, microdissected PCR products of case 13; and lane −, negative control.
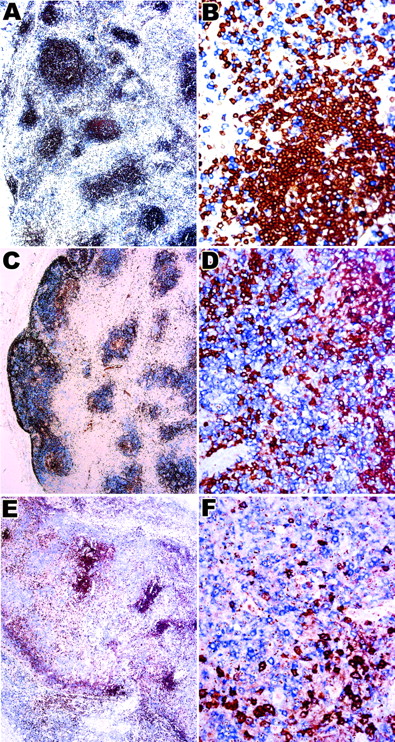
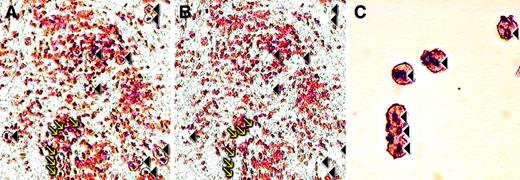
Single-cell microdissection
To investigate whether the CD10+ T cells were part of the neoplastic clone, we microdissected 30 to 50 CD10+cells from 5 cases (Figure 5) and performed PCR for TCR-γ gene. In each case, analysis of the PCR products from microdissected cells gave a dominant band(s) identical in size to that observed for the PCR performed on whole sections (Figure4). In contrast DNA extracted from areas lacking CD10+cells showed a polyclonal ladder or smear.
Microdissection of CD10+ cells in AITL.
(A) CD10 stained section before microdissection; the cells targeted for microdissection are indicated with arrows and arrowheads. (B) Same area after microdissection; the arrows and arrowheads indicate spaces left after cells have been removed. (C) High-power view of a cluster of CD10+ cells after microdissection. These are the cells/spaces indicated by arrows in the panels A and B. Original magnification A, B, × 200; C × 400.
Microdissection of CD10+ cells in AITL.
(A) CD10 stained section before microdissection; the cells targeted for microdissection are indicated with arrows and arrowheads. (B) Same area after microdissection; the arrows and arrowheads indicate spaces left after cells have been removed. (C) High-power view of a cluster of CD10+ cells after microdissection. These are the cells/spaces indicated by arrows in the panels A and B. Original magnification A, B, × 200; C × 400.
Cloning and sequencing of PCR products
The cloning and sequencing of the dominant bands isolated from PCR products of 3 cases showed a dominant clone with identical sequence, confirming the monoclonal nature of all cases examined. For each of the 3 cases the dominant clone from the whole sections and from the microdissected CD10+ cells were identical (Table3).
Results of cloning and sequencing of dominant bands obtained from PCR amplification of TCR-γ chain gene from whole lymph node sections and microdissected CD10+ cells
| Case no. . | PCR band . | Whole lymph node3-150 . | Microdissected CD10+cells3-151 . |
|---|---|---|---|
| 12 | Upper | 4/6 | 6/6 |
| Lower | 3/6 | 6/6 | |
| 26 | Upper | 3/5 | 4/6 |
| Lower | 11/13 | 8/8 | |
| 28 | Upper | 7/9 | 5/7 |
| Lower | 10/10 | Not done |
| Case no. . | PCR band . | Whole lymph node3-150 . | Microdissected CD10+cells3-151 . |
|---|---|---|---|
| 12 | Upper | 4/6 | 6/6 |
| Lower | 3/6 | 6/6 | |
| 26 | Upper | 3/5 | 4/6 |
| Lower | 11/13 | 8/8 | |
| 28 | Upper | 7/9 | 5/7 |
| Lower | 10/10 | Not done |
PCR indicates polymerase chain reaction; TCR-γ, T-cell receptor-γ.
The number of clones with identical TCR-γ chain gene sequence/total number of clones sequenced.
The number of clones with TCR-γ chain gene sequence identical to the dominant sequence obtained from the whole section PCR/total number of clones sequenced.
Discussion
The histologic diagnosis of AITL may be difficult as is shown by the large error rate (13 of 26; 50%) in referred cases in our series. This error rate is especially true in the early phase of the disease when preservation of follicles and only slight expansion of the paracortex are present. The expanded CD21+ FDC meshwork is very helpful in making the diagnosis3 but can be subtle. Molecular investigation and, in many instances, repeated biopsies may be required to reach a confident diagnosis. In this study we have shown that CD10 is a phenotypic marker that specifically identifies the tumor cells in 90% of AITLs, including, importantly, the early cases with pattern I histology. We believe that this marker provides an objective criterion for the diagnosis of AITL and should greatly assist in the diagnosis of this disorder.
Moreover, the presence of these cells distinguishes AITL from other more usual PTLs as none of the PTLs we have studied showed CD10 expression. Recently, de Leval et al21 have reported 3 cases of PTL with follicular growth pattern. In 2 of the cases there was CD10 expression by T cells. It is possible that these cases were also pattern I AITL without FDC proliferation.
The CD10-expressing T cells must be distinguished from germinal center B cells especially in cases with pattern I histology. They are typically seen at the margins of B-cell follicles as small-to-medium-sized lymphoid cells with crisp membrane staining in contrast to the larger follicle center B cells that stain more diffusely and with less intensity. In more advanced cases with pattern II or III histology the CD10+ T cells are more diffusely distributed as single cells and in small clusters. Double staining confirmed that these cells are CD3 and CD4+ T cells that accounted for a relatively small fraction of all T cells present in each case.
Several lines of evidence point to the CD10+ T cells being the malignant cells of AITL. First, an aberrant phenotype is itself considered a property of neoplastic T cells, although this phenotype usually takes the form of a loss of an antigen such as CD5 rather than expression of an antigen not usually associated with mature T cells.22 Second, the clusters of large cells with clear cytoplasm, which occur in some cases and which are widely regarded to represent the neoplastic clone in AITL,5 15 were consistently CD10+. Third, and most compelling, clonal analysis after microdissection has shown that the neoplastic clonally rearranged TCR genes are confined to the CD10+ population.
A subset of AITL cases contains more than one dominant T-cell clone with molecular or cytogenetic studies.6-8 23 Although we did not specifically address whether all dominant clones in a given case expressed CD10, all AITL cases with an oligoclonal T-cell population contained CD10+ T cells, suggesting that at least one of the dominant clones expressed CD10.
The reasons for the absence of CD10+ T cells in the small minority of cases (3 of 30 in the present series) that were otherwise entirely typical of AITL are unclear. Occasionally, cells that normally express CD10, such as germinal center B cells, fail to do so (A.D., unpublished observations, 2000). Sometimes this absence of CD10+ T cells is due to technical reasons, such as the fixation method, but more often it appears that the antigen has been down-regulated. The reasons for this down-regulation currently remain obscure.
The cases with the histologic patterns II and III fulfill the conventional criteria for the diagnosis of AITL. Thus, there was clinical evidence of a systemic disease characterized by generalized lymphadenopathy, hepatosplenomegaly, and anemia together with characteristic histologic features in lymph node biopsies. These features include effacement of lymph node architecture by a polymorphic infiltrate of lymphoid cells, proliferation of FDCs and small vessels, the presence of varying numbers of EBV-infected transformed B cells, and the presence of clusters of large transformed cells with clear cytoplasm. In addition there was molecular evidence for the presence of monoclonal or oligoclonal T-cell populations. Whether the cases falling into histologic pattern I can also be considered within the same spectrum is more controversial. The presence of hyperplastic follicle centers in otherwise typical AITL was first described by Ree et al,24 who reported progression of 2 such cases into typical AITL. We observed the CD10+ T cells in all histologic patterns, including the cases with hyperplastic germinal centres. These pattern I cases had the smallest number of CD10+ T cells, whereas pattern III cases had the most. In 2 cases, in which 2 consecutive biopsies were available, we observed progression from pattern II to pattern III. These observations strongly support that all 3 histologic patterns of AITL described in this study are part of the same disease process and that the histologic patterns I, II, and III form a biologic continuum.
In 5 cases in which microdissection was performed only the CD10+ T cells were clonal, and we failed to detect the presence of the clonal T cells in microdissected CD10−areas. Thus, the proportion of neoplastic T cells in AITL would appear to be small (5%-30%), and it appears that most of the lymph node enlargement is accounted for by reactive cells similar to what occurs in Hodgkin lymphoma. In this context it will be interesting to investigate the cytokine profile of the CD10+ T cells.
CD10 is a 100-kd, type II, integral membrane protein with neutral endopeptidase activity.25 The physiologic role of CD10 is thought to involve hydrolysis of polypeptides such as inflammatory mediators in the extracellular milieu. In peripheral lymphocytes, CD10 expression is limited to follicle center B cells that have a high rate of apoptosis. In a study, Cutrona et al26 have demonstrated expression of CD10 is also induced in T cells going through apoptosis in healthy or HIV-infected individuals. This apoptosis was seen only by flow cytometric analysis and not by immunohistochemistry, suggesting that the protein levels may be too low for detection on tissue sections. Those researchers have proposed that expression of CD10 may regulate apoptosis by interfering with negative or positive signals present in the extracellular environment. It is possible that aberrant CD10 expression in neoplastic T cells in AITL may be an indicator of disturbed apoptotic cell death. We observed that CD10+ neoplastic T cells have a low proliferation fraction. This is analogous to low-grade B-cell lymphomas such as the follicular lymphoma in which prevention of cell death by overexpression of antiapoptotic BCL2 protein rather than acquisition of high proliferative activity is considered to be the critical molecular event.27 An attractive hypothesis is that, analogous to follicular lymphoma, AITL is a biologically indolent/low-grade tumor-causing disease and that death is by immune deregulation rather than increased tumor load. This hypothesis is supported by the clinical observations suggesting that some patients do respond to immunosuppressive treatment.28-30
In conclusion, we have demonstrated that CD10 is a marker of neoplastic T cells in AITL. Early and accurate diagnosis of AITL can now be achieved by the simple expedient of immunostaining for CD10, a simple test that can be performed in any histopathology laboratory. More importantly, identification of a specific marker for the neoplastic T cells in AITL, for the first time, gives us the opportunity to investigate the biology of this disease with a view to devise novel therapeutic approaches.
We thank Professor S. Lakhani and the Ludwig Institute for Cancer Research for access to the Laser Capture Microscope and R. A. Hamoudi, Institute of Cancer Research, London, for assistance with sequencing.
Supported by a project grant from the Leukaemia Research Fund.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Ahmet Dogan, Department of Histopathology, Royal Free and University College Medical School, Rockefeller Bldg, University St, London WC1E 6JJ, United Kingdom; e-mail:a.dogan@ucl.ac.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal