Hodgkin- and Reed-Sternberg (HRS) cells microdissected from 41 classical Hodgkin lymphomas (cHL) of 40 patients comprising 8 lymphocyte-rich (cHL-LR), 16 nodular sclerosis (cHL-NS), 15 mixed-cellularity (cHL-MC), and 2 lymphocyte-depletion (cHL-LD) subtypes were analyzed by comparative genomic hybridization for recurrently imbalanced chromosomal subregions. Chromosomal gains most frequently involved chromosome 2p (54%), 12q (37%), 17p (27%), 9p and 16p (24% each), and 17q and 20q (20% each), whereas losses primarily affected chromosome 13q (22%). Using fluorescence in situ hybridization, amplification of the REL oncogene was demonstrated within a distinct 2p15-p16 amplicon. The high frequency of 2p overrepresentations including REL, particularly in cHL-NS (88%), suggests that an alternative mechanism of constitutive activation of nuclear factor NF-κB is a hallmark of HRS cells. Hierarchical cluster analysis of chromosomal imbalances revealed a closer relationship among cHL-NS than other subtypes. Furthermore, there is a tendency for different subtypes of cHL-MC tumors characterized by different ages at the time of tumor onset and gain of chromosome 17p. The imbalance pattern of cHL subtypes suggests that different molecular pathways are activated, with REL or other genes on chromosomal band 2p15-p16 playing a fundamental role in the pathogenesis of classical Hodgkin lymphoma.
Introduction
Hodgkin lymphoma (HL), which accounts for approximately one third of all malignant lymphomas, is characterized by the presence of only a small fraction of malignant cells. Neoplastic cells represented as mononucleated Hodgkin- and multinucleated Reed-Sternberg cells (HRS cells) are embedded in a varying infiltrate of reactive cells including B and T lymphocytes, eosinophils, plasma cells, and fibroblasts.1 According to the new World Health Organization (WHO) classification,2 4 well-defined histotypes of classical Hodgkin lymphoma (cHL) can be distinguished: lymphocyte-rich (cHL-LR), nodular-sclerosis (cHL-NS), mixed-cellularity (cHL-MC), and lymphocyte-depletion (cHL-LD). Paragranuloma (nodular lymphocyte predominant Hodgkin lymphoma [NLPHL]) has been shown to be clinically and immunophenotypically distinct and eventually to transform to large B-cell non-Hodgkin lymphoma. This indicates that NLPHL is essentially different from the cHL subtypes.
Because of the small number of malignant cells, cytogenetic analysis is particularly difficult in HL and, to date, has not revealed any specific chromosomal rearrangements. Detailed analysis by chromosome banding is further limited by the low mitotic index of neoplastic cells, frequently poor chromosome morphology, and complex karyotypic rearrangements. For these reasons, it is difficult to obtain sufficient numbers of karyotypes for evaluation that are representative of the malignant cell population.3,4 Alternatively, combined immunohistochemical and cytogenetic analyses by fluorescence in situ hybridization (FISH) have been applied. It could be demonstrated that chromosomal changes are almost exclusively restricted to CD30+ HRS cells. Furthermore, significant heterogeneity in terms of the copy number of single chromosomes was detected using this approach.5-8
Recently, comparative genomic hybridization (CGH) was applied in combination with universal polymerase chain reaction (PCR) technology for cytogenetic analyses of HRS cells.9-11 These analyses indicated higher rates of numerical aberrations of individual chromosomes than had previously been found by banding analysis, in which the identification of numerical changes is difficult because of the complex karyotypes of HRS cells. Gains and losses in more than 50% of the cHL tumors were identified on chromosomal arms 2p, 7q, and 16q,9,10 whereas in NLPHL, chromosomal arms 1q, 3p, 5q, and Xq were affected.11 Although the number of analyses is still low (20 cHLs and 20 NLPHLs to date), CGH has already allowed the identification of several imbalanced chromosomal subregions, indicating the localization of candidate genes that may be involved in the etiology of this disease.
To further define critical subregions in cHL, a series of 41 tumors was analyzed (a small subset of cases was reported recently10). To this end, collected pools of approximately 30 malignant HRS cells from single tumors were isolated using microdissection technology. Genomic DNA from the individual cell pools was subsequently amplified by universal PCR technology and analyzed by CGH. Hierarchical cluster analysis of the CGH data was performed to assess particular patterns of chromosomal imbalances associated with different histotypes of cHL.
Patients, materials, and methods
Patients, lymphoma materials, and cell lines
For isolation of HRS cells 41 lymph node biopsy specimens obtained from 40 unselected patients were drawn from our frozen tissue bank. All patients were enrolled in multicenter national Hodgkin trials approved by the local ethics committees. Tumors were derived from 18 male and 22 female patients. The median age at time of diagnosis was 37 years (range, 12-78 years). Lymphomas were classified according to the guidelines of the pathology panel of the WHO classification2 on the basis of routinely stained paraffin sections. HRS cells were phenotyped by immunohistology using antibodies to CD3, CD15, CD20, CD30, the joining chain (J-chain), and the epithelial membrane antigen. For validity testing of the degenerate-oligo–primed (DOP)–PCR–CGH approach, cell line Colo 320-HSR, derived from a colon carcinoma,12,13 and cell line MedB-1, derived from a primary (thymic) mediastinal B-cell lymphoma,14 were used.
HRS cell isolation
Small parts of the snap-frozen lymph node tissue were thawed and wiped onto a glass slide with a pair of tweezers. Slides were immediately stained with hemalum for 1 minute and were dehydrated in 40%, 70%, and 100% ethanol for 10 minutes each. After further treatment of the slides in xylol for 10 minutes, areas with a high density of cytologically discernible HRS cells were identified by microscopy. Approximately 30 HRS cells were picked as single cell isolates by laser capture microdissection (PixCell II, Arcturus, CA) and were further processed.
Degenerate-oligo–primed polymerase chain reaction
HRS cells were digested with 10 to 20 units/mg proteinase K (250 μg/mL) in 20 μL of 1× PCR buffer (2 mM MgCl2, 50 mM KCl, 10 mM Tris, pH 8.3, 0.1 mg/mL gelatin) for 10 to 15 hours at 50°C, with subsequent inactivation of the enzyme at 94°C for 15 minutes. An additional aliquot of proteinase K was added and incubated for 3 hours at 37°C, during which time it was constantly shaken. After further inactivation of the enzyme, a DOP-PCR reaction was performed according to the protocol of Telenius et al.15
Comparative genomic hybridization
CGH was performed as previously described.16,17Briefly, universally amplified genomic DNA from HRS cells was labeled with biotin by nick translation, after DOP primers had been removed using suitable columns. Because the control probe DNA isolated from peripheral blood lymphocytes or normal tonsils of healthy controls was differentially labeled with digoxigenin, 1 μg biotin-labeled tumor DNA, 1 μg digoxigenin-labeled control DNA, and 70 μg human Cot-1-DNA (BRL Life Sciences, Gaithersburg, MD) were cohybridized to slides with metaphase cells from the blood of a healthy donor. After hybridization, control and test DNAs were detected by rhodamine and fluorescein isothiocyanate (FITC), respectively. Images of 15 or fewer metaphase cells were recorded, and the ratio of FITC–rhodamine fluorescence intensities was calculated along each individual chromosome and averaged using dedicated software (CytoVision 3.52, Applied Imaging, Sunderland, United Kingdom; ISIS, Version 1.80, MetaSystems, Altlussheim, Germany). Ratio values of 1.25 and 0.75, which have been proven to provide robust criteria for diagnosing overrepresentation and underrepresentation, were used as upper and lower thresholds for the identification of chromosomal imbalances.18 Overrepresentations were considered high-level amplifications when the fluorescence ratio values exceeded 2.0 or when the FITC fluorescence showed strong focal signals and the corresponding ratio profile revealed an overrepresentation. Of the chromosomal regions recognized to be critical in CGH analyses, the centromeric regions, the Y chromosome, 1p32-p36, and chromosome 19 were not scored in the results for reasons specified elsewhere.19 The validity of the combined DOP-PCR–CGH approach was tested by several control experiments described below.
Combined immunophenotyping and genotyping
For combined immunophenotyping and genotyping of CD30+ HRS cells,20 frozen slides containing cryostat sections and imprints of primary cHLs were thawed, fixed for 10 minutes in acetone, and air dried. Immunostaining was performed using a CD30 monoclonal antibody (Dakopatts, Hamburg, Germany) followed by a detection cascade with AMCA-conjugated secondary antibodies. All steps were performed at room temperature with a 30-minute incubation period and subsequent washing. After fluorescence immunostaining, slides were incubated for 10 minutes in Carnoy fixative (methanol-acetic acid), washed in dH2O, and postfixed in 1% paraformaldehyde for 1 minute. Slides were then dehydrated through an ethanol series (70%, 85%, and 100%) and air-dried in the dark. FISH was performed with probes for centromere 2 (CEP-2 labeled with spectrum orange, Vysis) and pooled bacterial artificial chromosomes (BACs) 373L24 and 498O5 covering the RELoncogene (RZPD, Berlin, Germany). BACs were labeled with dUTP-spectrum green (Vysis) by random priming. Thirteen to 16 CD30+ HRS cells were evaluated in each tumor.
Hierarchical cluster analysis
To test for correlations between cytogenetic aberrations and clinical parameters including histologic cHL subtypes, age at tumor onset, and patient gender, a hierarchical cluster analysis of the CGH data was performed. Imbalances for 290 chromosomal subbands of all human metaphase chromosomes21—except 1p13-pter, 9q11, 16q11, 19, and Y (see above)—were used for an uncentered complete clustering. This was performed using the programs Cluster and TreeView, developed by Eisen et al22 (http://rana.lbl.gov/).
Results
Validity of combined DOP-PCR–CGH experiments
The sensitivity and specificity of the combined PCR–CGH approach were determined by analyzing defined pools of cells from 2 cell lines, Colo 320 and MedB-1. Both lines had been thoroughly characterized by chromosome banding and FISH.12-14 Results of CGH analysis with genomic DNA that had not previously been subjected to DOP-PCR (conventional CGH) were compared with those obtained from combined PCR–CGH analysis of defined numbers of cells. Conventional CGH revealed 16 imbalances of chromosomal arms in cell line Colo 320 and 6 imbalances in MedB-1. With pools of 30 isolated cells, 12 of 16 imbalances were detected in Colo 320, and 5 of 6 imbalances were detected in MedB-1. With smaller cell populations, the sensitivity decreased as indicated in Table 1. No false-positive results were obtained in any of the groups under these conditions. Based on these results, all subsequent CGH analyses were performed, starting with approximately 30 cells. The reproducibility of this approach was demonstrated by independent analyses of 2 tumors (cHL-2 and cHL-33) in 2 of the laboratories involved in this study; both laboratories yielded identical results.
Number of chromosomal imbalances detected in small populations of Colo320- and MedB-1 cells by combined DOP-PCR-CGH compared with conventional CGH
| Imbalances detected . | Colo 320 . | MedB-1 . |
|---|---|---|
| Conventional CGH | 16 | 6 |
| 30 cells | 12 | 5 |
| 10 cells | 12 | 4 |
| 5 cells | NA | 3 |
| 1 cell | 8 | 1 |
| Imbalances detected . | Colo 320 . | MedB-1 . |
|---|---|---|
| Conventional CGH | 16 | 6 |
| 30 cells | 12 | 5 |
| 10 cells | 12 | 4 |
| 5 cells | NA | 3 |
| 1 cell | 8 | 1 |
NA indicates no CGH evaluation possible because of poor hybridization quality.
Analysis of HRS cells by combined DOP-PCR–CGH
Pools of malignant HRS cells from 41 different cHL tumors were analyzed. Results are summarized in Figure1 and Table2. Imbalances were detected in all but 4 cHLs with an average number of 5 imbalances per tumor case (range, 0-16 imbalances). These included 158 gains (mean, 3.9 per tumor) and 46 losses (1.1 per tumor). Chromosomal losses most frequently affected chromosomal arms 13q (22%), Xp and Xq (12% each), and 1p, 4q and 17p (7% each). Overrepresentations were predominantly found on chromosomal arms 2p in 54%, on 12q in 37%, and on 17p in 27% of cases. Lower frequencies (less than 25%) were detected for 9p and 16p (24% each), 17q and 20q (20% each), and 9q, 22q and Xq (17% each). Detailed chromosomal subregions involved are indicated in Table 2. Distinct amplifications were found in 7 different tumors and involved chromosomal bands 2p14-p16, 3q21, 4p16, 4q23-q24, 5p15, 5p11-p13, and 12q13-q14.
Ideogram of gains and losses of chromosomal regions in 41 cases of cHL.
Lines on the right of each ideogram indicate gains, lines on the left show losses of chromosomal material. High-level DNA amplifications are symbolized by bold lines. cHL tumors are indicated by numbers.
Ideogram of gains and losses of chromosomal regions in 41 cases of cHL.
Lines on the right of each ideogram indicate gains, lines on the left show losses of chromosomal material. High-level DNA amplifications are symbolized by bold lines. cHL tumors are indicated by numbers.
CGH karyotypes of 41 classical Hodgkin lymphomas
| Case . | Sex . | Age (y) . | Subtype . | CGH karyotype . |
|---|---|---|---|---|
| 1 | M | 53 | cHL-LR | rev ish enh(8p11-p21, 8q11-q24) |
| 2 | F | 13 | cHL-LR | rev ish enh(8), dim(7q11-q21), amp(5p15), |
| 3 | M | 52 | cHL-LR | rev ish enh(1q41-q44, X), dim(6q13-q27) |
| 4 | F | 42 | cHL-LR | rev ish enh(3q13-q29, 4p16, 9p11-p13, 9q33-q34, 12p11-p13, 12q13, 12q24, 16p11-p13, 16q24, 17, 21q22, 22), dim(1p22-p31, Xq11-q28) |
| 5 | F | 43 | cHL-LR | — |
| 6 | F | 46 | cHL-LR | rev ish enh(2p13-p21, 12q23-q24) |
| 7 | M | 26 | cHL-LR | rev ish enh(X), amp(4p16) |
| 8 | M | 48 | cHL-LR | — |
| 9 | M | 33 | cHL-MC | — |
| 10 | M | 49 | cHL-MC | rev ish enh(1q23-q25, 2p13-p25, 2q11-q21, 2q34-q37, 4, 14q22-q32), dim(17p11-p13) |
| 11 | F | 21 | cHL-MC | rev ish enh(2p15-p25, 9p11-p24, 12q22-q24, 20q11-q13), dim(4q32-q35), amp(4q23-q24) |
| 12 | M | 63 | cHL-MC | rev ish enh(2p13-p16, 3p13-p24, 9p11-p24, 11p15, 12q11-q24, 14q11-q32, X) |
| 13 | F | 23 | cHL-MC | rev ish enh(2p13-p25), amp(5p11-p13) |
| 14 | F | 16 | cHL-MC | rev ish enh(4p16, 14q22-q32, 16p11-p13, 17, 20, 21q11-q22, 22q11-q13, X) dim(2q22-q33, 4q11-q34, 13q21-q22) |
| 15 | M | 13 | cHL-MC | rev ish enh(9q34, 12q24, 17p11-p13, 20q13, 22q11-q13) |
| 16 | F | 32 | cHL-MC | rev ish enh(9q34, 11p15, 16p11-p13, 17p11-p13, 17q24-q25, 21q22) |
| 17 | M | 38 | cHL-MC | rev ish enh(3, 15q11-q26), dim(9p22-p24, 12q24, 17p11-p13, X), amp12q13-q14 |
| 18 | M | 17 | cHL-MC | rev ish enh(2p11-p24, 2q35-q37, 4p16, 5p15, 5q35, 12q24, 16, 17, 20, 21q11-q21, 22q11-q13), dim(3p22-p23, 13q11-q21, Xq21-q27) |
| 19 | F | 25 | cHL-MC | rev ish enh(9p21-p24, 13q22-q34, X), dim(22q11-q13) |
| 20 | F | 29 | cHL-MC | rev ish enh(2p11-p25, 6p21-p22, 9q34, 16p11-p13, 16q22-q24, 17) dim(1p22-p31), amp(3q21) |
| 21 | F | 20 | cHL-MC | rev ish enh(1q42-q44, 4q28-4q35, 5q34-5q35, 7q11-7q36, 9p11-9p13, 12q24, 17p11-p12, 17q11-q25), dim(6p25, 8q11-8q24, 11p11-11p14, 11q14-q25, 13q11-q24, 14q32, 20p12-p13, Xp22) |
| 22 | M | 43 | cHL-MC | rev ish enh(2p14-p21, 7p15, 12q13, 16p13, 20q11-q13, 22q11-q12) |
| 23 | M | 15 | cHL-MC | — |
| 24 | F | 55 | cHL-NS | rev ish enh(5p15, 6p25, 7p21, 9p11-p24, 9q21-q32), dim(4q28-q35, 7q11-q36, 13q21), amp(2p16) |
| 25 | F | 32 | cHL-NS | rev ish enh(1q22-q32, 2p14-p25, 2q31-q37, 5q12-q35, 12q14-q21), dim(13q31-q34,18p11) |
| 26 | F | 16 | cHL-NS | rev ish enh(2p15-p16, 2q11-q31, 7p13-p15, 7q32-q36, 16p11-p13) |
| 27 | F | 37 | cHL-NS | rev ish enh(2p14-p25, 9q22-q34, 12q21-q24, 15q22-q26, 20q11-q13) |
| 28 | M | 14 | cHL-NS | rev ish enh(2p13-p25, 9p11-p24, 12q23-q24) |
| 29a | F | 38 | cHL-NS | rev ish enh(2p12-p24, Xq28) |
| 29b | F | 38 | cHL-NS | rev ish enh(2p13-p24, 12q24) |
| 30 | F | 40 | cHL-NS | rev ish enh(2p11-p24, 2q37, 7q36), dim(13q21-q22, 17q25) |
| 31 | F | 27 | cHL-NS | rev ish enh(2p11-p25, 5p11-p15, 12p13, 17, 18p11, 22q13), dim(1p22, 2q11-q37, 6p24-p25, 6q27, 9q31, 13q11-q34, 16q11-q24, Xp21-p22) |
| 32 | F | 46 | cHL-NS | rev ish enh(2p11-2p24, 3p13-3p26, 8q24, 9p24, 13q33-q34) |
| 33 | F | 37 | cHL-NS | rev ish enh(2p12-p21) |
| 34 | M | 27 | cHL-NS | rev ish enh(Xp22, Xq24) |
| 35 | M | 12 | cHL-NS | rev ish enh(2p11-p25, 2q11-q13), dim(X) |
| 36 | M | 38 | cHL-NS | rev ish enh(16p11, 17p11) |
| 37 | M | 40 | cHL-NS | rev ish enh(2p11-p25, 16p11-p12, 17p11-p13,17q11-q12), dim(13q14-q21) |
| 38 | M | 18 | cHL-NS | rev ish enh(1q41-q43, 2p11-p25, 9p11-p13, 9p23-p24, 12p11-p13, 12q12-q13, 12q22-q24, 17p11-p13, 18q11-q23, 20, 22q11-q13), dim(13q21-q22, X) |
| 39 | F | 78 | cHL-LD | rev ish dim(17p11-p13) |
| 40 | F | 23 | cHL-LD | rev ish enh(5p15, 9p11-24, 9q32-34, 12q24, 16p13, 20q11-q13) |
| Case . | Sex . | Age (y) . | Subtype . | CGH karyotype . |
|---|---|---|---|---|
| 1 | M | 53 | cHL-LR | rev ish enh(8p11-p21, 8q11-q24) |
| 2 | F | 13 | cHL-LR | rev ish enh(8), dim(7q11-q21), amp(5p15), |
| 3 | M | 52 | cHL-LR | rev ish enh(1q41-q44, X), dim(6q13-q27) |
| 4 | F | 42 | cHL-LR | rev ish enh(3q13-q29, 4p16, 9p11-p13, 9q33-q34, 12p11-p13, 12q13, 12q24, 16p11-p13, 16q24, 17, 21q22, 22), dim(1p22-p31, Xq11-q28) |
| 5 | F | 43 | cHL-LR | — |
| 6 | F | 46 | cHL-LR | rev ish enh(2p13-p21, 12q23-q24) |
| 7 | M | 26 | cHL-LR | rev ish enh(X), amp(4p16) |
| 8 | M | 48 | cHL-LR | — |
| 9 | M | 33 | cHL-MC | — |
| 10 | M | 49 | cHL-MC | rev ish enh(1q23-q25, 2p13-p25, 2q11-q21, 2q34-q37, 4, 14q22-q32), dim(17p11-p13) |
| 11 | F | 21 | cHL-MC | rev ish enh(2p15-p25, 9p11-p24, 12q22-q24, 20q11-q13), dim(4q32-q35), amp(4q23-q24) |
| 12 | M | 63 | cHL-MC | rev ish enh(2p13-p16, 3p13-p24, 9p11-p24, 11p15, 12q11-q24, 14q11-q32, X) |
| 13 | F | 23 | cHL-MC | rev ish enh(2p13-p25), amp(5p11-p13) |
| 14 | F | 16 | cHL-MC | rev ish enh(4p16, 14q22-q32, 16p11-p13, 17, 20, 21q11-q22, 22q11-q13, X) dim(2q22-q33, 4q11-q34, 13q21-q22) |
| 15 | M | 13 | cHL-MC | rev ish enh(9q34, 12q24, 17p11-p13, 20q13, 22q11-q13) |
| 16 | F | 32 | cHL-MC | rev ish enh(9q34, 11p15, 16p11-p13, 17p11-p13, 17q24-q25, 21q22) |
| 17 | M | 38 | cHL-MC | rev ish enh(3, 15q11-q26), dim(9p22-p24, 12q24, 17p11-p13, X), amp12q13-q14 |
| 18 | M | 17 | cHL-MC | rev ish enh(2p11-p24, 2q35-q37, 4p16, 5p15, 5q35, 12q24, 16, 17, 20, 21q11-q21, 22q11-q13), dim(3p22-p23, 13q11-q21, Xq21-q27) |
| 19 | F | 25 | cHL-MC | rev ish enh(9p21-p24, 13q22-q34, X), dim(22q11-q13) |
| 20 | F | 29 | cHL-MC | rev ish enh(2p11-p25, 6p21-p22, 9q34, 16p11-p13, 16q22-q24, 17) dim(1p22-p31), amp(3q21) |
| 21 | F | 20 | cHL-MC | rev ish enh(1q42-q44, 4q28-4q35, 5q34-5q35, 7q11-7q36, 9p11-9p13, 12q24, 17p11-p12, 17q11-q25), dim(6p25, 8q11-8q24, 11p11-11p14, 11q14-q25, 13q11-q24, 14q32, 20p12-p13, Xp22) |
| 22 | M | 43 | cHL-MC | rev ish enh(2p14-p21, 7p15, 12q13, 16p13, 20q11-q13, 22q11-q12) |
| 23 | M | 15 | cHL-MC | — |
| 24 | F | 55 | cHL-NS | rev ish enh(5p15, 6p25, 7p21, 9p11-p24, 9q21-q32), dim(4q28-q35, 7q11-q36, 13q21), amp(2p16) |
| 25 | F | 32 | cHL-NS | rev ish enh(1q22-q32, 2p14-p25, 2q31-q37, 5q12-q35, 12q14-q21), dim(13q31-q34,18p11) |
| 26 | F | 16 | cHL-NS | rev ish enh(2p15-p16, 2q11-q31, 7p13-p15, 7q32-q36, 16p11-p13) |
| 27 | F | 37 | cHL-NS | rev ish enh(2p14-p25, 9q22-q34, 12q21-q24, 15q22-q26, 20q11-q13) |
| 28 | M | 14 | cHL-NS | rev ish enh(2p13-p25, 9p11-p24, 12q23-q24) |
| 29a | F | 38 | cHL-NS | rev ish enh(2p12-p24, Xq28) |
| 29b | F | 38 | cHL-NS | rev ish enh(2p13-p24, 12q24) |
| 30 | F | 40 | cHL-NS | rev ish enh(2p11-p24, 2q37, 7q36), dim(13q21-q22, 17q25) |
| 31 | F | 27 | cHL-NS | rev ish enh(2p11-p25, 5p11-p15, 12p13, 17, 18p11, 22q13), dim(1p22, 2q11-q37, 6p24-p25, 6q27, 9q31, 13q11-q34, 16q11-q24, Xp21-p22) |
| 32 | F | 46 | cHL-NS | rev ish enh(2p11-2p24, 3p13-3p26, 8q24, 9p24, 13q33-q34) |
| 33 | F | 37 | cHL-NS | rev ish enh(2p12-p21) |
| 34 | M | 27 | cHL-NS | rev ish enh(Xp22, Xq24) |
| 35 | M | 12 | cHL-NS | rev ish enh(2p11-p25, 2q11-q13), dim(X) |
| 36 | M | 38 | cHL-NS | rev ish enh(16p11, 17p11) |
| 37 | M | 40 | cHL-NS | rev ish enh(2p11-p25, 16p11-p12, 17p11-p13,17q11-q12), dim(13q14-q21) |
| 38 | M | 18 | cHL-NS | rev ish enh(1q41-q43, 2p11-p25, 9p11-p13, 9p23-p24, 12p11-p13, 12q12-q13, 12q22-q24, 17p11-p13, 18q11-q23, 20, 22q11-q13), dim(13q21-q22, X) |
| 39 | F | 78 | cHL-LD | rev ish dim(17p11-p13) |
| 40 | F | 23 | cHL-LD | rev ish enh(5p15, 9p11-24, 9q32-34, 12q24, 16p13, 20q11-q13) |
Among the cHL subtypes, there were numerical differences in chromosomal aberrations. The average number of imbalances was highest in cHL-MC, as indicated for selected chromosomes shown in Table3. Gains of chromosomal arm 2p were highly characteristic for cHL-NS (88% of cases) but occurred less frequently in cHL-MC (47% of cases) and cHL-LR (13% of cases).
Frequency of chromosomal gains in cHL-subtypes (absolute numbers)
| Chromosomal arm . | 2p . | 5p . | 14q . | 8p . | 17q . | 20q . | 21q . |
|---|---|---|---|---|---|---|---|
| cHL-MC (n = 15) | 7 | 3 | 3 | 0 | 5 | 3 | 3 |
| cHL-NS (n = 16) | 14 | 1 | 0 | 0 | 2 | 2 | 0 |
| cHL-LR (n = 8) | 1 | 1 | 0 | 2 | 1 | 0 | 1 |
| Chromosomal arm . | 2p . | 5p . | 14q . | 8p . | 17q . | 20q . | 21q . |
|---|---|---|---|---|---|---|---|
| cHL-MC (n = 15) | 7 | 3 | 3 | 0 | 5 | 3 | 3 |
| cHL-NS (n = 16) | 14 | 1 | 0 | 0 | 2 | 2 | 0 |
| cHL-LR (n = 8) | 1 | 1 | 0 | 2 | 1 | 0 | 1 |
Hierarchical cluster analysis
To identify possible genotype–phenotype relationships, hierarchical cluster analysis based on CGH data was performed. Only tumors in which chromosomal imbalances had been identified were included in this analysis.
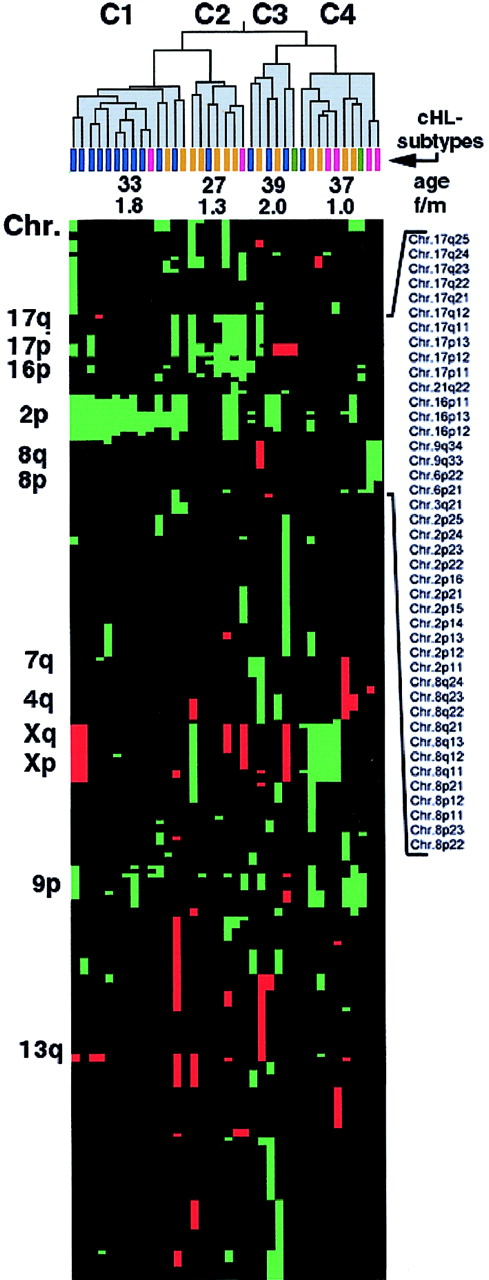
Figure 2 shows the resultant dendrogram of the clustered tumor cases. Pairs of tumors with the highest degree of similarity are connected and linked with further similar cases resulting in the generation of higher order clusters, with short or long “branches” indicating a higher or lower degree of similarity, respectively. Comparing the tumors in clusters C1 to C4, indicated in Figure 2, revealed a nonrandom distribution of cHL subtypes: 11 of 16 cHL-NS cases were attributed to a single cluster (C1), whereas 4 of 6 cHL-LR subtypes appeared in cluster C4. cHL-MC tumors were found distributed in clusters C2 to C4. This indicates a strong association of most of the cHL-NS tumors, with a pattern of chromosomal imbalances that is distinct from that in cHL-MC and cHL-LR.
Hierarchical cluster analysis of 37 cHLs with chromosomal imbalances.
Data are according to Eisen et al.22 The dendrogram was divided into clusters C1 to C4, which are indicated in gray. Histologic subtypes of the clustered cHL tumors are indicated by differently colored bars—cHL-NS in blue, cHL-MC in orange, cHL-LR in violet, and cHL-LD in green. Average values of age at time of diagnosis and the female-to-male ratio (f/m) in C1 to C4 are indicated. Below the dendrogram, all gains and losses of chromosomal subbands are represented by green and red squares, respectively. Selected chromosomal arms and subbands are indicated. Note the clustering of cHL-NS tumors in C1 compared with the dispersed distribution of cHL-MC tumors in clusters C2 to C4. In C2 a distinct group of mostly cHL-MC cases is visible, associated with gains of chromosome 17p and a low average age of the patients at the time of diagnosis.
Hierarchical cluster analysis of 37 cHLs with chromosomal imbalances.
Data are according to Eisen et al.22 The dendrogram was divided into clusters C1 to C4, which are indicated in gray. Histologic subtypes of the clustered cHL tumors are indicated by differently colored bars—cHL-NS in blue, cHL-MC in orange, cHL-LR in violet, and cHL-LD in green. Average values of age at time of diagnosis and the female-to-male ratio (f/m) in C1 to C4 are indicated. Below the dendrogram, all gains and losses of chromosomal subbands are represented by green and red squares, respectively. Selected chromosomal arms and subbands are indicated. Note the clustering of cHL-NS tumors in C1 compared with the dispersed distribution of cHL-MC tumors in clusters C2 to C4. In C2 a distinct group of mostly cHL-MC cases is visible, associated with gains of chromosome 17p and a low average age of the patients at the time of diagnosis.
The variously clustered patterns were also correlated with age at time of diagnosis. Thus, patients in clusters C1 and C2 were younger than those in C3 and C4 (Figure 2). The cluster containing the youngest patients was C2 (mean age, 27 years). In addition, this cluster was predominantly composed of tumors of the cHL-MC subtype and frequently showed overrepresentation of 17p. This is indicative of a possible correlation of 17p gains with early tumor onset within a subgroup of cHL-MC. Concerning the distribution of sexes in the 4 clusters, there was a predominance of female patients in clusters C1 and C3 (female-to-male ratio, 1.8 and 2.0, respectively), whereas in C2 and C4 an almost equal ratio was observed (female-to-male ratio, 1.3 and 1.0, respectively). C1 and C3 are largely composed of cHL-NS tumors, reflecting the predominance of females in this subtype.23
Analysis of increased copy number of REL
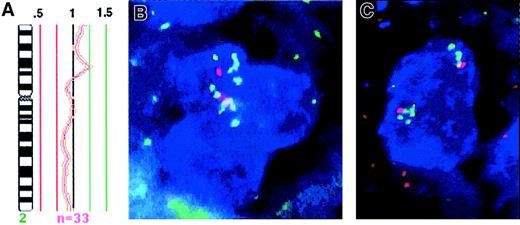
In 3 cHL tumors, in which CGH detected a chromosome 2p gain (cHL-24, cHL-31, and cHL-33), the copy number of the RELoncogene located within 2p14-p15 could be determined. This was performed by combined immunophenotyping and genotyping analysis of CD30+ HRS cells without prior knowledge of the CGH results. BAC clones spanning the complete REL gene were used in addition to a differentially labeled control probe specific for the chromosome 2 centromere. Consistent with CGH, cHL-31 showed increased copy numbers of the centromeric region and the REL locus (median, 5 copies each). Case cHL-33 displayed a median of 4 copies of the chromosome 2 centromeric region and a median of 6 copies of theREL gene, also coinciding well with the CGH results (Figure1). The signal pattern in this case suggested duplication within chromosomal arm 2p. In cHL-24, which according to CGH exhibits a distinct high-level amplification of chromosomal band 2p15-p16, a median number of 3 centromere-2 signals versus 9 REL signals was observed (Figure 3). This demonstrates that the REL locus is included within the distinct 2p15-p16 amplicon, which constitutes the consensus region of 2p gains detected in cHLs by CGH.
Amplification of REL on chromosome 2p.
(A) Mean ratio profile of chromosome 2 in cHL-24 derived from 33 chromosomes analyzed by CGH (central line = ratio value 1; green lines = ratio values of 1.25 and 1.5; red lines = values of 0.75 and 0.5). The profile shows a distinct peak at chromosomal region 2p15-p16, which reaches the diagnostic threshold of 1.25 and indicates high-level DNA amplification. This region corresponds to the mapping position of the REL oncogene. (B, C) FISH analysis of cHL-24 with a REL specific probe (green) and a centromeric specific probe for chromosome 2 (red) on CD30+HRS cells (blue). Three to 4 centromeric signals are detected in each cell. Multiple green signals indicate amplification ofREL, therefore revealing this oncogene as part of the 2p15-p16 amplicon detected by CGH. Original magnification, ×630.
Amplification of REL on chromosome 2p.
(A) Mean ratio profile of chromosome 2 in cHL-24 derived from 33 chromosomes analyzed by CGH (central line = ratio value 1; green lines = ratio values of 1.25 and 1.5; red lines = values of 0.75 and 0.5). The profile shows a distinct peak at chromosomal region 2p15-p16, which reaches the diagnostic threshold of 1.25 and indicates high-level DNA amplification. This region corresponds to the mapping position of the REL oncogene. (B, C) FISH analysis of cHL-24 with a REL specific probe (green) and a centromeric specific probe for chromosome 2 (red) on CD30+HRS cells (blue). Three to 4 centromeric signals are detected in each cell. Multiple green signals indicate amplification ofREL, therefore revealing this oncogene as part of the 2p15-p16 amplicon detected by CGH. Original magnification, ×630.
Discussion
In the current study we show that chromosomal imbalances can be detected with high sensitivity and specificity in pools of 30 microdissected cells. The potential of this combined DOP-PCR–CGH method has been demonstrated by several groups, though larger pools of cells were analyzed in most studies.9-11,24-29 The application of this powerful approach for the analysis of enriched HRS cells in cHL revealed highly characteristic chromosomal imbalances. The most typical change was an overrepresentation of chromosomal arm 2p, which was observed in approximately half of all cases analyzed and in 88% of the cHL-NS subtype. Such a characteristic abnormality has not been observed in previous chromosome banding studies. Our analysis also indicated distinct patterns of cytogenetic aberrations associated with different cHL subtypes. Finally, evidence was provided to show that the classical type of Hodgkin lymphoma clearly differs from NLPHL.11 None of the chromosomes most frequently imbalanced in NLPHL (eg, gains of 1q, 3p, 5q, and Xq and losses of 11q and 17p) was affected in cHL. On the other hand, only a few of the imbalances typically found in cHL (eg, 2p, 9p, and 13q) were observed in NLPHL. Hence, Hodgkin lymphoma cannot be subdivided based only on morphologic and immunologic parameters,23 it can be subdivided based on the genomic level.
To date, chromosome-banding analysis has contributed most information about cytogenetic aberrations in HRS cells. Among a number of nonrandom changes associated with HL, additional copies of chromosomes 2, 5, 9, and 12 were observed by several groups in up to 46% of cases.30-35 However, a wide range of other chromosomes was found to be affected by numerical changes, and the significance of individual aberrations was thus difficult to estimate. By measuring net chromosomal gains and losses in pools of HRS cells, a more specific pattern of recurrent chromosomal imbalances was detected that involved relatively few chromosomal arms or subregions (eg, 2p15-p16, 9p23-p24, 12q24, 13q14-q21, 16p and 17). Therefore, CGH results point to the localization of genes that might be critical for the etiology of cHL.
Gains of chromosome 2p, the most frequent aberration found in 54% of cHL cases, were only detected recurrently in diffuse large B-cell lymphoma and mediastinal B cell lymphoma (23% and 22%, respectively).17,36,37 This high frequency points to a gene important for cHL pathogenesis. The consensus region of chromosome 2p gains involved band 2p15-p16, which corresponds to the mapping position of the REL oncogene.38 Gains and high-level amplifications of this gene were also found in approximately 80% of cHL tumors by combined immunophenotyping and interphase cytogenetics (see accompanying article by Martı́n-Subero et al,39 page 1474). REL codes for a subunit of NF-κB transcription factors, which are associated with cell proliferation and tumor development,40-43 and it was reported to be constitutively activated in HRS cells.44Interestingly, mutations in the IκB gene, a potent inhibitor of NF-κB, have been described in HRS cells, which could account for NF-κB activation. However, most HL cell lines and HRS cells from primary tumors lack IκB mutations.45-47 The high frequency of REL overrepresentations on chromosome 2p15-p16 detected by CGH and FISH in HRS cells suggests an alternative mechanism leading to NF-κB activation in Hodgkin disease, namely a gene dose effect resulting in an increased expression of the REL gene.
Gains of chromosomal arm 9p were detected in 24% of cHLs. The smallest overrepresented regions included 9p11-p12 and 9p24. Interesting candidate genes within these chromosomal bands are the NF1Bgene encoding for a transcription factor and the tyrosine kinase geneJAK2, both of which are located on 9p24. JAK2 is involved in signal transduction of cytokine receptors48,49and was recently shown to be amplified in the HL-derived line HDLM-2 and in one case of NLPHL.10 Interestingly, both the JAK/STAT system and the NF-κB system were reported to activate a number of target genes synergistically.50 51
In HRS cells, 9p gains were detected in 25% of cHL tumors. The frequency of this aberration is topped by mediastinal B-cell lymphoma (MBL), in which 9p gains were detected by CGH in more than 50% of tumors. In contrast, in all other B-NHLs tested so far, cases with 9p gains account for just 2%.14 Both, cHL and MBL also exhibit frequent gains of chromosomal arm 2p, including theREL oncogene.14,37 These parallels support a recent hypothesis suggesting a pathogenic relationship between these 2 types of lymphoma. This hypothesis arose from the observation of rare cases of cHL–MBL of composite lymphomas.52 In addition, both share a number of clinical and immunologic features, including their frequent mediastinal origin and the lack of functional expression of HLA class I and immunoglobulin molecules.53-55
Concerning chromosomal region 12q24, further candidates include BCL7A56 and the gene for the HRS-specific intermediate filament, restin,57 whereas chromosome 16p13 harbors cyclin F.58 The RB1 gene is located within the recurrently deleted region 13q13-q21, and there is strong evidence of further tumor suppressor genes residing within this region.59-61
Facing the complexity of chromosomal imbalances we found in HRS cells, hierarchical cluster analysis was carried out to eventually detect genotype–phenotype relations. Four clusters emerged. Cluster C1 was predominantly attributed to cHL-NS. Clusters C2 and C4 were predominantly attributed to cHL-MC. Cluster C4 also comprised most of cHL-LR. Cluster C3 did not appear to be related to cHL subtypes.
The different clustering of cHL subtypes suggests nonrandom combinations of chromosomal imbalances. First, cHL-NS showed a particularly high frequency of 2p gains. Second, the major features of differences between cHL-MC clustering in C2 and C4 were age of onset and 17p+. Overrepresentation of 17p in cHL-MC was more frequent in younger patients than in older patients. These results have now to be confirmed in an independent cohort of cHL tumors. However, the different emerging clusters argue for different routes of pathogenesis in cHL, with 2p gains as an early event or a common feature.
We thank Axel Benner (DKFZ, Heidelberg) for discussion of statistical analysis, Iwona Nerbas for excellent technical assistance, and Caroline Higginson for editorial help.
Supported by the Tumorzentrum Heidelberg/Mannheim (I.I.2), the Deutsche Krebshilfe (70-2310-Ba), and the IZKF Kiel.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Peter Möller, Abteilung für Pathologie, Universität Ulm, Albert- Einstein-Allee 11, D-89081 Ulm, Germany; e-mail: peter.moeller@medizin.uni-ulm.de.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal