Antisense oligodeoxynucleotide (ODN) drugs might be more effective if their delivery was optimized and they were targeted to short-lived proteins encoded by messenger RNA (mRNA) species with equally short half-lives. To test this hypothesis, an ODN targeted to the c-mybproto-oncogene was developed and used to purge marrow autografts administered to allograft-ineligible chronic myelogenous leukemia patients. CD34+ marrow cells were purged with ODN for either 24 (n = 19) or 72 (n = 5) hours. After purging, Myb mRNA levels declined substantially in approximately 50% of patients. Analysis of bcr/abl expression in long-term culture-initiating cells suggested that purging had been accomplished at a primitive cell level in more than 50% of patients and was ODN dependent. Day-100 cytogenetics were evaluated in surviving patients who engrafted without infusion of unmanipulated “backup” marrow (n = 14). Whereas all patients were approximately 100% Philadelphia chromosome–positive (Ph+) before transplantation, 2 patients had complete cytogenetic remissions; 3 patients had fewer than 33% Ph+ metaphases; and 8 remained 100% Ph+. One patient's marrow yielded no metaphases, but fluorescent in situ hybridization evaluation approximately 18 months after transplantation revealed approximately 45% bcr/abl+ cells, suggesting that 6 of 14 patients had originally obtained a major cytogenetic response. Conclusions regarding clinical efficacy of ODN marrow purging cannot be drawn from this small pilot study. Nevertheless, these results lead to the speculation that enhanced delivery of ODN, targeted to critical proteins of short half-life, might lead to the development of more effective nucleic acid drugs and the enhanced clinical utility of these compounds in the future.
Introduction
Many believe that gene-targeted therapies, with their promise of high specificity and low toxicity, will lead to a revolution in cancer therapeutics.1-3 Numerous gene therapy strategies are under development, one of which uses reverse complementary (antisense) nucleic acids to inhibit gene expression at the messenger RNA (mRNA) level.4 Simply stated, delivering an antisense nucleic acid into a cell where the gene of interest is expressed should lead to hybridization between the antisense sequence and the targeted gene's mRNA. Stable mRNA-antisense duplexes cannot be translated and, depending on the chemical composition of the antisense molecule, can lead to the destruction of the mRNA by binding of endogenous nucleases, such as RNase H, or by intrinsic enzymatic activity engineered into the sequence.4-8 Although conceptually elegant, this approach for treating human malignancies remains of uncertain utility.9
In developing potential antisense nucleic acid compounds, we have targeted the mRNAs of genes encoding a variety of cytokines, cell surface receptors, and signal-transducing proteins,10-16but have concentrated our efforts on a relatively hematopoietic-specific transcription factor encoded by the c-myb proto-oncogene.17-21 This gene is the normal cellular homolog of v-myb, the transforming oncogene of avian myeloblastosis virus and avian leukemia virus E26.22,23 Located on chromosome arm 6q in humans, c-myb's transcript encodes an approximately 72-kd nuclear-binding protein (Myb) that recognizes the consensus sequence 5′-PyAAC(G/Py)G-3′.24 Myb family members play a major role in regulating the G1/S transition in cycling hematopoietic cells,18,25,26 and c-myb in particular functions as a transactivator of a number of important cellular genes, such as those encoding mim-1,27CD34,28 the Kit receptor,10 and myeloblastin.29
Myb's ability to transactivate genes required for cell proliferation and growth underlies its importance for normal hematopoietic cell development.17,30 Control of such critical functions also suggests a potential role for Myb in leukemic transformation.31-33 Myb might then represent a legitimate therapeutic target in patients with hematologic malignancies, but only if normal cells are more tolerant of transient Myb deprivation, such as that induced by antisense oligodeoxynucleotide (ODN), than their malignant counterparts. This has, in fact, been shown to be the case.19,20,34,35 Although the mechanism for this finding remains uncertain, we have speculated that Myb's role in regulating Kit-receptor expression10,36 may be a factor, since, as the Steel and White Spotting mouse37,38 and patients treated with STI571 demonstrate,39 normal hematopoietic cells tolerate loss of Kit-receptor function, while malignant cells subjected to similar deprivation are highly likely to undergo apoptosis.10 40
On the basis of the hypothesis that Myb plays a role in leukemogenesis, and the speculation that the very short half-life of c-myb's mRNA and protein41,42 would make it an ideal target for an antisense nucleic acid drug, we initiated a pilot bone marrow purging study in patients with chronic myelogenous leukemia (CML). CML was thought to be a particularly apt disease model for our investigations because of the ease with which normal and malignant cells could be distinguished. In addition, because c-myb plays a role in regulating c-myc expression43,44 and because Myc may play an important role in bcr/abl–mediated transformation,45 46 we thought Myb had enhanced legitimacy as a therapeutic target in this disease. Finally, we also believed that the purging strategy, which was under active investigation as a CML treatment, would allow us to determine the performance of an ODN in a clinically relevant model under conditions in which cellular delivery might be optimal. We suggest that the results reported here support the basic assumptions that motivated this study, ie, that an efficiently delivered, appropriately targeted ODN may ultimately prove useful for therapeutically motivated squelching of gene expression.
Patients, materials, and methods
Study subjects
The study was approved by the Institutional Review Board of the Hospital of the University of Pennsylvania, and all patients gave signed informed consent. Eligibility requirements included a CML diagnosis confirmed by cytogenetics, good performance status (Eastern Cooperative Oncology Group 0-1), and organ function adequate for marrow transplantation. Patients were ineligible if they had a known related HLA-identical marrow donor, had been previously treated with busulfan for longer than 3 months, had received interferon-α within 6 weeks of planned marrow harvest, or were younger than 18 or older than 61 years of age. Chronic phase patients treated in Philadelphia were also required to demonstrate sequence-specific sensitivity to the c-myb–targeted ODN (see below). Under these criteria, 21 patients were ultimately entered in the trial in Philadelphia. An additional 4 patients were identified, consented in accord with institutional requirements, and were treated on a similar protocol at the Hammersmith Hospital, London, United Kingdom. Of the total 25 patients, 20 were in first chronic phase and 5 in accelerated phase CML. Patient characteristics, including age, sex, time from diagnosis to transplantation, and prior treatment, are described in Table 1.
Characteristics of patients in the study
| . | 24-hour purge (n = 20) . | 72-hour purge (n = 5) . |
|---|---|---|
| Total, no. (%) | 18 (90) | 2 (40) |
| 2 (10) | 3 (60) | |
| Age, y | ||
| Mean | 44.6 | 50.8 |
| SD | 8.0 | 7.9 |
| Minimum | 31.0 | 37.0 |
| Maximum | 59.0 | 57.0 |
| Gender, no. (%) | ||
| Males | 11 (55) | 1 (20) |
| Females | 9 (45) | 4 (80) |
| Prior therapy, no. (%) | ||
| Hydrea | 20 (100) | 5 (100) |
| IFN | 16 (80) | 4 (80) |
| Busulfan | 0 (0) | 1 (20) |
| Ida/Ara-C | 1 (6) | 1 (20) |
| ICE | 0 (0) | 1 (20) |
| Time since diagnosis, y | ||
| Mean | 3.1 | 2.2 |
| SD | 2.9 | 1.1 |
| Minimum | 0.6 | 1.0 |
| Maximum | 13.2 | 3.6 |
| . | 24-hour purge (n = 20) . | 72-hour purge (n = 5) . |
|---|---|---|
| Total, no. (%) | 18 (90) | 2 (40) |
| 2 (10) | 3 (60) | |
| Age, y | ||
| Mean | 44.6 | 50.8 |
| SD | 8.0 | 7.9 |
| Minimum | 31.0 | 37.0 |
| Maximum | 59.0 | 57.0 |
| Gender, no. (%) | ||
| Males | 11 (55) | 1 (20) |
| Females | 9 (45) | 4 (80) |
| Prior therapy, no. (%) | ||
| Hydrea | 20 (100) | 5 (100) |
| IFN | 16 (80) | 4 (80) |
| Busulfan | 0 (0) | 1 (20) |
| Ida/Ara-C | 1 (6) | 1 (20) |
| ICE | 0 (0) | 1 (20) |
| Time since diagnosis, y | ||
| Mean | 3.1 | 2.2 |
| SD | 2.9 | 1.1 |
| Minimum | 0.6 | 1.0 |
| Maximum | 13.2 | 3.6 |
IFN indicates interferon; ICE, ifosfamide, carboplatin, and etoposide.
Oligodeoxynucleotides
ODNs were supplied by Lynx Therapeutics (Hayward, CA) as a sterile sodium salt, endotoxin-free, lyophilized powder. Compounds were fully phosphorothioated, were 24 nucleotides in length, and corresponded to either the sense (5′-GCC CGA AGA CCC CGG CAC AGC ATA-3′), or antisense (5′-TAT GCT GTG CCG GGG TCT TCG GGC-3′) orientation of codons 2 through 9 of the c-myb mRNA.
ODN-sensitivity testing
Patient-derived marrow cells were evaluated for sequence-specific sensitivity to the Myb-targeted antisense ODN as previously reported.20 In brief, patient-derived granulocyte-macrophage colony-forming units (CFU-GMs) were cloned in semisolid media after sham incubation (untreated control) or after incubation in equivalent concentrations (150 μg/mL total) of either Myb sense or Myb antisense ODN. Sequence-specific sensitivity to the antisense compound was defined as a 50% or greater decrease in CFU-GM colonies when compared with growth of untreated cells, or sense ODN–treated cells in those cases in which the sense ODN also inhibited colony formation. Sequence-specific sensitivity to the c-myb antisense compound was also said to exist if antisense-treated, but not sense-treated, cells demonstrated either a complete or a nearly complete decline in Myb or Bcr/abl mRNA when assayed simultaneously by means of a previously well-described reverse-transcriptase polymerase chain reaction (RT-PCR) reaction.12,20,36 47 Patients treated in the United Kingdom did not undergo sensitivity testing prior to transplantation.
Bone marrow harvest
Marrow was surgically harvested under general anesthesia. Patients were required to be off hydroxyurea and interferon for longer than 1 week and longer than 6 to 12 weeks, respectively. A maximum of 2 L marrow was aspirated, and a minimum of 3.5 × 108/kg nucleated bone marrow cells was required to proceed with processing and transplantation. If insufficient cells were obtained, patients underwent a second bone marrow harvest 1 month later. After filtration, a portion of the buffy coat (0.5 to 1 × 108 cells/kg) was immediately cryopreserved and stored as backup in case the patient did not engraft with the ODN-purged marrow. CD34+ cells, isolated from the remaining buffy coat (at least 3 × 108/kg) by means of a CEPRATE SC device (Baxter, Deerfield, IL), were then used for purging.
Ex vivo purging and cryopreservation
ODNs were dissolved in TC 199 (100 μg/mL) containing 2% (in London), or 5% (in Philadelphia) autologous serum and then added to the marrow cell suspension (2 × 106 cells/mL). Cells were incubated for 18 hours (5% CO2 at 37°C), followed by another addition of ODN (final ODN concentration: 150 μg/mL, or approximately 18 μM). Incubation was continued for an additional 6, or in some cases 54, hours. Cells were then washed and cryopreserved with the use of a programmable freezer. A minimum collection of 0.5 × 106 CD34+ cells/kg was required after purging to proceed with transplantation. If insufficient cells were obtained, patients could undergo a second bone marrow harvest. In some cases, the effect of purging on bcr/abl status of CFU-GM colonies derived from long-term culture-initiating cells (LTCICs) was also determined. In these instances, CD34+ cells were obtained from actual patient samples just prior to, and just after, purging with the Myb antisense ODN. These specimens were cultured simultaneously for LTCICs, and their CFU-GM progeny assayed for Myb and bcr/abl expression, as previously reported.48
Transplantation conditioning regimen
Patients were prepared for transplantation with busulfan (4 mg/kg/d × 4 days) and cyclophosphamide (60 mg/kg/d × 2 days) (in Philadelphia) or busulfan alone (16 mg/kg/d × 4 days) (in London). At the discretion of the treating physician, patients received prophylactic phenytoin. Bladder irrigation was used during cyclophosphamide treatment. Autologous marrow reinfusion was initiated 24 to 36 hours after chemotherapy. Filgrastim (5 μg/kg/d) was begun on the day of marrow infusion and continued until the absolute granulocyte count was at least 1 × 109/L for 3 consecutive days. Intravenous hydration, antiemetics, antibiotics, irradiated blood products, and intravenous nutrition were administered as needed.
Infusion of backup marrow
If absolute granulocyte count was lower than 0.1 × 109/L by day 30 after transplantation, a bone marrow aspirate and biopsy was performed. If the marrow was aplastic, or if cellularity was less the 20%, the patient's unmanipulated backup marrow could be administered at the discretion of the treating physician. Backup marrow could be infused prior to day 30, if clinically indicated, or withheld until day 42 at the discretion of the attending physician. At that time, clinical lack of engraftment and a hypocellular marrow triggered mandatory backup-marrow infusion.
Evaluation of toxicity and measurement of purge effect
Patients were evaluated for toxicity by means of the National Cancer Institute common toxicity criteria and by response. Hematologic recovery was defined as unsupported neutrophil and platelet counts exceeding 0.5 × 109/L and 20 × 109/L, respectively. All patients, save one who died of sepsis on day 13, were evaluable for hematologic recovery. Patients who did not receive backup-marrow infusion were evaluable for cytogenetic response at day 100.
Statistical analysis
Kaplan-Meier analysis, performed by means of SPSS statistical software (SPSS, Chicago, IL), was used to estimate time to hematologic engraftment. Patients who received backup bone marrow were censored for neutrophil and platelet engraftment because the decision to infuse backup marrow was based on the lack of engraftment. One additional patient was censored for platelet engraftment owing to progressive disease on day 43, which prevented the further evaluation of platelet recovery.
Results
Marrow harvesting
Between 1994 and 1999, a total of 43 patients were evaluated for ODN sensitivity in Philadelphia, and of these, 31 patients demonstrated sequence-specific sensitivity to the anti-Myb ODN. All eligibility criteria were subsequently met by 21 of these patients and they were enrolled in the study in Philadelphia. An additional 4 patients entered the study in London, United Kingdom. Of 25 total patients, 20 were in chronic phase, and 5 had accelerated disease. Sufficient numbers of purged CD34+ marrow cells were obtained from 24 patients. These individuals subsequently received transplants. One patient (in Philadelphia) could not receive a transplant because of inadequate cell yield after 2 bone marrow harvests.
Effect of Myb-targeted ODN on marrow cell growth and c-myb gene expression
Pharmacodynamic data, generated from protocol-eligibility testing and actual prepurge and postpurge studies, are shown in Tables2 (24-hour purge) and 3 (72-hour purge). It is useful to note that in only 1 of 14 cases was a truly significant decrease in colony inhibition observed in comparison with the screen result. In this patient (patient no. 2), the screen demonstrated a 69% inhibition of CFU-GMs, whereas only 22% inhibition was observed in the postpurge specimen. Accordingly, the screen and postpurge results were consistent, as 13 of 14 patients had colony inhibition of at least 50% by both assays. Importantly, false-negative sensitivity was observed in only 1 of 14 patients.
Laboratory correlates on 24-hour purge patients
| Patient no. . | CFU-GM inhibition . | Myb mRNA, prepurge/postpurge . | Bcr/abl mRNA . | ||
|---|---|---|---|---|---|
| Screen specimen, % . | Purge specimen, % . | Prepurge/postpurge . | LTCICs . | ||
| 1 | 83 | 60 | +/− | +/− | NE |
| 2 | 56 | 22 | +/± | +/+ | NE |
| 3 | 44 | 44 | +/− | +/± | NE |
| 4 | No growth | 99.9 | +/+ | +/− | NE |
| 5 | 80 | 50 | +/+ | +/+ | NE |
| 6 | 59 | 50 | NE | +/+ | − |
| 7 | 85 | 85 | NE | NE | NE |
| 8 | 55 | 54 | +/+ | +/+ | + |
| 14 | 72 | 76 | +/+ | +/+ | ± |
| 15 | 77 | 88 | +/− | +/+ | + |
| 16 | 81 | 100 | +/− | +/± | − |
| 17 | 65 | 71 | +/± | +/+ | + |
| 18 | 54 | 78 | +/+ | +/+ | + |
| 19 | 46 | 47 | +/+ | −/− | − |
| 21 | 55 | 71 | +/− | +/± | ± |
| London 1 | NE | NE | NE | +/+ | NE |
| London 2 | NE | NE | NE | +/+ | NE |
| London 3 | NE | NE | NE | +/+ | NE |
| London 4 | NE | NE | NE | +/+ | NE |
| Patient no. . | CFU-GM inhibition . | Myb mRNA, prepurge/postpurge . | Bcr/abl mRNA . | ||
|---|---|---|---|---|---|
| Screen specimen, % . | Purge specimen, % . | Prepurge/postpurge . | LTCICs . | ||
| 1 | 83 | 60 | +/− | +/− | NE |
| 2 | 56 | 22 | +/± | +/+ | NE |
| 3 | 44 | 44 | +/− | +/± | NE |
| 4 | No growth | 99.9 | +/+ | +/− | NE |
| 5 | 80 | 50 | +/+ | +/+ | NE |
| 6 | 59 | 50 | NE | +/+ | − |
| 7 | 85 | 85 | NE | NE | NE |
| 8 | 55 | 54 | +/+ | +/+ | + |
| 14 | 72 | 76 | +/+ | +/+ | ± |
| 15 | 77 | 88 | +/− | +/+ | + |
| 16 | 81 | 100 | +/− | +/± | − |
| 17 | 65 | 71 | +/± | +/+ | + |
| 18 | 54 | 78 | +/+ | +/+ | + |
| 19 | 46 | 47 | +/+ | −/− | − |
| 21 | 55 | 71 | +/− | +/± | ± |
| London 1 | NE | NE | NE | +/+ | NE |
| London 2 | NE | NE | NE | +/+ | NE |
| London 3 | NE | NE | NE | +/+ | NE |
| London 4 | NE | NE | NE | +/+ | NE |
CFU-GM indicates granulocyte-macrophage colony-forming units; mRNA, messenger RNA; LTCICs, long-term culture-initiating cells; NE, not evaluable.
In the actual purge specimens, 24-hour exposure to the c-myb–targeted ODN inhibited CFU-GMs a mean (± SD) of 66% ± 22% (range, 22%-100%) when compared with control, or sense-ODN–treated cell growth. The 72-hour exposure inhibited CFU-GMs by 75% ± 16% (range, 55%-91%). In the Philadelphia cohort, Myb mRNA levels were routinely measured before and after ODN exposure by semiquantitative RT-PCR. After 24-hour exposure to the ODN, c-myb mRNA was undetectable in 5 of 13 evaluable cases and markedly diminished in 2 of 13 cases. The 72-hour ODN exposure had similar effects in 4 of 5 cases. Overall, c-myb mRNA became undetectable in 8 of 18 evaluable samples and clearly diminished compared with control in 3 of 18 cases (total = 61%). We did not measure Myb protein levels in treated cells because of the limited numbers of cells available for analysis, but given the extremely short half-life of Myb protein, it is highly unlikely that Myb protein existed in measurable amounts in cells where the mRNA had been concomitantly decreased.49 The β-actin mRNA levels were measured simultaneously and found to be unchanged, suggesting that c-myb mRNA downregulation was sequence specific. Effect of the ODN on Myb mRNA levels was not studied in the London patients. Of note, prepurge and postpurge bcr/abl mRNA levels were also measured at both sites. After 24- and 72-hour exposure, bcr/abl mRNA was diminished in 6 of 13 and 1 of 5 evaluable cases, respectively. No changes in postpurge bcr/abl mRNA levels were observed in the London samples.
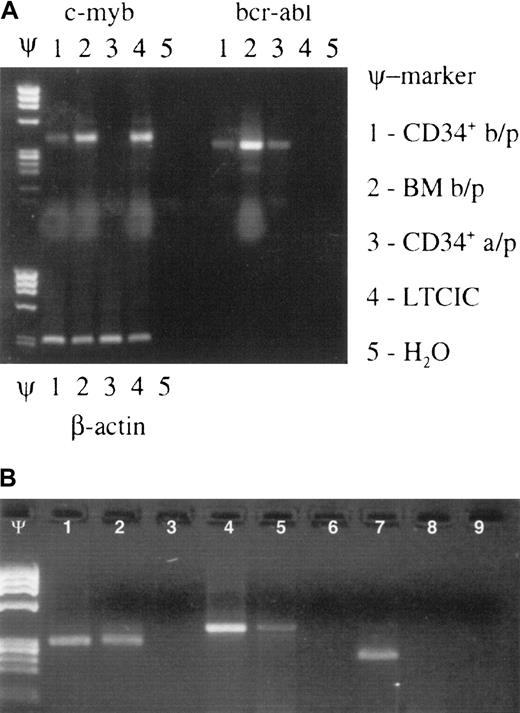
The above data were gathered on CD34+ cells. Since the percentage of true stem cells in this population is likely to be no more than 1% and since stem cells are ultimately responsible for long-term engraftment, we sought to determine the effect of the Myb-targeted ODN on the more primitive, stemlike cell populations present within the CD34+ cell fraction. For this purpose, we initiated LTCIC cultures from anti-Myb ODN–treated CD34+ cells. In all cases, untreated CD34+cells from the same patient were subjected to the same LTCIC culture procedure as a control. We reasoned that if primitive bcr/abl–expressing cells had been eliminated from the purged population, cells derived from an LTCIC culture should be bcr/abl−. Accordingly, after 10 days of culture, cells were withdrawn for CFU-GM assays, and resulting colonies were assayed for bcr/abl expression. Samples were assayed from 9 patients whose cells had been exposed to oligonucleotide for 24 hours (Table 2) and from 5 patients whose cells had been exposed to oligonucleotide for 72 hours (Figure 1; Table 3). Bcr/abl mRNA was either downregulated or undetectable in LTCICs derived from 5 of 9 and 4 of 5 patients, respectively. Although the number of 72-hour purging samples was too small to allow statistical analysis, the results suggested that a 72-hour purge might be more effective at eliminating primitive bcr/abl clones than one carried out for 24 hours. This interpretation must be viewed as speculative, however, since we did not perform limiting dilution assays for precise LTCIC quantitation. In addition, others have noted that long-term culture has an intrinsic ability to purge CML marrow,50,51 although prolonged culture is usually required to achieve this effect.52 With regard to this concern, however, it is important to note that untreated and antisense-treated cells from the same patient were simultaneously subjected to the same LTCIC procedure (Figure 1B). Since all untreated LTCICs gave rise to CFU-GMs that expressed bcr/abl, it seems most unlikely that the purging effect was due only to the long-term culture. Although the 72-hour ODN exposure appeared to enhance in vitro purging, the effect of the longer purge on day-100 cytogenetics was unevaluable because 2 patients in this group died in the peritransplantation period and 3 required backup-marrow infusion.
Effect of Myb ODN purging on brc/abl–expressing primitive hematopoietic stem/progenitor cells.
Myb ODN purging preferentially affects malignant (brc/abl–expressing) primitive hematopoietic stem/progenitor cells. (A) Ethidium bromide–stained agarose gel containing c-myb, bcr-abl, and β-actin mRNA RT-PCR products derived from patient no. 16's CD34+cells before marrow purging (b/p); whole bone marrow cells (BM) before purging; CD34+ cells after purging (a/p); LTCICs; and water (H2O) after exposure to the c-myb–targeted ODN for 72 hours. Lanes containing molecular weight markers are indicated by the symbol Ψ. (B) Ethidium bromide–stained agarose gel demonstrating β-actin (lanes 1-3), c-myb (lanes 4-6), and bcr/abl mRNA (lanes 7-9) expression in CFU-GM colonies derived from a single patient's untreated (lanes 1, 4, 7) or anti-Myb ODN–purged (lanes 2, 5, 8) LTCICs. Respective H2O control reactions are shown in lanes 3, 6, and 9. Molecular weight markers: Ψ.
Effect of Myb ODN purging on brc/abl–expressing primitive hematopoietic stem/progenitor cells.
Myb ODN purging preferentially affects malignant (brc/abl–expressing) primitive hematopoietic stem/progenitor cells. (A) Ethidium bromide–stained agarose gel containing c-myb, bcr-abl, and β-actin mRNA RT-PCR products derived from patient no. 16's CD34+cells before marrow purging (b/p); whole bone marrow cells (BM) before purging; CD34+ cells after purging (a/p); LTCICs; and water (H2O) after exposure to the c-myb–targeted ODN for 72 hours. Lanes containing molecular weight markers are indicated by the symbol Ψ. (B) Ethidium bromide–stained agarose gel demonstrating β-actin (lanes 1-3), c-myb (lanes 4-6), and bcr/abl mRNA (lanes 7-9) expression in CFU-GM colonies derived from a single patient's untreated (lanes 1, 4, 7) or anti-Myb ODN–purged (lanes 2, 5, 8) LTCICs. Respective H2O control reactions are shown in lanes 3, 6, and 9. Molecular weight markers: Ψ.
Laboratory correlates on 72-hour purge patients
| Patient no. . | CFU-GM inhibition . | Myb mRNA, prepurge/postpurge . | Bcr/abl mRNA . | ||
|---|---|---|---|---|---|
| Screen specimen, % . | Purge specimen, % . | Prepurge/postpurge . | LTCICs . | ||
| 9 | 62 | 61 | +/+ | +/+ | + |
| 10 | 68 | 55 | +/− | +/+ | − |
| 11 | 57 | 91 | +/− | +/+ | ± |
| 12 | 81 | 90 | +/± | +/+ | − |
| 13 | 52 | 77 | +/− | +/± | − |
| Patient no. . | CFU-GM inhibition . | Myb mRNA, prepurge/postpurge . | Bcr/abl mRNA . | ||
|---|---|---|---|---|---|
| Screen specimen, % . | Purge specimen, % . | Prepurge/postpurge . | LTCICs . | ||
| 9 | 62 | 61 | +/+ | +/+ | + |
| 10 | 68 | 55 | +/− | +/+ | − |
| 11 | 57 | 91 | +/− | +/+ | ± |
| 12 | 81 | 90 | +/± | +/+ | − |
| 13 | 52 | 77 | +/− | +/± | − |
See Table 1 for abbreviations.
Engraftment
Results from the 2 centers were considered separately because of differences in transplantation-conditioning regimens. The first 8 patients in Philadelphia received marrow purged for 24 hours. The next 5 patients received marrow purged for 72 hours on the basis of studies, described above, that suggested this would yield a “cleaner” product. Engraftment concerns in the latter cohort prompted a return to the original purging protocol in the remaining 7 patients. The London patients all received 24-hour–purged marrow.
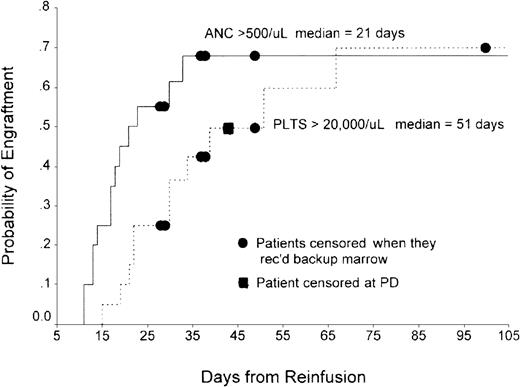
In Philadelphia, the total number of CD34+ cells/kg infused ranged from 6.38 × 105 to 6.08 × 106(n = 19; median, 1.35 × 106). Nineteen of 20 patients were evaluable for hematopoietic recovery. The exception was a patient who died of sepsis 13 days after transplantation. By Kaplan-Meier analysis, median time to recovery of absolute neutrophil count (ANC) exceeding 500/μL was 21 days (range, 11-33 days) and median time to recovery of platelets to more than 20 000 was 51 days (range, 15-67 days) (Figure 2). Patients who engrafted were infused with a mean of 1.45 ± 1.35 × 106/kg CD34+ cells versus 1.07 ± 1.36 × 106/kg CD34+ cells for those who did not engraft (P = .71). In London, median time to an ANC exceeding 500/μL was 34 days (range, 28-38 days), while median time to platelet count exceeding 20 000/μL was 66 days (range, 39-141 days).
Time to neutrophil and platelet recovery in patients undergoing transplantation in Philadelphia with ODN-purged marrow.
Probability of engraftment of neutrophils and platelets from time of marrow reinfusion are shown on the ordinate and abcissa, respectively.
Time to neutrophil and platelet recovery in patients undergoing transplantation in Philadelphia with ODN-purged marrow.
Probability of engraftment of neutrophils and platelets from time of marrow reinfusion are shown on the ordinate and abcissa, respectively.
Patients whose physicians deemed their blood counts inadequate were eligible to receive unpurged backup marrow. Two of 4 patients treated in London required such rescue. Table 4reviews the data on hematopoietic recovery of all Philadelphia patients who received backup-marrow infusions. Two of 15 patients who received 24-hour–purged marrow (13%) had delayed engraftment and required backup-marrow infusion on days 28 and 39, respectively. The patient who received backup marrow on day 28 remained aplastic and died on day 56 of fungal sepsis. The other patient also died of sepsis with progressive disease on day 163. An additional patient received backup marrow on day 150 because of persistent thrombocytopenia. It is of interest to note that none of the 3 patients who received backup-marrow infusions in this group had significant changes in their peripheral blood cell counts, with follow-up ranging from 28 to 124 days. This suggests the possibility that poor engraftment may have been due to intrinsic marrow cell and/or microenvironment problems in these individuals and not the result of the purging procedure.
Hematologic recovery after backup infusion (Philadelphia patients)
| Patient no. . | Purge, h . | Day of backup . | Day after backup . | Comments . | Day-100 cytogenetics, % . | |
|---|---|---|---|---|---|---|
| ANC above 500 . | Platelet count above 20 000 . | |||||
| 4 | 24 | 28 | Not recovered | Not recovered | Died day 56 | NE |
| 10 | 72 | 29 | 64 | 71 | — | 52 |
| 12 | 72 | 49 | 22 | 451 | — | 100 |
| 13 | 72 | 38 | 17 | 45 | — | 100 |
| 16 | 24 | 39 | Not recovered | Not recovered | PD, died day 163 | NE |
| 21 | 24 | > 100 | Recovered prior to backup | Not recovered | PD > 33 days after backup | 100 |
| Patient no. . | Purge, h . | Day of backup . | Day after backup . | Comments . | Day-100 cytogenetics, % . | |
|---|---|---|---|---|---|---|
| ANC above 500 . | Platelet count above 20 000 . | |||||
| 4 | 24 | 28 | Not recovered | Not recovered | Died day 56 | NE |
| 10 | 72 | 29 | 64 | 71 | — | 52 |
| 12 | 72 | 49 | 22 | 451 | — | 100 |
| 13 | 72 | 38 | 17 | 45 | — | 100 |
| 16 | 24 | 39 | Not recovered | Not recovered | PD, died day 163 | NE |
| 21 | 24 | > 100 | Recovered prior to backup | Not recovered | PD > 33 days after backup | 100 |
ANC indicates absolute neutrophil count; NE, not evaluable, PD, platelet dependent.
Three of 5 patients treated with bone marrow purged for 72 hours were evaluable for engraftment. The number of CD34+ cells infused ranged from 8.2 × 105 to 6.6 × 106/kg. All 3 had prolonged aplasia and required administration of unpurged backup marrow on days 29, 38, and 49, respectively. In contrast to patients who received 24-hour–purged marrow, all 3 patients demonstrated marked improvement of their peripheral blood counts after backup-marrow infusion. Time to an ANC exceeding 500/μL ranged from 17 to 64 days, while time to platelet recovery (greater than 20 000/μL) ranged from 45 to 451 days following reinfusion of backup unpurged marrow.
Day-100 and follow-up cytogenetics
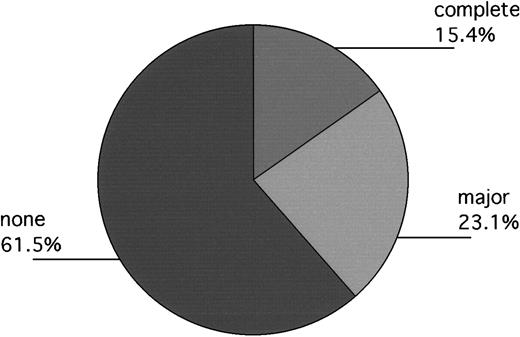
In Philadelphia, 13 patients engrafted without backup-marrow infusion, but 1 had clinical evidence of disease progression prior to day 100. Accordingly, 12 patients were evaluated for day-100 cytogenetics. Of these, 1 had a complete response; 3 had fewer than 33% Ph+ metaphases (11%, 12%, 17%); and 7 remained 100% Ph+. One patient's marrow persistently yielded no, or single, metaphases, but subsequent fluorescent in situ hybridization (FISH) evaluation approximately 18 months after transplantation revealed 40% bcr/abl+ cells, suggesting that major cytogenetic responses had probably been obtained in 5 of 13 patients. Equally interesting, of evaluable patients who required backup marrow (n = 4), 3 had 100% Ph+ cells at day 100, while the fourth patient had 52% Ph+ cells (Table 4). In London, 1 of the 2 evaluable patients had a complete cytogenetic response documented on day 100 as well. The other patient remained 100% Ph+ (Figure3).
Day-100 cytogenetics of evaluable patients (Philadelphia and London) not receiving backup-marrow infusion (n = 13).
Responses labeled “complete,” “major,” and “none” indicate patients with 0%, 1% to fewer than 33%, and 100%, Ph+cells, respectively. One additional patient, not included in the Figure, had no metaphases at day 100, but subsequent FISH evaluation at 18 months after transplantation revealed approximately 40% bcr/abl+ cells, suggesting that a major cytogenetic response had mostly likely occurred previously.
Day-100 cytogenetics of evaluable patients (Philadelphia and London) not receiving backup-marrow infusion (n = 13).
Responses labeled “complete,” “major,” and “none” indicate patients with 0%, 1% to fewer than 33%, and 100%, Ph+cells, respectively. One additional patient, not included in the Figure, had no metaphases at day 100, but subsequent FISH evaluation at 18 months after transplantation revealed approximately 40% bcr/abl+ cells, suggesting that a major cytogenetic response had mostly likely occurred previously.
Toxicity
In spite of the experience with backup marrow discussed above, we cannot exclude the possibility of some damage to engrafting cells by the ODN. No other untoward effects directly attributed to the ODN were discerned. All patients experienced variable degrees of stomatitis, and all but one required intravenous antibiotics for neutropenic fevers, effects that were clearly attributable to the chemotherapy used for transplantation conditioning. Other grade 3 or higher nonhematologic toxicities noted were as follows: gram-positive bacteremia in 3 patients, gastrointestinal bleeding in 2 patients, hyperbilirubinemia in 3 patients, fungemia in 1 patient, hypocalcemia in 3 patients, deep venous thrombosis in 1 patient. The 24-hour purge patient who received backup marrow on day 28 remained aplastic and died on day 56 of fungal sepsis. The other 24-hour purge patient who required backup also died of sepsis with progressive disease on day 163. Two of 5 patients treated with bone marrow purged for 72 hours succumbed in the peritransplantation period. One death was the result of sepsis on day 13, and one was secondary to sepsis and acute respiratory distress syndrome on day 28.
Long-term follow-up
Out of interest, long-term follow-up is presented for the 12 patients who did not receive backup marrow within the first 100 days after transplantation (Table 5). An additional patient who received backup marrow on day 150 because of persistent thrombocytopenia was excluded from the analysis. One member of this cohort remains in complete hematologic remission at longer than 79 months on prophylactic interferon (approximately 5 million units 3 times a week. Four patients remain on treatment (2 on hydroxyurea; 2 on STI571). Six patients were able to undergo allogeneic transplantation (5 matched unrelated donors [MUDs]; 1 matched related donor) at 24 to 45 months after autotransplantation with Myb ODN purged marrow. Three MUD transplantation patients died of complications related to the procedure, while 2 MUD recipients are alive at longer than 14 and 24 months after MUD transplantation (more than 59 and 68 months, respectively, after autotransplantation). The one related donor transplantation took place longer than 24 months after auto–bone marrow transplantation when a sibling donor who had previously declined to donate changed his mind. This recipient also died of allotransplantation-related complications. Finally, one patient developed progressive disease approximately 12 months after autotransplantation and subsequently died in blast crisis.
Long-term follow-up of patients who did not receive backup-marrow infusions (Philadelphia patients)
| Status and patient no. . | No. patients . | Follow-up, mo . |
|---|---|---|
| In hematologic remission, no. 1 | 1 | Prophylactic IFN, 79 + pABMT |
| On treatment with disease | 4 | |
| No. 7 | 71 + pABMT (hydroxyurea) | |
| No. 14 | 50 + pABMT (STI571) | |
| No. 15 | 51 + pABMT (STI571) | |
| No. 19 | 14 + pABMT | |
| Underwent allogeneic transplantation | 6 | |
| No. 2 | MUD 24 mo pABMT, died pMUD | |
| No. 3 | MUD 25 mo pABMT, died pMUD | |
| No. 6 | MUD 30 mo pABMT, died pMUD | |
| No. 5 | MUD 44 mo pABMT, alive, 24 mo pMUD | |
| No. 8 | MUD 45 mo pABMT, alive, 14 mo pMUD | |
| No. 17 | Allo 24 + mo pABMT, died pAllo5-150 | |
| Progressive disease, No. 18 | 1 | 12 + mo pABMT (deceased) |
| Status and patient no. . | No. patients . | Follow-up, mo . |
|---|---|---|
| In hematologic remission, no. 1 | 1 | Prophylactic IFN, 79 + pABMT |
| On treatment with disease | 4 | |
| No. 7 | 71 + pABMT (hydroxyurea) | |
| No. 14 | 50 + pABMT (STI571) | |
| No. 15 | 51 + pABMT (STI571) | |
| No. 19 | 14 + pABMT | |
| Underwent allogeneic transplantation | 6 | |
| No. 2 | MUD 24 mo pABMT, died pMUD | |
| No. 3 | MUD 25 mo pABMT, died pMUD | |
| No. 6 | MUD 30 mo pABMT, died pMUD | |
| No. 5 | MUD 44 mo pABMT, alive, 24 mo pMUD | |
| No. 8 | MUD 45 mo pABMT, alive, 14 mo pMUD | |
| No. 17 | Allo 24 + mo pABMT, died pAllo5-150 | |
| Progressive disease, No. 18 | 1 | 12 + mo pABMT (deceased) |
IFN indicates interferon; p, after; BMT, bone marrow transplantation; ABMT, auto bone marrow transplantation; MUD, transplantation from matched unrelated donor; Allo, allotransplantation.
This patient had an HLA-matched sibling who declined to donate BM for transplant but subsequently agreed to become a donor.
Cytogenetic responses of the 4 US patients who recovered with Ph− hematopoiesis lasted from more than 100 to more than 1100 days: patient no. 1, more than 1100 days; patient no. 2, more than 100 but fewer than 190 days; patient no. 3, more than 700 days; patient no. 8, more than 279 but fewer than 531. The London patient was reported to be 90% Ph+ at 12.5 months.
Discussion
The concept of inhibiting gene expression with antisense nucleic acids developed from studies initiated almost a quarter century ago.53,54 In the ensuing years, thousands of investigations using this strategy have been conducted, some yielding results of considerable importance in their field (reviewed in Gewirtz et al4). Despite the fact that the mechanism whereby these molecules modulate gene expression is not always certain,9,55-57 clinical development of antisense compounds has proceeded to the point where several oligonucleotide drugs have entered phase I/II and, in a few cases, phase III trials. Reports of such trials in patients with advanced cancers58-63 and human immunodeficiency virus (HIV)64 65 have recently been published. Although the oligonucleotides' sequence and chemical backbones varied, all compounds were well tolerated. Nevertheless, therapeutic activity was not observed in HIV patients, and clinical responses in patients with a variety of cancers were both uncommon and modest in scope.
One approach to improve the effectiveness of antisense nucleic acid drugs as anticancer agents is to combine them with more traditional therapeutic modalities.9 While this may well prove useful, we continue to explore strategies designed to promote more reliable and efficient mRNA squelching with antisense compounds. This pilot study was designed in accord with this goal and we feel that useful leads may be derived from its results. These are discussed below.
First, we think antisense compounds might be well suited to purging applications since it seems intuitive that delivery to cells in ex vivo suspension culture, as in marrow purging, is likely to be more efficient than to cells growing in a tumor mass. While we did not measure ODN uptake in the purged cells directly, previous reports from our laboratory have shown that hematopoietic cell lines growing in suspension have predictable uptake of these materials under conditions similar to those used in this study.66 Evidence for effective delivery includes the finding that c-myb mRNA expression was largely diminished or essentially undetectable in approximately 50% of the cases in which the purging procedure was carried out. We cannot explain why downregulation of c-mybexpression was not observed in a higher percentage of samples, but one could speculate that individual patient cell differences in oligonucleotide uptake or pretreatment levels of c-myb mRNA might be factors. We will address these issues in future studies. Alternative delivery strategies, including the use of various lipid transfecting agents, liposomes, cationic polyelectrolytes, and physical methods for membrane permeabilization, could reasonably be expected to improve delivery still further.67-69 At the same time, results reported here suggest that enhanced delivery may also increase cell toxicity, at least with the anti-Myb ODN, and potentially with other ODNs that target genes required by both normal and malignant cells. In this regard, it is worth noting that the 72-hour purge appeared more efficient than the 24-hour exposure, but unacceptable lowering of the compound's therapeutic index was suggested by the prolonged marrow aplasia observed in 3 patients.
Second, we believe that mRNA and protein half-life should be a consideration in target gene selection. The c-myb mRNA, as well as its encoded protein, has an estimated half-life of approximately 30 to 50 minutes,41,42 and this could be an important factor in the apparent efficiency of mRNA targeting in our system.70 While inadequate cell numbers prevented us from measuring Myb protein levels in treated samples, we have previously shown that mRNA and protein levels are correlated.49Further, it is clear that in the absence of mRNA to translate, Myb protein levels must decline rapidly. In contrast to Myb protein, bcl-2 protein, for example, has a half-life that has been estimated to be approximately 14 hours,71 and Raf and Ras proteins have half-lives estimated to exceed 24 hours.72 73 Attempts to eliminate these proteins from cells by means of oligonucleotides may prove more difficult.
Third, sequence accessibility within the targeted mRNA may also be a factor in antisense drug design. Folding of mRNA in vivo, as well as interactions with proteins in its intracellular environment, have been cited as determinants of oligonucleotide hybridization potential.4,74,75 Accordingly, efficacy of the anti-Myb oligonucleotide may relate in part to an ability to efficiently hybridize with its target. The means to test this possibility in a formal manner are presently being developed.57 76
In our view, 2 smaller ODN purging studies (3 and 8 patients, respectively) carried out on CML patients appear to support these points.68,77 These trials differed from our own in a number of respects, but the most fundamental differences were that both used oligonucleotides directed against the bcr/abl oncogene. If the data provided in those reports are compared with those reported here, the anti-Myb ODN appeared to have greater activity against its target gene. This may be due to the fact that the bcr/abl protein has an estimated half-life longer than 48 hours.78 In addition, some experimental data suggest that the bcr/abl breakpoint may be buried in the mRNA's tertiary structure and therefore less accessible for hybridization.74 The patient data suggest that the anti-Myb purge may also have been more efficient since, in the larger series, 6 of 8 patients had almost complete cytogenetic relapse by day 90 after transplantation. This could have clinical ramifications as well since a number of investigators have suggested that cytogenetic response by at least 3 months is a function of the number of normal CD34+ cells in the purged marrow.79-81 It is also worth noting that of the 4 evaluable patients in our study who received unmanipulated backup marrow, 3 had 100% Ph+metaphases at day 100, while 1 was 52% Ph+.
The above discussion assumes that we have demonstrated sequence-specific inhibition of c-myb gene expression. With this in mind, we acknowledge that our clinical results would probably be viewed as more convincing if a sense ODN purge arm had been included in our study design. We considered, and subsequently rejected, this possibility as unethical because of multiple studies from our laboratory that failed to demonstrate any consistent ability of the c-myb sense-ODN sequence to inhibit the growth of normal or malignant hematopoietic cells.17-20,34,82 We emphasize again, however, that in addition to our previously reported work, 21 of 24 patients reported did have preclinical sensitivity testing that included exposure to the corresponding sense-sequence oligonucleotide. Only patients whose cells displayed an antisense-specific inhibition of growth were included in the Philadelphia series. Accordingly, we strongly believe that the preclinical testing results, combined with numerous previous studies, leave little doubt that sequence-specific downregulation of c-myb was obtained. Nevertheless, we cannot rule out the possibility that non–sequence-dependent effects may also have played a role in the response we observed.55,56 ODNs are polyanionic molecules that may bind proteins through charge interactions or by assuming structural motifs that allow specific but unintended (aptameric) protein associations. It has been suggested that oligonucleotide sequences containing 4 contiguous guanine residues are inherently cytotoxic,83,84 perhaps owing to their ability to form complex intramolecular and intermolecular structures around metal cations such as Na+ and K+. The anti-Myb ODN tested in this trial contains such a motif, but since we have shown that oligonucleotides containing 4 contiguous guanines are nontoxic to hematopoietic cells, we do not believe that this type of non–sequence-dependent effect can explain our results.47
With regard to the possible clinical utility of the intervention we are reporting, 2 major caveats apply. First, the very small number of patients involved in this pilot study simply do not allow us to draw meaningful conclusions with regard to clinical efficacy of ODN purging or to attempt to make correlations between Myb downregulation and recovery of Ph− hematopoiesis. The latter issue in particular is complex. The assay for Myb mRNA was performed only once, immediately following the purge. As noted above, it is certainly possible that some cells destined to die still had PCR-detectable message, and the 72-hour purge data (Table 3) appear to support this. In addition, we would have preferred to carry out the Myb assays on a more highly purified, more stemlike population of CD34+cells since they are the true target for any therapeutic intervention. Limited cell numbers available for the transplantations precluded this from being done.
Second, we cannot compare the efficacy of this purging strategy with any of a number of other previously reported techniques.79,81,85-87 Indeed, this was not our purpose. Rather, we wished only to develop a model that might allow us to determine if ODN could be used for gene targeting in a useful clinical manner. While a number of the previously cited studies suggest that there may be clinical benefit for CML patients associated with the use of marrow purging, this has not been proved.88,89 Given the recent clinical success of the abl kinase inhibitor STI571 in treating patients with CML,90,91 it is unlikely that this question will be rigorously tested any time in the near future. It is worth noting, however, that autografting remains an active area of study for patients with acute myelogenous leukemia,92,93myeloma,94 and lymphoma.95 96 Since c-myb is a rational therapeutic target in these diseases, and the ODN approach to squelching Myb expression appears effective, it is conceivable that autografting with c-myb–purged marrow may be of clinical utility in patients with these diseases. Studies are being designed to test this hypothesis.
We gratefully acknowledge the technical assistance of Ms Deborah Magee, MT (ASCP), SSB, and the editorial assistance of E. R. Bien and Miriam Goodrum.
Supported in part by National Institutes of Health grants PO1 CA72765 and R01-CA66731 (A.M.G.) and a Translational Research Grant from the Leukemia and Lymphoma Society (A.M.G.). A.M.G. is a Distinguished Clinical Scientist of the Doris Duke Charitable Foundation.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Alan M. Gewirtz, Rm 713 BRB II/III, University of Pennsylvania School of Medicine, 421 Curie Blvd, Philadelphia, PA, 19104; e-mail: gewirtz@mail.med.upenn.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal