Abstract
Activating transcription factor (ATF) 4 is a ubiquitous basic leucine-zipper transcription factor that is a member of the ATF/cyclic adenosine monophosphate responsive element–binding (CREB) protein family. To determine the in vivo function of ATF4, the ATF4 gene in murine embryonic stem cells was deleted and homozygous mutant mice were generated. ATF4 null fetuses were severely anemic because of an impairment in fetal-liver definitive hematopoiesis; the hematocrit in 15.5-day mutant fetuses was 0.15, whereas that in controls was 0.35. The fetal livers in homozygous ATF4 mutants were pale and hypoplastic. In vitro culture of fetal-liver cells showed fewer hematopoietic progenitors per embryo and a dramatic decrease in the size of progenitor colonies. Culture of primary murine embryonic fibroblasts showed a proliferative defect. These results suggest that ATF4 is critical, in a cell-autonomous manner, for normal cellular proliferation, especially for the high-level proliferation required during fetal-liver hematopoiesis.
Introduction
The activating transcription factor (ATF)/cyclic adenosine monophosphate responsive element–binding (CREB) family consists of transcription factors that bind the cyclic adenosine monophosphate (cAMP) response element (CRE) in vitro through highly related, carboxy-terminal basic leucine-zipper (bZip) dimerization domains. ATF4 was initially isolated in a human λgt11 library screen by using the ATF binding site (GTGACGTACAG) as a probe.1 ATF4 was also independently isolated through λgt11 library screens as CREB-22 and TAXREB673 by using the CRE (TGACGTCA) and the Tax-responsive enhancer element (TGACGTCT), respectively, of the long-terminal repeat (LTR) of human T-cell leukemia virus type 1 (HTLV-1 ) as probes.
Murine ATF4 is a 381–amino acid protein containing 3 acidic regions similar to acidic transcriptional activation domains in other transcription factors and a carboxy-terminal bZip motif involved in DNA binding and dimerization. The murine and human complementary DNAs (cDNAs) are 85% homologous in nucleotide sequence in the region encompassing the open reading frame, and they encode proteins that are 80% similar overall, with a 98.6% amino acid identity in the bZip region.4 The ATF4 gene is present as a single copy on mouse chromosome 15.5 Northern blotting with ATF4 cDNA as a probe detected a single 1.7-kilobase (kb) band in mouse liver, spleen, kidney, heart, lung, muscle, thymus, testis, and brain.2,6 ATF binding sites are present in the promoters of a wide variety of genes. Most are thought to be involved in responses to environmental stimuli (eg, mitogens, phorbol esters, viral infection, and peptide hormones that elevate cAMP levels) and include transforming growth factor β2, somatostatin, vasoactive intestinal peptide, heat shock protein 70, c-fos, phosphoenolpyruvate carboxykinase, proenkephalin, 3-hydroxy-3-methylglutaryl coenzyme A reductase, DNA polymerase β, HTLV-1 LTR, and E1A-inducible adenovirus genes.1 7-11
ATF4 has been shown to form a homodimer in vitro and to heterodimerize with Jun, Fos, and Fra-1,12,13 HTLV-1 Tax,14GPE1-binding protein,15 IGEBP1,16 nuclear factor–interleukin 6 (NF–IL-6) (CCAAT/enhancer-binding protein [C/EBP] β),6 NF-E2–related factor (Nrf) 2,17 and c-maf.18,19 ATF4 appears to function in a variety of different pathways. The protein can act as a negative regulator of CRE-dependent transcription.2 ATF4 and NF–IL-6 synergistically activate somatostatin and enkephalin gene expression.6 ATF4 is induced rapidly in fibroblasts in response to anoxia,20 and ATF4 translation is increased by activation of the eukaryote initiation factor 2 kinases PERK and GCN2, resulting in induction of the downstream gene C/EBP-homologous protein (GADD153).21,22 The Drosophila ATF4 homologuecrc is required for normal molting and metamorphosis.23 The Aplysia ATF4 homologue is involved in memory formation, acting as a repressor of long-term facilitation by functionally competing with the activator CREB1.24 In rat and human neurons, nuclear localization of ATF4 is regulated by activation of γ-aminobutyric acid type B receptor.25 26
Tanaka et al19 and Hettmann et al27independently described the effect of ATF4 inactivation on the developing eye in mice. They found that a deficiency in ATF4 results in a defect in embryonic lens formation leading to severe microphthalmia in adults. The effect of inactivation of ATF4 in murine tissues other than the eye has not been determined.
We isolated ATF4 in a yeast 2-hybrid screen with locus control region factor 1 (LCR-F1). LCR-F1 is a bZip transcription factor identified in 2 independent screens for erythroid proteins capable of binding the tandem activator protein 1–like sites (also designated NF-E2 sites) in deoxyribonuclease I hypersensitive site 2 of the human β-globin LCR.28,29 Although LCR-F1 (also designated Nrf1,28 transcription factor 11,30 and Nfe2L131) is ubiquitously expressed, it activates high-level β-globin expression specifically in erythroid cells in transient-transfection experiments.29 Inactivation of LCR-F1 through homologous recombination on an outbred Black Swiss mice background resulted in arrest of most embryos at 6.5 days postcoitus (dpc) during the late egg-cylinder stage and failure to form a primitive streak and mesoderm.32 When the mutation was backcrossed into the C57BL/6J inbred mouse strain, homozygous mutant embryos had no mesoderm block but did have a severe fetal anemia due to a defect in definitive hematopoiesis33 (unpublished data, August 1999). Interestingly, the fetal anemia resolved by 18.5 dpc (unpublished data, August 1999).
In the current study, we examined the function of ATF4 by deleting the ATF4 coding region by means of homologous recombination. We found that, like LCR-F1 mutant mice, ATF4-deficient mice are severely anemic during fetal development, apparently because of an impairment in definitive hematopoiesis.
Materials and methods
Construction of the ATF4 targeting vector
The murine ATF4 gene was isolated from a 129/SvJ mouse genomic bacterial artificial chromosome (BAC) library (Genome Systems, St Louis, MO) by using full-length murine ATF4 cDNA as a probe. Positive clones were restriction mapped and partly sequenced. Southern blot analysis was done to verify that no rearrangements were present. A 14.8-kb region of the BAC clone containing the ATF4 gene was subcloned. From this, a 5′ homology region consisting of a 9.9-kbBamHI-SmaI fragment and a 3′ homology region consisting of a 1.3-kb BamHI-SacI fragment were used to construct a targeting vector in pNTK.34 35 The targeting vector was designed to replace a 2.4-kb region containing the 2 exons that constitute the entire ATF4 coding region after the first codon, as well as the polyadenylation signal, with a neomycin phosphotransferase resistance cassette (neo) in the same orientation as the ATF4 gene.
Transfection of embryonic stem cells and generation of ATF4-deficient mice
The murine ATF4 gene was disrupted by homologous recombination in both R1 and HM1 embryonic stem (ES) cells. Electroporated ES cells were subjected to positive and negative selection with G418 and ganciclovir, and surviving clones were screened by polymerase chain reaction (PCR) analysis with a 5′ primer within neo (5′-ATATTGCTGAAGAGCTTGGCGGC-3′) and a 3′ primer external to the 3′ homology region (5′-AAGTGGCAAGGGAAGGACTGACC-3′). Correct targeting was confirmed by Southern blot analysis of genomic DNA. The 5′ end was determined by digestion with XbaI and SalI and probing of the blot with a 2.6-kb HindIII-HindIII fragment from within the 5′ homology region (Figure1A), which resulted in an 11.4-kb endogenous and a 12.2-kb homologous recombinant band. The 3′ end was determined by digestion of DNA with EcoRV and probing of the blot with a 550–base pair (bp) BamHI-BglII fragment from within the 3′ homology region (Figure 1A), which resulted in a 3.3-kb endogenous and a 2.2-kb homologous recombinant band. Additional insertions of the targeting construct were ruled out by digestion of DNA with NcoI or NdeI and probing of the blot with a 630-bp PstI-XbaI fragment from the coding region of neo, which detected only single bands of 2.0 kb and 8.6 kb, respectively, as predicted. One in 15 clones resistant to G418 and ganciclovir was found to be correctly targeted.
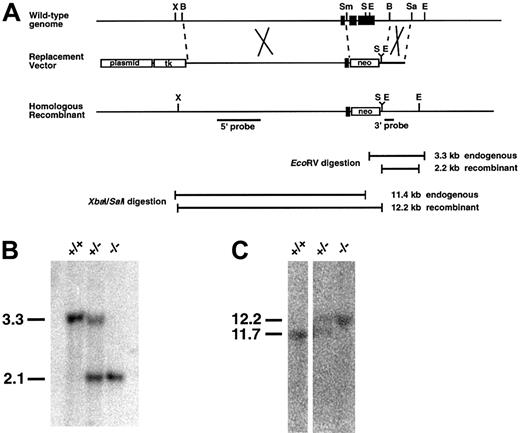
Targeted disruption of the ATF4 gene.
(A) Diagram of the murine ATF4 gene, the replacement vector, and the homologous recombinant allele. The exons of the ATF4 gene are represented by solid boxes. Plasmid sequences are indicated by open boxes. A 9.9-kb BamHI-SmaI 5′ homology fragment and a 1.3-kb BamHI-SacI 3′ homology fragment were used to construct a gene-replacement targeting vector in pNTK containing the neomycin phosphotransferase cassette (neo). Broken lines indicate the regions of homology used for homologous recombination. The sites at the ends of both homology fragments were removed as they were subcloned. An EcoRV site was added at the 3′ end of the 5′ homology fragment. SalI andEcoRV sites were added to the 5′ end of the 3′ homology fragment. Locations of the 5′ and 3′ probes used for Southern blot analysis are indicated by labeled lines below the maps. The locations and sizes of the fragments produced by restriction-enzyme digestions used in Southern blot analysis are indicated below the maps: B indicates BamHI; E, EcoRV; Sa,SacI; S, SalI; Sm, SmaI; and X, XbaI. (B) Typical Southern blot analysis of progeny from heterozygote matings after EcoRV restriction-enzyme digestion and hybridization with the 3′ probe shown in panel A. Positions of diagnostic bands are indicated at left. The probe detected bands of 3.3 and 2.2 kb corresponding to the sizes predicted for the wild-type and homologous recombinant alleles, respectively. Genotypes are shown above each lane. (C) Typical Southern blot analysis of progeny from heterozygote matings afterXbaI-SalI double restriction-enzyme digestion and hybridization with the 5′ probe shown in panel A. The probe detects bands of 11.4 and 12.2 kb corresponding to the sizes predicted for the wild-type and homologous recombinant alleles, respectively.
Targeted disruption of the ATF4 gene.
(A) Diagram of the murine ATF4 gene, the replacement vector, and the homologous recombinant allele. The exons of the ATF4 gene are represented by solid boxes. Plasmid sequences are indicated by open boxes. A 9.9-kb BamHI-SmaI 5′ homology fragment and a 1.3-kb BamHI-SacI 3′ homology fragment were used to construct a gene-replacement targeting vector in pNTK containing the neomycin phosphotransferase cassette (neo). Broken lines indicate the regions of homology used for homologous recombination. The sites at the ends of both homology fragments were removed as they were subcloned. An EcoRV site was added at the 3′ end of the 5′ homology fragment. SalI andEcoRV sites were added to the 5′ end of the 3′ homology fragment. Locations of the 5′ and 3′ probes used for Southern blot analysis are indicated by labeled lines below the maps. The locations and sizes of the fragments produced by restriction-enzyme digestions used in Southern blot analysis are indicated below the maps: B indicates BamHI; E, EcoRV; Sa,SacI; S, SalI; Sm, SmaI; and X, XbaI. (B) Typical Southern blot analysis of progeny from heterozygote matings after EcoRV restriction-enzyme digestion and hybridization with the 3′ probe shown in panel A. Positions of diagnostic bands are indicated at left. The probe detected bands of 3.3 and 2.2 kb corresponding to the sizes predicted for the wild-type and homologous recombinant alleles, respectively. Genotypes are shown above each lane. (C) Typical Southern blot analysis of progeny from heterozygote matings afterXbaI-SalI double restriction-enzyme digestion and hybridization with the 5′ probe shown in panel A. The probe detects bands of 11.4 and 12.2 kb corresponding to the sizes predicted for the wild-type and homologous recombinant alleles, respectively.
Two R1 and 2 HM1 independently targeted ES-cell clones were injected into the inner cell mass of donor blastocysts. High-percentage chimeras were generated and bred to Black Swiss mice (Taconic Farms, Germantown, NY). ATF4+/− offspring were obtained from both the R1 and the HM1 ES-cell line. Heterozygous mice were phenotypically normal and were intercrossed to obtain ATF4−/− mice. No difference in phenotype was observed between homozygotes from the 2 cell lines. Therefore, the data presented here were obtained from one of the R1 ES-cell lines.
The neo cassette was not removed from the mutant mice. Although this marker gene could affect expression of linked genes, a search of the National Center for Biotechnology Information and Celera Genomics (Rockville, MD) human and mouse genomic databases determined that the closest gene is mannoside acetylglucosaminyltransferase 3 (Mgat3), which is located 42 kb from the ATF4 gene. There is no other gene within 100 kb of ATF4. Targeted inactivation of the Mgat3 gene was previously shown to have no phenotypic effect.36 In particular, Mgat3−/− mice have no hematologic defects. Thus, it is unlikely that the presence of the resistance cassette in the homologous recombinant affects the ATF4−/− phenotype.
Genotyping of ATF4-deficient mice
DNA was extracted from 1-cm mouse tail clippings by using standard methods. Mice were genotyped by Southern blot analysis as described above or by PCR analysis. The primers used to amplify the targeted allele were a 5′ primer within the neo coding region (5′-ATATTGCTGAAGAGCTTGGCGGC-3′) and a 3′ primer from the 3′ homology region (5′-AGCCTGCCAGCCATTTTACATCCC-3′). The primers used to amplify the endogenous locus flanked the bZip region of ATF4 (5′ primer, 5′-AGC- AAAACAAGACAGCAGCCACTA-3′; and 3′ primer, 5′-ACTCTCTTC- TTCCCCCTTGCCTTA-3′).
Hematologic analysis of embryonic and adult blood
An incision was made in the extraembryonic membranes, with care taken to produce as little damage as possible to yolk sac blood vessels. The membranes were retracted but left attached to the placenta, and the uterine artery and vein were not severed. The embryo was dried with the tip of a Kimwipe (Kimberly-Clark, Dallas, TX). Blood was collected with a Microcaps micropipette (Drummond Scientific, Broomall, PA) from the carotid arteries of decapitated embryos. Blood smears were prepared by using the wedge technique followed by air drying and Wright-Giemsa staining. Reverse-phase high-performance liquid chromatography (HPLC) analysis of the globin chains was done on lysates of washed erythrocytes by using a Dynamax HPLC system (Rainin, Palo Alto, CA) with a Vydac C4 column as described previously.37 Individual globin chains were quantitated with Dynamax HPLC Method Manager software. In adult mice, blood was collected from the tail vein. Reticulocyte counts were determined by incubating blood in 100 ng/mL thiazole orange (Aldrich Chemical, Milwaukee, WI) for 15 minutes at 37°C and sorting on a fluorescence-activated cell-sorter machine (FACSCalibur; Becton Dickinson, Franklin Lakes, NJ) and analysis with CellQuest software (Becton Dickinson). Complete blood counts were done with a Hemavet 1500R automated hematology system (CDC Technologies, Oxford, CT).
In situ hybridization
Embryos from the mating of wild-type Black Swiss mice were removed at 11.5 dpc and fixed in 4% paraformaldehyde overnight at 4°C. Fixed embryos were then dehydrated and embedded in paraffin. Six-micron sections were cut and either stained with hematoxylin and eosin for histologic analysis or used for RNA in situ hybridization. In situ probes consisted of α-phosphorus 32-uridine triphosphate (UTP)–labeled full-length antisense murine ATF4 cDNA or the corresponding labeled sense control. In situ hybridization was done as described previously and was followed by counterstaining with hematoxylin.32
Hematopoietic colony assays
Single-cell suspensions from 15.5-dpc fetal livers were prepared in Iscoves modified medium supplemented with 2% fetal-calf serum (FCS). Total counts were determined by diluting cells in medium; nucleated cell counts were determined by diluting cells in 3% acetic acid with methylene blue. To quantitate colonies of erythroid colony-forming unit (CFU-Es), fetal-liver cells were plated in triplicate in 1 ml methylcellulose-based medium containing 150 U/mL recombinant erythropoietin (MethoCult M3334; Stem Cell Technologies, Vancouver, BC, Canada) at a density of 1.7 × 104nucleated fetal-liver cells/35-mm Petri dish. Cells were incubated in a fully humidified atmosphere with 5% carbon dioxide (CO2) at 37°C. The number of CFU-E colonies was determined after 36 to 48 hours of culture. Colonies of erythroid burst-forming units (BFU-Es), granulocyte-macrophage colony-forming units (CFU-GM), and granulocyte-erythrocyte-megakaryocyte-macrophage colony-forming units (CFU-GEMMs) were quantitated by plating 5.0 × 104nucleated fetal-liver cells in 1 ml complete methylcellulose medium (MethoCult M3434). BFU-E colonies were counted at 7 days of culture and CFU-GM and CFU-GEMM colonies at 10 and 14 days, respectively. Assays of bone marrow progenitors in adult mice were done as described above with the following changes. Cells were collected by flushing the marrow cavities of the tibias and femurs with medium, and the cells were filtered through sterile nylon mesh. The number of nucleated cells was determined, and cells were plated at densities of 3.0 × 104 and 1.5 × 104 nucleated cells per plate, respectively, for CFU-E assays and assays of BFU-E, CFU-GM, and CFU-GEMM.
Mouse embryonic fibroblast culture
For culture of mouse embryonic fibroblasts (MEFs), 15.5-dpc embryos were obtained from the mating of ATF4 heterozygous mice. The embryo heads were processed for genotyping, and the liver, heart, lungs, and spleen were discarded. The remainder of the embryo was minced and incubated in 0.25% trypsin at 4°C for 12 hours and for 20 minutes at 37°C, with periodic disruption by pipetting. Remaining aggregates were discarded and the fibroblasts collected by centrifugation. Cells were maintained in Dulbecco modified Eagle medium supplemented with 15% FCS in 5% CO2 at 37°C. At passage 2, the MEFs were replated on 60-mm plates at a density of 2 × 104 cells/plate. Every 24 hours, duplicate plates for each embryo were trypsinized, and the number of MEFs per plate was determined.
Statistical analysis
Statistical analysis was done with InStat software (GraphPad Software, San Diego, CA). Significance was determined by using the Kruskal-Wallis nonparametric analysis of variance and the Dunn multiple comparison test. All errors shown represent the SEM.
Results
Targeted inactivation of the ATF4 gene in mice
The murine ATF4 gene was disrupted by homologous recombination in both R1 and HM1 ES cells. The targeting vector was designed to replace the entire ATF4 coding region after the first codon, as well as the polyadenylation signal, with a neomycin resistance cassette (Figure 1A).
Electroporated ES cells were subjected to G418 and ganciclovir selection, and surviving clones were screened by PCR with a 3′ primer external to the homology region. Correct targeting was confirmed in these ES cells by Southern blot analysis of genomic DNA digested withEcoRV and probing of the blot with the internal 3′ probe indicated in Figure 1A. This resulted in the predicted 3.3-kb and 2.1-kb bands for the wild-type and homologous recombinant alleles, respectively (Figure 1B). HindIII digestion of DNA and probing of the blot with a fragment external to the 3′ homology confirmed the correct 3′ end of the homologous allele (data not shown). Similarly, the 5′ end of the homologous recombinant allele was verified by double restriction-enzyme digestion with XbaI andSalI enzymes followed by Southern blot analysis using the 5′ probe indicated in Figure 1A. Bands corresponding to wild-type (11.4-kb) and homologous recombinant (12.2-kb) alleles were observed as predicted (Figure 1C). Southern blotting analysis using the neomycin cassette probe detected only a single band, indicating that no random integration occurred in these clones (data not shown).
To verify that the entire coding region of ATF4 was deleted, we conducted PCR amplification with primers within the ATF4 coding region. No PCR product was observed in the homozygous deletion mutant (data not shown). One in 15 clones resistant to G418 and ganciclovir was found to be correctly targeted. Euploidy was verified for 2 R1 and 2 HM1 targeted ES-cell clones, and these were injected into 3.5-dpc mouse blastocysts. High-percentage chimeras were generated and bred to Black Swiss mice. Heterozygous ATF4 mutant offspring were obtained from chimeras of both the R1 and the HM1 ES-cell line.
Heterozygous mice were phenotypically normal. In particular, they had no significant changes in embryonic or adult hematopoiesis or eye formation. Heterozygous mice were intercrossed to obtain homozygous mice. These offspring were genotyped using PCR analysis, and findings were confirmed by Southern blot analysis. The expected Mendelian frequency of genotypes was observed at 17.5 dpc. Some neonatal deaths occurred; the ratio of wild-type to heterozygous to homozygous offspring at weaning was 1:2:0.4 (numbers of mice, 41:81:17). Homozygous pups generally died during the first hour after birth, although excess mortality occurred throughout the first 3 weeks of life.
Most homozygous males were infertile. Among those that were fertile, the rate at which they impregnated females was greatly decreased, and they had a reduced period of fertility. Homozygous females were infertile despite having grossly normal ovaries and histologically normal ovarian follicles. When bred to wild-type males, homozygous females did form cervical mucus plugs indicating successful mating, although at a rate lower than normal. However, no homozygous female carried a litter to term and few supported pregnancies to 6.5 dpc.
ATF4−/− embryos are anemic
Primitive erythrocytes are produced initially in the extraembryonic blood islands of the yolk sac and are characterized by the presence of a nucleus and expression of the embryonic forms of the β-globin chains. From 12.5 to 16.5 dpc, the fetal liver is the predominant site of erythropoiesis. The definitive (adult) erythrocytes produced in the fetal liver undergo enucleation before release into the circulation and express the adult β chain. Finally, erythropoiesis shifts to the bone marrow and spleen, where it remains in adults.
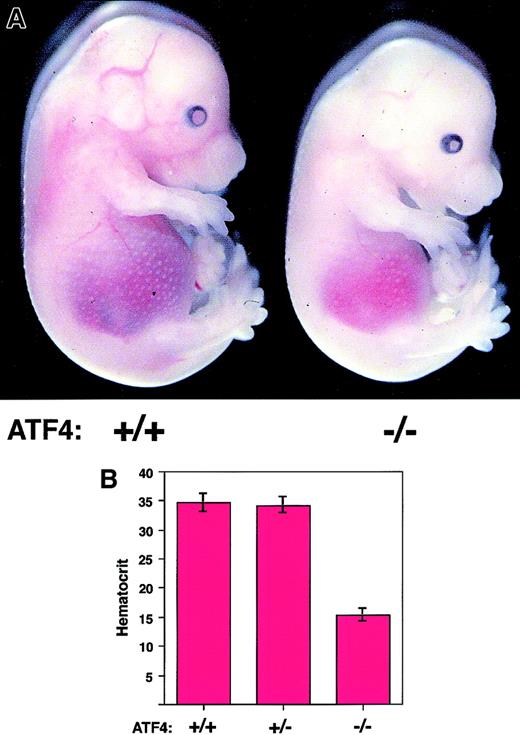
From 13.5 to 16.5 dpc, ATF4−/− embryos in this study were considerably paler than their littermates (Figure2A). To determine whether this was due to anemia, hematocrit values for viable embryos were obtained. At 13.5 dpc, the average hematocrit of ATF4−/− embryos was decreased 1.4 fold compared with that of their littermates. Mean hematocrit values of homozygous embryos were 2.2-fold lower at 14.5 and 15.5 dpc and 1.6-fold lower at 16.5 dpc than those of their littermates (Table 1 and Figure 2B). All decreases in hematocrit from 13.5 to 16.5 dpc were significant (P < .0001). Thus, during the period of fetal-liver hematopoiesis, ATF4−/− embryos were severely anemic.
ATF4−/− embryos are anemic.
(A) Gross appearance of 15.5-dpc embryos. The ATF4−/−embryo (right) is paler and smaller than a wild-type littermate (left). The pupils of ATF4−/− embryos are smaller. The gross appearance is otherwise normal. (B) Hematocrit values for ATF4+/+ (n = 35), ATF4+/− (n = 51), and ATF4−/− (n = 29) 15.5-dpc embryos. Error bars represent SEM. The mean hematocrit values for ATF4−/− 15.5-dpc embryos were significantly lower (P < .0001) than those for either ATF4+/+ or ATF4+/−embryos.
ATF4−/− embryos are anemic.
(A) Gross appearance of 15.5-dpc embryos. The ATF4−/−embryo (right) is paler and smaller than a wild-type littermate (left). The pupils of ATF4−/− embryos are smaller. The gross appearance is otherwise normal. (B) Hematocrit values for ATF4+/+ (n = 35), ATF4+/− (n = 51), and ATF4−/− (n = 29) 15.5-dpc embryos. Error bars represent SEM. The mean hematocrit values for ATF4−/− 15.5-dpc embryos were significantly lower (P < .0001) than those for either ATF4+/+ or ATF4+/−embryos.
Hematologic variables in mice embryos, according to genotype
| Variable and age (dpc) . | Genotype . | Fold difference from ATF4−/− values . | |||
|---|---|---|---|---|---|
| ATF4+/+ . | ATF4+/− . | ATF4−/− . | ATF4+/+ . | ATF4+/− . | |
| Hematocrit, % | |||||
| 13.5 | 0.2 ± 0.1 | 0.21 ± 0.1 | 0.15 ± 0.03 | 1.4 L | 1.4 L |
| 14.5 | 0.22 ± 0.02 | 0.22 ± 0.2 | 0.1 ± 0.01 | 2.1 L | 2.2 L |
| 15.5 | 0.35 ± 0.02 | 0.34 ± 0.01 | 0.15 ± 0.01 | 2.3 L | 2.2 L |
| 16.5 | 0.44 ± 0.04 | 0.47 ± 0.02 | 0.28 ± 0.02 | 1.6 L | 1.7 L |
| 17.5 | 0.42 ± 0.02 | 0.42 ± 0.02 | 0.43 ± 0.02 | 1.0 | 1.0 |
| 18.5 | 0.54 ± 0.01 | 0.5 ± 0.01 | 0.45 ± 0.03 | 1.2 L | 1.1 L |
| Newborn | 0.51 ± 0.01 | 0.51 ± 0.01 | 0.49 ± 0.02 | 1.0 | 1.0 |
| Nucleated blood cells, % | |||||
| 15.5 | 6.8 ± 0.8 | 6.4 ± 0.9 | 14.2 ± 1.5 | 2.1 H | 2.2 H |
| Embryonic α, % | |||||
| 15.5 | 5.1 ± 0.6 | 3.9 ± 0.8 | 10.7 ± 1.6 | 2.1 H | 2.7 H |
| Embryonic β, % | |||||
| 15.5 | 14.9 ± 2.9 | 12.0 ± 1.7 | 32.3 ± 3.7 | 2.2 H | 2.7 H |
| Variable and age (dpc) . | Genotype . | Fold difference from ATF4−/− values . | |||
|---|---|---|---|---|---|
| ATF4+/+ . | ATF4+/− . | ATF4−/− . | ATF4+/+ . | ATF4+/− . | |
| Hematocrit, % | |||||
| 13.5 | 0.2 ± 0.1 | 0.21 ± 0.1 | 0.15 ± 0.03 | 1.4 L | 1.4 L |
| 14.5 | 0.22 ± 0.02 | 0.22 ± 0.2 | 0.1 ± 0.01 | 2.1 L | 2.2 L |
| 15.5 | 0.35 ± 0.02 | 0.34 ± 0.01 | 0.15 ± 0.01 | 2.3 L | 2.2 L |
| 16.5 | 0.44 ± 0.04 | 0.47 ± 0.02 | 0.28 ± 0.02 | 1.6 L | 1.7 L |
| 17.5 | 0.42 ± 0.02 | 0.42 ± 0.02 | 0.43 ± 0.02 | 1.0 | 1.0 |
| 18.5 | 0.54 ± 0.01 | 0.5 ± 0.01 | 0.45 ± 0.03 | 1.2 L | 1.1 L |
| Newborn | 0.51 ± 0.01 | 0.51 ± 0.01 | 0.49 ± 0.02 | 1.0 | 1.0 |
| Nucleated blood cells, % | |||||
| 15.5 | 6.8 ± 0.8 | 6.4 ± 0.9 | 14.2 ± 1.5 | 2.1 H | 2.2 H |
| Embryonic α, % | |||||
| 15.5 | 5.1 ± 0.6 | 3.9 ± 0.8 | 10.7 ± 1.6 | 2.1 H | 2.7 H |
| Embryonic β, % | |||||
| 15.5 | 14.9 ± 2.9 | 12.0 ± 1.7 | 32.3 ± 3.7 | 2.2 H | 2.7 H |
Values are means ± SEM. The age and numbers of mice of each genotype (+/+:+/−:−/−) assayed to determine the hematocrit values were as follows: 13.5 days postcoitus (dpc), 5:3:6; 14.5 dpc, 11:14:8; 15.5 dpc, 35:51:29; 16.5 dpc, 7:15:3; 17.5 dpc, 7:20:6; 18.5 dpc, 16:25:8; and newborn, 40:31:14. The percentages of nucleated blood cells and embryonic β chains were determined from 12:15:14 and 3:4:4 embryos, respectively, at 15.5 dpc.
L indicates lower; H, higher.
ATF4−/− embryos have elevated levels of primitive erythrocytes
In ATF4−/− embryos at 15.5 dpc, the percentage of nucleated erythrocytes was increased 2.1 fold compared with that in wild-type embryos (Table 1). The morphologic features of nucleated erythrocytes from homozygous embryos were the same as those from wild-type controls (Figure 3A). Because cellular and nuclear volumes of primitive erythrocytes are closely correlated with developmental age38 and presumably the length of time the erythrocytes have been in circulation, this similarity in morphologic characteristics suggests that these nucleated erythrocytes represent primitive yolk sac–derived erythrocytes that have persisted in circulation rather than ongoing yolk sac hematopoiesis. HPLC analysis of lysates of peripheral blood obtained from 15.5-dpc embryos was done to determine the level of embryonic α (ζ) and β globin (βh1 and εy) relative to adult α- and β-globin chains. In the homozygous 15.5-dpc embryos, the percentage of embryonic α-globin chains was increased 2.1 fold and the percentage of embryonic β-globin chains was increased 2.2 fold compared with values in their wild-type littermates (Table 1 and Figure3B). Thus, the relative increase in the number of nucleated erythrocytes in homozygous mutants resulted from persistence of primitive yolk sac–derived erythrocytes rather than premature release of nucleated definitive erythrocytes into circulation due to anemia, that is, stress erythropoiesis.
Primitive erythrocytes persist in the circulation of 15.5-dpc ATF4−/− embryos.
(A) Blood smears from 15.5-dpc embryos stained with Wright-Giemsa stain. The blood smear from a ATF4−/− embryo (right) shows a greater than 2-fold increase in the percentage of nucleated erythrocytes compared with the sample from its wild-type littermate (left). Magnification 1000 ×. (B) HPLC analysis of globin chains. Blood from the 15.5-dpc ATF4−/−embryo (right) had an increased level of embryonic globin chains (βh1, εy, and ζ) compared with blood from its wild-type littermate (left). The percentages of α globin chains that were embryonic (ζ) were 10.7% ± 1.6% in the ATF4−/− embryo and 3.9% ± 0.8% and 5.1% ± 0.6%, respectively, in the wild-type and heterozygous embryos. The percentages of β globin chains that were embryonic (βh1 and εy) were 32.3% ± 3.7% in the ATF4−/−embryo and 14.9% ± 2.9% and 12.0% ± 1.7%, respectively, in the wild-type and heterozygous embryos.
Primitive erythrocytes persist in the circulation of 15.5-dpc ATF4−/− embryos.
(A) Blood smears from 15.5-dpc embryos stained with Wright-Giemsa stain. The blood smear from a ATF4−/− embryo (right) shows a greater than 2-fold increase in the percentage of nucleated erythrocytes compared with the sample from its wild-type littermate (left). Magnification 1000 ×. (B) HPLC analysis of globin chains. Blood from the 15.5-dpc ATF4−/−embryo (right) had an increased level of embryonic globin chains (βh1, εy, and ζ) compared with blood from its wild-type littermate (left). The percentages of α globin chains that were embryonic (ζ) were 10.7% ± 1.6% in the ATF4−/− embryo and 3.9% ± 0.8% and 5.1% ± 0.6%, respectively, in the wild-type and heterozygous embryos. The percentages of β globin chains that were embryonic (βh1 and εy) were 32.3% ± 3.7% in the ATF4−/−embryo and 14.9% ± 2.9% and 12.0% ± 1.7%, respectively, in the wild-type and heterozygous embryos.
The fetal liver is pale and hypoplastic in ATF4−/−mouse embryos
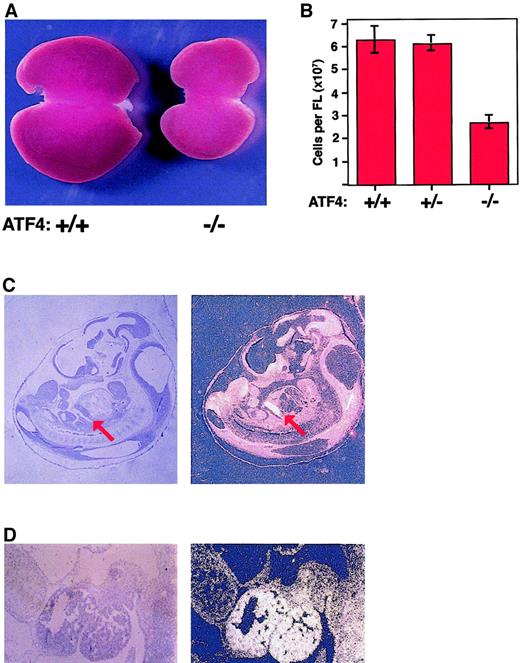
The livers of 15.5-dpc homozygous embryos were considerably smaller and paler than those of their littermates (Figure4A). The gross morphologic features of homozygous fetal livers were normal, and sections stained with hematoxylin and eosin showed normal cellular architecture. However, far fewer definitive erythrocytes were visible in the hepatic vasculature, a finding consistent with the low hematocrit values and decreased numbers of definitive erythrocytes in peripheral blood smears. Touch and cytospin preparations of homozygous fetal livers showed a decrease in the number of enucleated cells compared with controls and an increase in the number of more primitive erythroid precursors. Terminal deoxynucleotidyl transferase–mediated deoxy-UTP end-labeling assays detected no increase in apoptosis in mutant fetal livers (data not shown).
ATF4−/− fetal livers are hypoplastic, and the ATF-4 gene is expressed at high levels in wild-type fetal livers.
(A) Gross appearance of livers from wild-type mice (left) and ATF4−/− littermates (right) at 15.5 dpc. The liver from the homozygous mutant mouse is smaller and paler. (B) The mean total number of cells in the fetal liver of ATF4−/− mice at 15.5 dpc was significantly lower than that in littermates. The mean number of cells in ATF4−/− fetal liver at 15.5 dpc was 2.70 × 107 ± 0.29 × 107 (n = 15). The values for ATF4+/+ (n = 13) and ATF4 +/−(n = 19) embryos were 6.31 × 107 ± 0.59 × 107 and 6.15 × 107 ± 0.35 × 107, respectively. Thus, ATF4−/− embryos had a decrease in cells of 2.3 fold and 2.1 fold, respectively, compared with values in ATF4+/− and ATF4+/+ embryos (P < .0001 for both comparisons). There was no significant difference between ATF4+/+ and ATF4+/− embryos. Error bars represent SEM. (C) In situ hybridization of a midsagittal section from a 11.5-dpc wild-type embryo with an antisense ATF4 cDNA probe (left, bright-field image; and right, dark-field image). Signal from the hybridization appears bright in the dark-field image. The red arrow indicates the position of the fetal liver. Magnification 10 ×. (D) Higher magnification of results from in situ hybridization for ATF4 in a sagittal section of fetal liver from a wild-type embryo. Magnification 100 ×.
ATF4−/− fetal livers are hypoplastic, and the ATF-4 gene is expressed at high levels in wild-type fetal livers.
(A) Gross appearance of livers from wild-type mice (left) and ATF4−/− littermates (right) at 15.5 dpc. The liver from the homozygous mutant mouse is smaller and paler. (B) The mean total number of cells in the fetal liver of ATF4−/− mice at 15.5 dpc was significantly lower than that in littermates. The mean number of cells in ATF4−/− fetal liver at 15.5 dpc was 2.70 × 107 ± 0.29 × 107 (n = 15). The values for ATF4+/+ (n = 13) and ATF4 +/−(n = 19) embryos were 6.31 × 107 ± 0.59 × 107 and 6.15 × 107 ± 0.35 × 107, respectively. Thus, ATF4−/− embryos had a decrease in cells of 2.3 fold and 2.1 fold, respectively, compared with values in ATF4+/− and ATF4+/+ embryos (P < .0001 for both comparisons). There was no significant difference between ATF4+/+ and ATF4+/− embryos. Error bars represent SEM. (C) In situ hybridization of a midsagittal section from a 11.5-dpc wild-type embryo with an antisense ATF4 cDNA probe (left, bright-field image; and right, dark-field image). Signal from the hybridization appears bright in the dark-field image. The red arrow indicates the position of the fetal liver. Magnification 10 ×. (D) Higher magnification of results from in situ hybridization for ATF4 in a sagittal section of fetal liver from a wild-type embryo. Magnification 100 ×.
To quantitate the decrease in fetal-liver size, the number of cells in fetal livers was determined. The mean number of cells per liver in ATF4−/− 15.5-dpc embryos was decreased 2.3 fold, and the number of nucleated fetal-liver cells was decreased 2.1 fold compared with values in wild-type controls (Figure 4B and data not shown). The decreases in the number of fetal-liver cells in homozygous embryos were significant (P < .0001) compared with values in both wild-type and heterozygous embryos.
ATF4 is expressed at a high level in fetal livers of wild-type mice
ATF4 is expressed ubiquitously in adult mice and in many cell lines.2,3,6 Reverse transcriptase–PCR amplification detected ATF4 expression in murine blastocysts and ES cells (data not shown). In situ hybridization of 6.5- to 8.5-dpc embryos detected uniform expression throughout the embryo, in agreement with previous findings.39
If ATF4 is critical for fetal-liver hematopoiesis, one would expect the gene to be expressed in fetal livers of wild-type mice. In normal mice, the liver rudiment is first apparent at 9 dpc as an evagination of the gut into the septum transversum. Erythroblasts are first visible in the liver at 9 dpc, definitive hematopoiesis is detectable at 10 dpc, and the first long-term repopulating hematopoietic stem cells are observed at 11 dpc.40 41 This is associated with a rapid expansion of erythroid lineage cells in the fetal liver and a definitive adult pattern of globin expression.
In this study, in situ hybridization detected ATF4 expression throughout 11.5-dpc embryos but at a much higher level in the liver than in other tissues (Figure 4C and D). A high level of expression was also present in the developing eye, in agreement with previous results.19 No signal above background level was detected with the sense-oriented control probe (data not shown). The high level of ATF4 expression in fetal livers of wild-type mice is consistent with an important role for this protein in fetal-liver hematopoiesis.
Hematopoietic progenitor colonies in ATF4−/− 15.5-dpc embryos are smaller and fewer in number per fetal liver
Impairment of fetal-liver hematopoiesis could result from either an intrinsic defect in hematopoietic progenitors or failure of the ability of the fetal-liver microenvironment to support erythropoiesis. To characterize the erythropoietic defect in ATF4−/−embryos, we conducted in vitro progenitor assays. Compared with values in wild-type embryos, no significant difference was found in the number of colonies/105 nucleated fetal-liver cells, suggesting that a similar fraction of the fetal liver in ATF4−/−embryos and wild-type embryos is composed of hematopoietic cells. However, the number of CFU-E, BFU-E, and CFU-GM colonies per ATF4−/− fetal liver were reduced 2.2, 2.1, and 2.1 fold, respectively, and the number of CFU-GEMM colonies was reduced 2.9 fold compared with values in wild-type littermates (Figure5A). The ratio of BFU-Es to CFU-Es was normal in ATF4−/− embryos, indicating that there was no defect in the terminal differentiation of erythroid progenitors.
Hematopoietic progenitor colonies from ATF4−/− 15.5-dpc embryos are smaller and reduced in number per fetal liver.
(A) In vitro differentiation of 15.5-dpc fetal-liver cells from wild-type (n = 18), ATF4+/− (n = 19), and ATF4−/− (n = 22) embryos. The number of CFU-E, BFU-E, CFU-GM, and CFU-GEMM colonies per fetal liver were all significantly reduced in ATF4−/− embryos. There was no significant difference between the number of colonies in wild-type and ATF4+/− embryos. (B) Hematopoietic progenitor colonies. BFU-E colonies were photographed on day 8 and CFU-GEMM colonies on day 10 of in vitro differentiation. Magnification 100 ×. (C) The number of cells per plate and per colony after 12 days of in vitro differentiation of 5 × 104 nucleated 15.5-dpc fetal-liver cells. The average size of ATF4−/−hematopoietic progenitors was significantly smaller than those from wild-type and ATF4+/− embryos. There was no significant difference between the size of colonies from wild-type embryos and those from ATF4+/− embryos.
Hematopoietic progenitor colonies from ATF4−/− 15.5-dpc embryos are smaller and reduced in number per fetal liver.
(A) In vitro differentiation of 15.5-dpc fetal-liver cells from wild-type (n = 18), ATF4+/− (n = 19), and ATF4−/− (n = 22) embryos. The number of CFU-E, BFU-E, CFU-GM, and CFU-GEMM colonies per fetal liver were all significantly reduced in ATF4−/− embryos. There was no significant difference between the number of colonies in wild-type and ATF4+/− embryos. (B) Hematopoietic progenitor colonies. BFU-E colonies were photographed on day 8 and CFU-GEMM colonies on day 10 of in vitro differentiation. Magnification 100 ×. (C) The number of cells per plate and per colony after 12 days of in vitro differentiation of 5 × 104 nucleated 15.5-dpc fetal-liver cells. The average size of ATF4−/−hematopoietic progenitors was significantly smaller than those from wild-type and ATF4+/− embryos. There was no significant difference between the size of colonies from wild-type embryos and those from ATF4+/− embryos.
A striking decrease in colony size was observed for BFU-Es, CFU-GMs, and CFU-GEMMs (Figure 5B). To quantitate this reduction, the number of colonies, the number of cells per plate, and the number of cells per colony were determined at 10 days of culture in methylcellulose medium supplemented with growth factors that promote the growth of all myeloid lineages. There was no significant difference in the total number of colonies per plate. However, for fetal livers from ATF4−/− embryos, the number of cells per plate and the average number of cells per colony were reduced 5.8 and 5.9 fold, respectively, compared with control values (Figure 5C). These results suggest that the anemia in ATF4−/− embryos was due to a cell-autonomous defect in the ability of hematopoietic progenitors to proliferate in response to growth factors rather than to an abnormality in the fetal-liver microenvironment.
Adult ATF4−/− mice are mildly anemic and have smaller hematopoietic progenitor colonies
The mean hematocrit value in adult homozygous mutant mice was decreased 1.1 fold compared with the value in both wild-type and heterozygous controls (P < .05). Consistent with this mild anemia, complete blood counts showed a significant decrease in red blood cell count, hemoglobin concentration, and mean corpuscular hemoglobin in ATF4−/− adults compared with controls but no significant change in reticulocyte count or mean corpuscular hemoglobin concentration (Table 2). Interestingly, leukocyte and platelet counts of ATF4−/−mice were not significantly different from those of controls (Table 2and data not shown).
Hematologic variables in adult mice, according to genotype
| Variable . | Genotype . | ||
|---|---|---|---|
| ATF4+/+ . | ATF4+/− . | ATF4−/− . | |
| Hematocrit* | 0.45 ± 0.02 | 0.44 ± .01 | 0.40 ± .01 |
| Hemoglobin, g/L* | 129 ± 6 | 124 ± 4 | 103 ± 5 |
| RBC count, 1012/L* | 8.7 ± 0.3 | 8.5 ± 0.3 | 7.7 ± 0.3 |
| MCH, pg* | 14.4 ± 0.5 | 14.6 ± 0.3 | 13.3 ± 0.2 |
| MCHC, g/L | 302 ± 7 | 292 ± 6 | 290 ± 4 |
| MCV, fL* | 47.9 ± 1.0 | 49.7 ± 0.7 | 45.8 ± 0.8 |
| RDW* | 19.2 ± 0.7 | 19.3 ± 0.4 | 20.6 ± 0.3 |
| Reticulocytes | 0.02 ± 0.003 | 0.03 ± 0.002 | 0.03 ± 0.003 |
| WBC count, 109/L | 7.1 ± 0.7 | 8.8 ± 0.6 | 10.3 ± 1.0 |
| Variable . | Genotype . | ||
|---|---|---|---|
| ATF4+/+ . | ATF4+/− . | ATF4−/− . | |
| Hematocrit* | 0.45 ± 0.02 | 0.44 ± .01 | 0.40 ± .01 |
| Hemoglobin, g/L* | 129 ± 6 | 124 ± 4 | 103 ± 5 |
| RBC count, 1012/L* | 8.7 ± 0.3 | 8.5 ± 0.3 | 7.7 ± 0.3 |
| MCH, pg* | 14.4 ± 0.5 | 14.6 ± 0.3 | 13.3 ± 0.2 |
| MCHC, g/L | 302 ± 7 | 292 ± 6 | 290 ± 4 |
| MCV, fL* | 47.9 ± 1.0 | 49.7 ± 0.7 | 45.8 ± 0.8 |
| RDW* | 19.2 ± 0.7 | 19.3 ± 0.4 | 20.6 ± 0.3 |
| Reticulocytes | 0.02 ± 0.003 | 0.03 ± 0.002 | 0.03 ± 0.003 |
| WBC count, 109/L | 7.1 ± 0.7 | 8.8 ± 0.6 | 10.3 ± 1.0 |
Values are means ± SEM. The numbers of adult mice of each genotype (+/+:+/−:−/−) assayed to determine the hematocrit and reticulocyte values were 22:48:49 and 6:26:18, respectively.
RBC indicates red blood cell; MCH, mean corpuscular hemoglobin; MCHC, mean corpuscular hemoglobin concentration; MCV, mean corpuscular volume; RDW, red cell distribution width; and WBC, white blood cell.
Significant difference between ATF4−/− mice and heterozygous controls. There was no significant difference between ATF4+/+ and ATF+/− adults in any assay.
The erythropoietic defect in ATF4−/− adults was characterized further with use of in vitro progenitor assays. There was no significant difference between ATF4−/− adult mice and control adults in the number of CFU-E, BFU-E, CFU-GM, or CFU-GEMM colonies (data not shown). However, the colony size was greatly reduced, with the mean number of cells per plate and cells per progenitor colony both reduced 8 fold and 7.5 fold compared with values in wild-type and heterozygous controls, respectively (data not shown). Thus, the defect in hematopoietic progenitor proliferation persisted in adult ATF4−/− mice.
Primary MEFs from ATF4−/− 15.5-dpc embryos have a proliferation defect
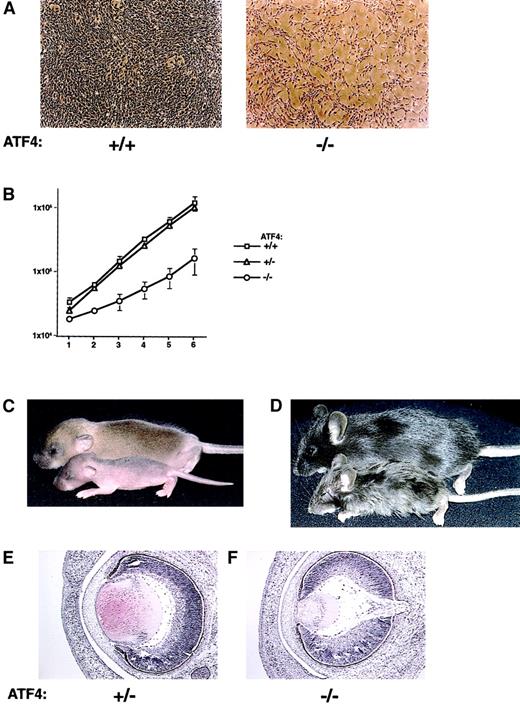
To determine whether the defect in hematopoietic progenitor proliferation represented a failure in normal cellular proliferation, the proliferation rate of primary MEFs was determined. MEFs from 15.5-dpc embryos from heterozygous matings were plated at a density of 2 × 104 cells/60-mm plate. The number of cells per plate was determined every 24 hours. ATF4−/− fibroblast cultures were considerably less dense at each time point despite having been plated at the same initial concentration as wild-type and heterozygous cultures (Figure6A). ATF4−/− fibroblasts divided at a greatly reduced rate, showing a mean doubling time of 45.2 hours, a 2.1-fold increase compared with the doubling time of 21.9 hours observed for both wild-type and heterozygous embryos (Figure 6B). ATF4−/− MEFs became contact inhibited at a density similar to those of controls and did not reach the end of their proliferative capacity sooner than those of wild-type controls (data not shown). These results suggest that the homozygous cells did not prematurely senesce and that ATF4−/− cells had a cell-autonomous defect in normal cellular proliferation.
Loss of ATF4 activity affects primary MEF proliferation, postnatal growth, and embryonic lens formation.
(A) In vitro proliferation defect in primary MEFs from ATF4−/− embryos. Passage-2 MEFs derived from 15.5-dpc embryos were plated at a density of 2 × 104 cells/60-mm plate. Representative regions at 5 days of culture of cells from wild-type (left) and ATF4−/− (right) embryos are shown. Magnification 40 ×. (B) Quantitation of the MEF growth rate from 15.5-dpc embryos. Fibroblasts were derived from ATF4+/+(n = 3), ATF4+/− (n = 7), and ATF4−/−(n = 5) 15.5-dpc embryos. Error bars represent SEM. (C) Seven-day-old ATF4+/+ and ATF4−/− mice. The ATF4−/− mouse (bottom) is smaller and shows a delay in hair growth compared with its wild-type littermate (top). (D) ATF4+/− and ATF4−/− littermates 4 weeks after birth. The ATF4−/− mouse (bottom) is runted and has severe microphthalmia. The heterozygote (top) is normal in appearance. (E) Histologic study of lens from 16-dpc embryos. The lens from the ATF4−/− embryo is considerably smaller and consists of primary lens fiber cells with no apparent formation of secondary lens fiber cells. Magnification 100 ×.
Loss of ATF4 activity affects primary MEF proliferation, postnatal growth, and embryonic lens formation.
(A) In vitro proliferation defect in primary MEFs from ATF4−/− embryos. Passage-2 MEFs derived from 15.5-dpc embryos were plated at a density of 2 × 104 cells/60-mm plate. Representative regions at 5 days of culture of cells from wild-type (left) and ATF4−/− (right) embryos are shown. Magnification 40 ×. (B) Quantitation of the MEF growth rate from 15.5-dpc embryos. Fibroblasts were derived from ATF4+/+(n = 3), ATF4+/− (n = 7), and ATF4−/−(n = 5) 15.5-dpc embryos. Error bars represent SEM. (C) Seven-day-old ATF4+/+ and ATF4−/− mice. The ATF4−/− mouse (bottom) is smaller and shows a delay in hair growth compared with its wild-type littermate (top). (D) ATF4+/− and ATF4−/− littermates 4 weeks after birth. The ATF4−/− mouse (bottom) is runted and has severe microphthalmia. The heterozygote (top) is normal in appearance. (E) Histologic study of lens from 16-dpc embryos. The lens from the ATF4−/− embryo is considerably smaller and consists of primary lens fiber cells with no apparent formation of secondary lens fiber cells. Magnification 100 ×.
ATF4−/− mice have growth retardation and microphthalmia
We found that ATF4−/− embryos were smaller than their wild-type and heterozygous littermates. From 13.5 dpc through birth, the mean length of ATF4−/− embryos was reduced 1.1 fold and the mean weight was reduced 1.3-fold compared with control values. After birth, the weight of ATF4−/− mice remained lower than that of control mice. At 3 weeks of age, the difference in weight was maximal: the weight of ATF4−/− mice was 2.5-fold lower than that of controls. The weight of ATF4−/− adult mice ultimately remained approximately 1.7-fold lower than that of controls.
Hair growth also appeared to be delayed in ATF4−/− mice. In wild-type and heterozygous pups, hair growth had resulted in darkened skin by the first day after birth, whereas homozygous mice remained pink through the third day after birth. By one week of age, homozygous pups had only fine sparse hair, which was distinct from the normal coat of wild-type and heterozygous littermates (Figure 6C). The delay in hair growth resolved by adulthood, although the fur of many homozygous mice had a ruffled appearance (Figure 6D).
Adult ATF4 homozygous mutants were severely microphthalmic, with no recognizable lens, anterior chamber, iris, or vitreous body. Development of the eye in ATF4−/− embryos was normal until 14.5 dpc, when the lens normally undergoes rapid cellular proliferation and enlargement, with formation of secondary lens fiber cells. From 14.5 to 16.0 dpc, the lens in ATF4−/− embryos failed to increase in size and did not develop cells with the morphologic characteristics of secondary lens fiber cells. By 16.0 dpc, vacuolization was visible in the lens fiber cells, with the anterior region of the lens particularly affected (Figure 6E). Subsequent cell death resulted in complete degeneration of the lens by adulthood. These observations are in agreement with previous results.19,27The changes in the eye were likely due to failure of proper lens formation, since the phenotype closely approximates that observed in genetic ablation of lens fiber cells.42-45
The phenotypic changes in ATF4−/− mice may have been caused by a requirement for ATF4 for normal proliferation in the affected tissues, as we observed in the in vitro cultures of ATF4−/− MEFs. The rapidly proliferating cells in the fetal liver, developing lens, and hair follicle may be particularly sensitive to the lack of ATF4. Also, ATF4 may have a cell-specific function in each of these tissues.
Discussion
In studies in mice, we found that homozygous inactivation of the ATF4 gene results in severe fetal anemia. The hematocrit of 15.5-dpc mutant fetuses was 0.15, whereas that in controls was 0.35. The percentage of nucleated erythrocytes was increased 2 fold at this stage of development, and this increase was accompanied by an increase in embryonic globin chains, indicating that these cells represented persistence of primitive yolk sac–derived erythrocytes rather than premature release of nucleated definitive erythrocytes into the circulation.
At 15.5 dpc, fetal livers in homozygous mutant fetuses were paler and much smaller than those in controls. Because most of the fetal liver at this stage is composed of erythroid elements, this finding also indicates the presence of a defect in fetal-liver hematopoiesis. In situ hybridization studies in wild-type mice showed that ATF4 is expressed at a high level in the fetal liver, a result consistent with an important role for ATF4 in fetal-liver hematopoiesis.
In vitro methylcellulose colony assays of ATF4−/−fetal-liver cells showed a greater than 2-fold reduction in the number of hematopoietic progenitors per fetal liver and a 5.9-fold reduction in colony size. Because exogenous growth factors provided in the medium did not prevent this defect, these results suggest that ATF4 is essential, in a cell-autonomous manner, for normal fetal-liver hematopoiesis.
ATF4 inactivation resulted in severe fetal anemia but relatively normal steady-state erythropoiesis in adults. This phenotype has been described in other mouse mutants, such as the “flexed,” E2F4, and signal transducer and activator of transcription (STAT) 5a−/−STAT5b−/− mutants.46-48Inactivation of E2F4, the predominant E2F family member, resulted in a phenotype of runted, pale embryos with a transient fetal anemia. Hematocrit values of E2F4−/− embryos were decreased 30% to 40% from 13.5 to 18.5 dpc, whereas adults were not anemic and had normal reticulocyte counts. Unlike ATF4 knockouts, E2F4 embryos had an increased level of apoptosis in cells of the hematopoietic lineage as well as changes in erythrocyte morphologic features.46Importantly, a 2- to 3-fold increase in the number of fetal-liver erythroid progenitors (CFU-Es and BFU-Es) was found in E2F4−/− embryos, and this represented an adaptive response to the anemia. No similar response was observed in ATF4−/− embryos despite the presence of severe anemia. In addition, unlike inactivation of ATF4, inactivation of E2F4 did not affect the proliferation of MEFs in culture, and E2F4 was found to be completely dispensable in the control of cellular proliferation.
STAT5a−/−STAT5b−/− embryos were severely anemic at 13.5 dpc, with hematocrit values reduced to 60% of those in wild-type littermates. Like ATF4−/− fetuses, STAT5a−/−STAT5b−/− mutants had a higher percentage of nucleated embryonic erythrocytes. In contrast to findings in ATF4−/− mice, more adult-type nucleated erythroblasts were present in the fetal circulation of STAT5 homozygous mutants than in controls, a result consistent with an adaptive response to anemia. STAT5a−/−STAT5b−/− embryos also had a reduced number of fetal-liver erythroid progenitors. However, unlike ATF4−/− embryos, STAT5 null embryos had a reduced ratio of CFU-Es to BFU-Es, indicating the presence of a reduction in the net growth of erythroid progenitors during terminal differentiation.47 As expected from the role of STAT5 in erythropoietin receptor signaling, a higher level of apoptosis was observed in STAT5a−/−STAT5b−/− fetal livers. In contrast, no increase in apoptosis was detected in fetal livers of ATF4−/− embryos.
Although ATF4−/− mice embryos were severely anemic, adult homozygous mice had only a mild microcytic anemia, with a mean hematocrit of 0.4, whereas that in controls was 0.44. Complete blood counts in homozygous mice revealed small but significant reductions in hemoglobin, erythrocyte count, mean corpuscular hemoglobin, and mean corpuscular volume and an increase in red cell distribution width. The reticulocyte count was not significantly different from values in controls, indicating that homozygous mutants did not have an increase in erythrocyte production in response to the anemia. There was no significant difference between homozygous mutants and controls in leukocyte or platelet counts, suggesting that erythropoiesis was more severely affected than other components of hematopoiesis, possibly because of the larger number and more rapid turnover of erythrocytes. In progenitor assays with bone marrow cells from adult mice, no differences in the numbers of each colony type were found, but a severe defect in the ability of hematopoietic progenitors to proliferate, similar to that observed in progenitor assays of fetal-liver cells, was observed.
The greater severity of anemia in embryos compared with adults was most likely due to the low level of hematopoietic reserve capacity during the fetal-liver stage of hematopoiesis. Adults have considerable erythropoietic reserve capacity, and the rate of erythropoiesis can be up-regulated 10 fold in response to stress such as hemorrhage or anemia.49 The rate of fetal erythropoiesis is several-fold higher than the steady-state erythropoietic rate in adults.50,51 During the period of fetal-liver hematopoiesis, the developing embryo has little hematopoietic reserve capacity.50-52 Thus, anemias during this stage may reveal factors required for maximal levels of hematopoiesis that are unnecessary for normal levels of adult hematopoiesis. Adult mice with such defects may be deficient in response to erythropoietic stress. Interestingly, adult ATF4−/− mice appeared to be more sensitive to the hemolytic agent phenylhydrazine in preliminary studies.
Several other proteins, including XBP-1, Rb-1, and c-myb, are required for normal fetal-liver hematopoiesis; and homozygous inactivation of each results in pale, runted embryos with fetal-liver hypoplasia, a relative increase in the number of nucleated erythrocytes, and a severe anemia at 15.5 dpc caused by inhibition of definitive erythropoiesis.53-55 Like ATF4, these proteins are expressed in many tissues but at the highest level in fetal liver. Unlike inactivation of ATF4, homozygous inactivation of these proteins results in embryonic death between 13.5 and 15.5 dpc. As was observed in ATF4−/− mice, in vitro differentiation of Rb−/− and c-myb−/− fetal-liver cells resulted in a decrease in the number of colonies per fetal liver in homozygous mutants.56-58 Interestingly, mice chimeric for wild-type and homozygously inactivated Rb do not undergo embryonic death but have lens abnormalities due to severely disrupted formation of lens fiber cells.59 It has been suggested that c-myb is also critical for proliferation of several different cell types, including fibroblasts, in addition to its role in hematopoiesis.60 Thus, inactivation of these genes and ATF4 may affect similar downstream target genes.
We previously isolated ATF4 in a yeast 2-hybrid screen by using LCR-F1 as bait. A knockout mutation of LCR-F1 resulted in severe fetal anemia in the C57BL/6J inbred mouse strain33 (unpublished data, August 1999). In both LCR-F1−/− and ATF4−/− fetuses, hematocrit levels were reduced approximately 2 fold at 15.5 dpc, fetal livers were pale, and the number of hematopoietic progenitors per fetal liver was reduced by 2 fold. These results suggest that ATF4 and LCR-F1 heterodimerize and regulate genes that are essential for normal fetal-liver hematopoiesis.
In addition to hematologic defects, ATF4-deficient embryos had defects in lens formation, postnatal hair growth, and body size. ATF4 may play a distinct role in each of the developmental pathways involved in these features. However, it is most probable that a defect in proliferation links all of the components of this phenotype, because the fetal liver, embryonic lens, and hair follicle are all sites of rapid proliferation. Consistent with this hypothesis, primary fibroblasts from ATF4−/− embryos had a greatly reduced rate of in vitro proliferation, with a doubling time 2-fold greater than that of controls. This in vitro proliferation defect, the increased expression of ATF4 in the rapidly proliferating fetal liver and lens, and the short in vivo half-life of the ATF4 protein6 are consistent with a critical role for ATF4 in the regulation of cellular proliferation—for example, through activating or repressing expression of genes involved in cell-cycle regulation. Importantly, the level of ATF4 protein increases after growth stimulation of quiescent cells, with maximal levels occurring during late G1, and ATF4 affects cyclin A promoter activity in transient-transfection assays.61 Interestingly, ATF4 expression and activity are induced in breast cancer cells by the growth factor heregulin β1.62
No increase in apoptosis has been observed in the fetal livers of ATF4−/− mice, but apoptosis was found to be prominent in the developing lens in such mice.19,27 ATF4 may function normally in lens fiber cells to prevent apoptosis. Alternatively, the failure to form a correctly sized and patterned lens at the proper developmental stage may lead to induction of apoptosis. Thus, the increase in apoptosis observed in the developing lens may be the result of improper formation of this structure rather than its cause. Consistent with a role for ATF4 in the regulation of proliferation, overexpression of this protein in the developing lens results in hyperproliferation of lens fiber cells.19Similarly, although the embryonic lens in double homozygous p53/ATF4−/− mice does not undergo apoptosis, the lens in adult ATF4−/− mice remains smaller and contains fewer lens fiber cells than the lens in controls.27 The relative restriction of cell types affected in embryos is most likely due to the ability of other genes to compensate in vivo for the loss of ATF4 activity or to the differential sensitivity of tissues to a decreased rate of cellular proliferation.
In summary, a knockout mutation of ATF4 resulted in severe fetal anemia. At 15.5 dpc, the hematocrit of ATF4 null fetuses was 0.15, whereas that in controls was 0.35. Fetal livers were pale and hypoplastic, and the number of hematopoietic progenitors of multiple lineages was decreased more than 2 fold. In addition to the reduction in progenitor numbers, the size of BFU-E, CFU-GM, and CFU-GEMM colonies was dramatically reduced. These results suggest that ATF4 is essential for the normal, high-level proliferation required for fetal-liver hematopoiesis.
We thank Jin-Xiang Ren for all ES-cell injections, Dr Vladimir Divoky for assistance with hematopoietic colony assays, Dr Thomas Ryan for assistance with hematologic assays, and Dr Peter Detloff and members of the Townes laboratory for critical review of the manuscript.
Supported in part by grant HL35559 from the National Institutes of Health.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Tim M. Townes, Dept of Biochemistry, University of Alabama at Birmingham, 537 Kaul Genetics Bldg, 720 20th St S, Birmingham, AL 35294; e-mail: ttownes@uab.edu.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal