Abstract
Given that the FcγRIIIa receptor 158V allotype displays a higher affinity for human immunoglobulin G1 and increased antibody-dependent cellular cytotoxicity, the aim of this study was to determine the influence of that FCGR3A polymorphism on the therapeutic response to rituximab, an anti-CD20 humanized immunoglobulin G1 increasingly used in the treatment of non-Hodgkin lymphomas. TheFCGR3A-158V/F genotype was determined in 49 patients having received rituximab for a previously untreated follicular non-Hodgkin lymphoma. The clinical response and the disappearance of theBCL2-JH gene rearrangement in both peripheral blood and bone marrow were evaluated at 2 months (M2) and at 1 year (M12). The study population consisted of 20% FCGR3A-158V homozygous patients, 35% FCGR3A-158F homozygous patients, and 45% heterozygous patients (FCGR3A-158F carriers). The objective response rates at M2 and M12 were 100% and 90%, respectively, inFCGR3A-158V homozygous patients compared with 67% (P = .03) and 51% (P = .03), respectively, in FCGR3A-158F carriers. A disappearance of theBCL2-JH gene rearrangement in both peripheral blood and marrow was observed at M12 in 5 of 6 of homozygousFCGR3A-158V patients compared with 5 of 17 ofFCGR3A-158F carriers (P = .03). The homozygous FCGR3A-158V genotype was confirmed to be the single parameter associated with clinical and molecular responses by multivariate analysis. This study showed an association between theFCGR3A genotype and clinical and molecular responses to rituximab. This finding will certainly give rise to new pharmacogenetic approaches to the management of patients with non-Hodgkin lymphomas.
Introduction
Rituximab (Mabthera, Rituxan) is a chimeric anti-CD20 immunoglobulin G1 (IgG1) monoclonal antibody consisting of human γ1 and κ constant regions linked to murine variable domains.1 Over the last few years, rituximab has considerably modified the therapeutic strategy for B lymphoproliferative malignancies, particularly non-Hodgkin lymphomas (NHLs). Rituximab, alone or in combination with chemotherapy, was shown to be effective in the treatment of both low-intermediate2-8 and high-grade NHL.6,9Unfortunately, 30% to 50% of patients with low-grade NHL exhibit no clinical response to rituximab.4,5 It has been suggested that the level of CD20 expression on lymphoma cells,2 the presence of high tumor burden at the time of treatment,6or low serum rituximab concentrations2 10 may explain the lack of efficacy of rituximab in some patients. Nevertheless, the actual causes of treatment failure remain largely unknown.
In vitro studies suggest that rituximab induces lymphoma cell lysis in vitro through antibody-dependent cell-mediated cytotoxicity (ADCC),11,12 complement-dependent cytotoxicity,11,13,14 or direct signaling leading to apoptosis.15,16 ADCC is an important effector mechanism in the eradication of intracellular pathogens and tumor cells. It requires leukocyte receptors for the Fc portion of IgG (FcγR), whose function is to link the IgG-sensitized antigens to FcγR-bearing cytotoxic cells and to trigger the cell activation mechanisms. The role of ADCC is still controversial,13,14 but the implication of FcγR in the antitumor effects of anti-CD20 antibodies against human lymphoma cell lines has been demonstrated in murine models.17-19Three classes of FcγR (FcγRI, FcγRII, and FcγRIII) and their subclasses are encoded by 8 genes in humans, all located on the long arm of chromosome 1. Some of those genes display a functional allelic polymorphism generating allotypes with different receptor properties. Those polymorphisms have been identified as genetic factors that increase susceptibility to autoimmune or infectious diseases.20-22 One of those genetic factors is a gene dimorphism in FCGR3A, which encodes FcγRIIIa with either a phenylalanine (F) or a valine (V) at amino acid position 158.23,24 This residue directly interacts with the lower hinge region of IgG1, as recently shown by IgG1-FcγRIII cocrystallization.25 It has been clearly demonstrated that human IgG1 binds more strongly to homozygous FcγRIIIa-158V natural killer (NK) cells than to homozygous FcγRIIIa-158F or heterozygous NK cells.23,24 Because FcγRIIIa is expressed on both NK cells and macrophages, which are the most important natural cytotoxic effectors, we have formulated the hypothesis that FCGR3Agene dimorphism may influence the response to rituximab. Genotyping ofFCGR3A was therefore performed on patients with previously untreated follicular NHL who had received rituximab alone, a particular situation in which the response rate is very high.5FCGR2A-131H/R was also determined as a control because that gene colocalizes with FCGR3A on chromosome 1q22 and encodes the macrophage FcγRIIa receptor.
Patients, materials, and methods
Patients and treatment
Clinical trial design, eligibility criteria, and end-point assessment have been previously reported.5 In brief, patients were eligible for inclusion in this study if they had previously untreated follicular CD20+ NHL according to the REAL classification.26 Patients were required to present with stage II to IV disease according to the Ann-Arbor classification and at least one measurable disease site. All patients were required to have low tumor burden according to the GELF criteria.27 A total of four 375 mg/m2 doses of rituximab (Roche, Neuilly, France) were administered by intravenous infusion (days 1, 8, 15, 22). The management of infusion and adverse events has already been reported.5 The study protocol was approved by an ethics committee, and all patients gave their informed consent.
Monitoring and end points
Baseline evaluation included clinical examination, chest x-ray, unilateral bone marrow biopsy, and computed tomography of the chest, abdomen, and pelvis. Response was assessed by an independent panel of radiologists who reviewed all the computed tomography scans of the included patients.
The primary efficacy end point was the objective response rate, ie, the proportion of patients achieving either complete remission (CR), unconfirmed CR (CRu), or partial response (PR) according to the criteria recently proposed by an international expert committee.28 Clinical response was evaluated at days 50 and 78. Only the maximum response was taken into account, and that assessment time point was named M2. All patients were evaluated for progression at 1 year (M12). Patients in CR or CRu with disappearance of bone marrow infiltration at M2 and reappearance of lymphoma cells in bone marrow at M12 were considered “progressive”; patients in PR with negative bone marrow biopsy at M2 and positive biopsy at M12 were considered in PR.
Molecular analysis of the BCL2-JH gene rearrangement was performed by polymerase chain reaction (PCR), as previously described,5 on a lymph node obtained at diagnosis and on both peripheral blood and bone marrow at diagnosis, M2, and M12.
FCGR3A-158V/F genotyping
Of the 50 patients included in the clinical trial, one patient was excluded after histologic review. Forty-nine patients were therefore available for FCGR3A genotype analysis. All samples were analyzed in the same laboratory, and the DNA was extracted using standard procedures. DNA was isolated from peripheral blood (n = 46) or bone marrow (n = 3). Genotyping ofFCGR3A-158V/F polymorphism was performed as described by Koene et al23 using a nested PCR followed by allele-specific restriction enzyme digestion. Briefly, 2FCGR3A-specific primers (5′-ATATTTACAGAATGGCACAGG-3′, 5′-GACTTGGTACCCAGGTTGAA-3′) (Eurobio, Les Ulis, France) were used to amplify a 1.2 kilobase fragment containing the polymorphic site. The initial PCR assay was performed with 1.25 μg genomic DNA, 200 ng of each primer, 200 μM of each deoxyribonucleoside triphosphate (dNTP) (MBI Fermentas, Vilnius, Lithuania), and 1 U Taq DNA polymerase (Promega, Charbonnière, France) as recommended by the manufacturer. This first PCR consisted of 10 minutes at 95°C, then 35 cycles (each consisting of steps at 95°C for 1 minute, 57°C for 1.5 minutes, and 72°C for 1.5 minutes), and 8 minutes at 72°C to achieve complete extension. The second PCR used primers (5′-ATCAGATTCGATCCTACTTCTGCAGGGGGCAT-3′, 5′-ACGTGCTGAGCTTGAGTGATGGTGATGTTCAC-3′) (Eurobio) amplifying a 94 base pair (bp) fragment and creating an NlaIII restriction site only in the FCGR3A-158V allele. This nested PCR was performed with 1 μL of the amplified DNA, 150 ng of each primer, 200 μM of each dNTP, and 1 U of Taq DNA polymerase. The first cycle consisted of 5 minutes at 95°C, then 35 cycles (each consisting of steps at 95°C for 1 minute, 64°C for 1 minute, and 72°C for 1 minute), and 9.5 minutes at 72°C to complete extension. The amplified DNA (10 μL) was then digested with 10 U NlaIII (New England Biolabs, Hitchin, England) at 37°C for 12 hours and separated by electrophoresis on 8% polyacrylamide gel. After staining with ethidium bromide, DNA bands were visualized under UV light. For homozygous FCGR3A-158F patients, only one undigested band (94 bp) was visible. Three bands (94 bp, 61 bp, and 33 bp) were seen in heterozygous individuals, whereas for homozygous FCGR3A-158V patients only 2 digested bands (61 bp and 33 bp) were obtained.
FCGR2A-131H/R genotyping
Genotyping of FCGR2A-131H/R consisted of PCR followed by an allele-specific restriction enzyme digestion, according to Liang et al.29 The sense primer (5′-GGAAAATCCCAGAAATTCTCGC-3′) (Eurobio) was modified to create a BstUI restriction site, in case of an R allele, while the antisense primer (5′-CAACAGCCTGACTACCTATTACGCGGG-3′) (Eurobio) was modified to carry a second BstUI restriction site that served as an internal control. PCR amplification was performed in a 50 μL reaction with 1.25 μg genomic DNA, 170 ng of each primer, 200 μM of each dNTP, 0.5 U Taq DNA polymerase, and the manufacturer's buffer. The first cycle consisted of 3 minutes at 94°C followed by 35 cycles (each consisting of 3 steps at 94°C for 15 seconds, 55°C for 30 seconds, and 72°C for 40 seconds) and 7 minutes at 72°C to complete extension. The amplified DNA (7 μL) was then digested with 20 UBstUI (New England Biolabs) at 60°C for 12 hours. Further analysis was performed as described for FCGR3A genotyping. The FCGR2A-131H and -131R alleles were visualized as 337 bp and 316 bp DNA fragments, respectively.
Statistical analysis
The clinical and laboratory characteristics and the clinical and molecular responses of the patients in the different genotypic groups were compared using the Fisher exact test. A logistic regression analysis including sex, age (> or ≤ 60 years), number of extranodal sites involved (≥ or < 2), bone marrow involvement,BCL2-JH rearrangement status at diagnosis, andFCGR3A genotype was used to identify independent prognostic variables influencing the clinical and molecular responses. Progression-free survival was calculated using the method of Kaplan and Meier30 and was measured from the start of treatment until progression, relapse, or death. Comparison of the progression-free survival by FCGR3A genotype was performed using the log-rank test. The significance level was P < .05.
Results
Clinical response
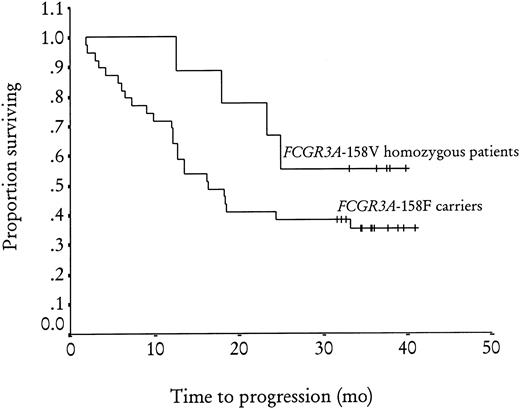
Of the 49 patients tested for the FCGR3A-158V/F polymorphism, 10 (20%) and 17 (35%) were homozygous forFCGR3A-158V and FCGR3A-158F, respectively, and 22 (45%) were heterozygous. The 3 groups were not different in terms of sex, disease stage, bone marrow involvement, number of extranodal sites involved, or presence of BCL2-JH rearrangement in peripheral blood and bone marrow at diagnosis (Table1). No difference was found when homozygous FCGR3A-158V patients were compared withFCGR3A-158F carriers (FCGR3A-158F homozygous and heterozygous patients) or when homozygous FCGR3A-158F patients were compared with FCGR3A-158V carriers (FCGR3A-158V homozygous and heterozygous patients). The objective response rate at M2 was 100% (CR + CRu = 40%), 70% (CR + CRu = 29%), and 64% (CR + CRu = 18%) inFCGR3A-158V homozygous, FCGR3A-158F homozygous, and heterozygous patients, respectively (P = .09). A significant difference in objective response rate was observed betweenFCGR3A-158V homozygous patients and FCGR3A-158F carriers, with a 67% (CR + CRu = 23%) objective response rate for this latter group (relative risk = 1.5; 95% [confidence interval] CI, 1.2-1.9; P = .03) (Table2). No difference was observed betweenFCGR3A-158F homozygous patients and FCGR3A-158V carriers. At M12, the objective response rate was 90% (CR + CRu = 70%), 59% (CR + CRu = 35%), and 45% (CR + CRu = 32%) in FCGR3A-158V homozygous, FCGR3A-158F homozygous, and heterozygous patients, respectively (P = .06). The difference in objective response rate was still present 1 year after treatment between the FCGR3A-158V homozygous group and FCGR3A-158F carriers, with a 51% (CR + CRu = 33%) objective response rate for this latter group (relative risk = 1.7; 95% CI, 1.2-2.5; P = .03). The logistic regression analysis showed that the homozygousFCGR3A-158V genotype was the only predictive factor for clinical response both at M2 (P = .02) and at M12 (P = .01). The progression-free survival at 3 years (median follow-up 35 months; range 31-41) (Figure1) was 56% in FCGR3A-158V homozygous patients and 35% in FCGR3A-158F carriers (nonsignificant). Of the 45 patients analyzed forFCGR2A-131H/R polymorphism, 9 (20%) and 13 (29%) were homozygous for FCGR2A-131R and FCGR2A-131H, respectively, while 23 (51%) were heterozygous. There was no difference in the characteristics at inclusion or clinical response to rituximab treatment for these 3 groups or for homozygousFCGR2A-131H patients and FCGR2A-131R carriers or for homozygous FCGR2A-131R patients andFCGR2A-131H carriers (data not shown).
Characteristics of patients by FCGR3A-158V/F polymorphism
| . | FCGR3A-158VV . | FCGR3A-158VF . | FCGR3A-158FF . | P* . |
|---|---|---|---|---|
| No. (%) | 10 (20) | 22 (45) | 17 (35) | |
| Sex | ||||
| M | 3 | 12 | 10 | NS |
| F | 7 | 10 | 7 | |
| Disease stage | ||||
| II-III | 3 | 6 | 6 | NS |
| IV | 7 | 16 | 11 | |
| Bone marrow involvement | ||||
| Yes | 7 | 16 | 9 | NS |
| No | 3 | 6 | 8 | |
| Extranodal sites involved | ||||
| Less than 2 | 8 | 20 | 13 | NS |
| 2 or more | 2 | 2 | 4 | |
| BCL2-JHrearrangement in peripheral blood | 8 | 12 | 11 | NS |
| BCL2-JH rearrangement in bone marrow | 7 | 12 | 11 | NS |
| . | FCGR3A-158VV . | FCGR3A-158VF . | FCGR3A-158FF . | P* . |
|---|---|---|---|---|
| No. (%) | 10 (20) | 22 (45) | 17 (35) | |
| Sex | ||||
| M | 3 | 12 | 10 | NS |
| F | 7 | 10 | 7 | |
| Disease stage | ||||
| II-III | 3 | 6 | 6 | NS |
| IV | 7 | 16 | 11 | |
| Bone marrow involvement | ||||
| Yes | 7 | 16 | 9 | NS |
| No | 3 | 6 | 8 | |
| Extranodal sites involved | ||||
| Less than 2 | 8 | 20 | 13 | NS |
| 2 or more | 2 | 2 | 4 | |
| BCL2-JHrearrangement in peripheral blood | 8 | 12 | 11 | NS |
| BCL2-JH rearrangement in bone marrow | 7 | 12 | 11 | NS |
NS indicates nonsignificant.
Statistical comparisons of the 3 groups, of homozygousFCGR3A-158V patients versus FCGR3A-158F carriers, and of homozygous FCGR3A-158F patients versusFCGR3A-158V carriers.
Clinical response to rituximab by FCGR3A-158V/F polymorphism
| . | FCGR3A-158VV . | FCGR3A-158F carriers . | P* . |
|---|---|---|---|
| Clinical response at M2 | |||
| Objective response (%) | 10 (100) | 26 (67) | .03 |
| CR | 3 | 7 | |
| CRu | 1 | 2 | |
| PR | 6 | 17 | |
| No response (%) | 0 (0) | 13 (33) | |
| No change | 0 | 10 | |
| Progressive disease | 0 | 3 | |
| Clinical response at M12 | |||
| Objective response (%) | 9 (90) | 20 (51) | .03 |
| CR | 6 | 11 | |
| CRu | 1 | 2 | |
| PR | 2 | 7 | |
| No response (%) | 1 (10) | 19 (49) | |
| No change | 0 | 2 | |
| Progressive disease | 1 | 17 |
| . | FCGR3A-158VV . | FCGR3A-158F carriers . | P* . |
|---|---|---|---|
| Clinical response at M2 | |||
| Objective response (%) | 10 (100) | 26 (67) | .03 |
| CR | 3 | 7 | |
| CRu | 1 | 2 | |
| PR | 6 | 17 | |
| No response (%) | 0 (0) | 13 (33) | |
| No change | 0 | 10 | |
| Progressive disease | 0 | 3 | |
| Clinical response at M12 | |||
| Objective response (%) | 9 (90) | 20 (51) | .03 |
| CR | 6 | 11 | |
| CRu | 1 | 2 | |
| PR | 2 | 7 | |
| No response (%) | 1 (10) | 19 (49) | |
| No change | 0 | 2 | |
| Progressive disease | 1 | 17 |
Statistical comparison of homozygousFCGR3A-158V patients versus FCGR3A-158F carriers. Data concerning the 3 genotype subgroups are given in the text.
Adjusted Kaplan-Meier estimates of progression-free survival after rituximab treatment byFCGR3A-158V/F genotype (P = .2).
Adjusted Kaplan-Meier estimates of progression-free survival after rituximab treatment byFCGR3A-158V/F genotype (P = .2).
Molecular response
At diagnosis, BCL2-JH rearrangement was detected in both peripheral blood and in bone marrow in 30 (64%) patients, enabling further follow-up. Twenty-five patients (6FCGR3A-158V homozygous patients and 19FCGR3A-158F carriers) and 23 patients (6FCGR3A-158V homozygous patients and 17FCGR3A-158F carriers) were analyzed for BCL2-JHrearrangement in both peripheral blood and bone marrow at M2 and at M12 (Table 3). At M2, a cleaning ofBCL2-JH rearrangement was observed in 3 of 6 of theFCGR3A-158V homozygous patients and in 5 of 19 of theFCGR3A-158F carriers (nonsignificant). In contrast, the rate of BCL2-JH rearrangement cleaning at M12 was higher (5 of 6) in the FCGR3A-158V homozygous patients than in theFCGR3A-158F carriers (5 of 17) (relative risk = 2.8; 95% CI, 1.2-6.4; P = .03). The logistic regression analysis showed that the FCGR3A-158V homozygous genotype was the only factor associated with a greater probability of exhibitingBCL2-JH rearrangement cleaning at M12 (P = .04). The single homozygous FCGR3A-158V patient still presenting with BCL2-JH rearrangement in peripheral blood and bone marrow at M12 was in CR 23 months after rituximab treatment. In contrast, the molecular responses at M2 and M12 were not influenced by the FCGR2A-131H/R polymorphism (data not shown).
Molecular response to rituximab at M2 and at M12 by FCGR3A-158V/F polymophism
| . | FCGR3A-158VV . | FCGR3A-158F carriers . | P . |
|---|---|---|---|
| Molecular response at M2 | NS | ||
| Cleaning of BCL2-JHrearrangement | 3 | 5 | |
| Persistent BCL2-JHrearrangement | 3 | 14 | |
| Molecular response at M12 | .03 | ||
| Cleaning of BCL2-JHrearrangement | 5 | 5 | |
| Persistent BCL2-JHrearrangement | 1 | 12 |
| . | FCGR3A-158VV . | FCGR3A-158F carriers . | P . |
|---|---|---|---|
| Molecular response at M2 | NS | ||
| Cleaning of BCL2-JHrearrangement | 3 | 5 | |
| Persistent BCL2-JHrearrangement | 3 | 14 | |
| Molecular response at M12 | .03 | ||
| Cleaning of BCL2-JHrearrangement | 5 | 5 | |
| Persistent BCL2-JHrearrangement | 1 | 12 |
NS indicates nonsignificant.
Discussion
Because of the increasing use of rituximab in B-cell lymphoproliferative malignancies, enhanced understanding of treatment failures and of the mode of action of rituximab is required. Given the expected role of NK cell and macrophage FcγRIIIa in rituximab-dependent cellular cytotoxicity against lymphoma cells, we genotyped FCGR3A in follicular NHL patients with well-defined clinical and laboratory characteristics and treated with rituximab alone.5 In particular, all the patients included in this study had a low tumor burden NHL and a molecular analysis ofBCL2-JH at diagnosis and during follow-up. TheFCGR3A allele frequencies in this population were similar to those of a general Caucasian population.23 24 Our results show an association between the FCGR3A genotype and the response to rituximab. Indeed, homozygous FCGR3A-158V patients, who account for one fifth of the population, had a greater probability of experiencing clinical response, with 100% and 90% objective response rates at M2 and M12, respectively. Moreover, 5 of the 6 FCGR3A-158V homozygous patients analyzed forBCL2-JH rearrangement showed molecular response at M12, compared with 5 of the 17 FCGR3A-158F carriers.FCGR3A-158V homozygosity was the only factor associated with the clinical and molecular responses. However, these higher clinical and molecular responses were still insufficient to significantly improve the progression-free survival in homozygousFCGR3A-158V patients.
This is the first report of an easily assessable genetic predictive factor for both clinical and molecular responses to rituximab. However, the genetic association does not demonstrate that the mode of action of rituximab involves FcγRIIIa. The association observed betweenFCGR3A genotype and response to rituximab might be due to another genetic polymorphism in linkage disequilibrium. Those polymorphisms could be located in FCGR3A itself, such as the triallelic FCGR3A-48L/H/R polymorphism,31 or in other FcγR-coding genes, because FCGR3A is located on the long arm of chromosome 1, which includes the 3 FCGR2 genes and FCGR3B.32 A linkage disequilibrium has been reported between FCGR2A and FCGR3B.33However, the fact that FCGR2A-131H/R polymorphism was not associated with a better response to rituximab strongly supports the fact that a gene very close to FCGR3A or FCGR3Aitself is directly involved.
Several in vitro studies argue in favor of direct involvement ofFCGR3A-158V/F polymorphism. First, Koene et al23have shown that the previously reported differences in IgG binding among the 3 FcγRIIIa-48L/H/R isoforms31 are a consequence of the linked FcγRIIIa-158V/F polymorphism, and several teams have demonstrated that NK cells from individuals homozygous for theFCGR3A-158V allotype have a higher affinity for human complexed IgG1 and are more cytotoxic toward IgG1-sensitized targets.23,24,34 In conjunction with our present results, those functional differences strongly suggest thatFCGR3A-158V homozygous patients have a better response to rituximab because of a better in vivo binding of that chimeric human IgG1 to FcγRIIIa. Secondly, NK cell- and macrophage-mediated ADCC is one of the mechanisms triggered by anti-CD20 antibodies in vitro8,11,12 as well as in murine models in vivo,17-19 and rituximab-mediated apoptosis is amplified by FcγR-expressing cells.15,16 Of all FcγR, FcγRIIIa is the only receptor shared by NK cells and macrophages. We thus postulate that FCGR3A-158V patients show a better response to rituximab because they have better ADCC activity against lymphoma cells. The fact that more than 50% of the FCGR3A-158F carriers nonetheless present a clinical response to rituximab could be explained by lower, but still sufficient, ADCC activity or, more likely, by other mechanisms operating in vivo such as complement-dependent cytotoxicity, complement-dependent cell-mediated cytotoxicity,11,13,14 and/or apoptosis.15 16ADCC could then be viewed as an additional mechanism in the response to rituximab that is particularly effective in FCGR3A-158V homozygous patients.
The in vitro studies suggest a “gene-dose” effect with a level of IgG1 binding to NK cells from FCGR3A heterozygous donors intermediate between that observed with NK cells fromFCGR3A-158V and FCGR3A-158F homozygotes.23 However, the clinical response of heterozygous patients appears similar to that of FCGR3A-158F homozygous patients. Further studies with larger groups of patients will be required to conclude against a “gene-dose” effect in vivo.
Because FcγRIIIa is strongly associated with a better response to rituximab, it needs to be taken into account in the development of new drugs targeting the CD20 antigen. For example, it may be possible to use engineered rituximab to treat FCGR3A-158F–carrier patients with B-cell lymphomas. Indeed, by modifying various residues in the IgG1 lower hinge region, Shields et al have recently obtained IgG1 mutants that bind more strongly to FcγRIIIa-158F than native IgG1.34
Taken together, those results will enable new therapeutic strategies against B lymphoproliferative disorders based upon prior determination of the patient's FCGR3A genotype. Because this polymorphism has the same distribution in various ethnic populations, including blacks and Japanese, such a strategy may be applied worldwide.23,35 36 Furthermore, such a pharmacogenetic approach may also be applied to other intact humanized IgG1 antibodies used in the treatment of B-cell malignancies, such as Campath-1H, or those used in the treatment of other malignancies, such as trastuzumab (Herceptin). Even more generally, this approach may apply to other intact humanized IgG1 developed to deplete target cells.
The authors thank Dr S. Iochman for her technical help in molecular biology assays and Prof G. Thibault and Prof G Paintaud for their critical review of the manuscript.
Supported by grants from the Fondation Langlois and the Comité de l'Indre de la Ligue Nationale Contre le Cancer.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Hervé Watier, Laboratoire d'Immunologie, Centre Hospitalier Universitaire, 2 boulevard Tonnellé, 37044 Tours Cedex, France; e-mail: watier@med.univ-tours.fr.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal