Comparative genomic hybridization studies have shown gains in chromosome region 2p as the most common imbalance in classical Hodgkin lymphoma (cHL). The minimal region of gain contained 2 candidate oncogenes, REL and BCL11A. This study examined the involvement of REL and BCL11A loci in 44 primary cases of cHL by combined immunophenotyping and interphase cytogenetics (FICTION). A median 2p13 copy number above the tetraploid range was detected in 24 (55%) cases. Adjustment for centromere 2 copy number indicated gains of 2p13 in 11 of 31 cHLs (35%) with 8 (26%) high-level amplifications. One cHL displayed selective amplification of the REL locus not affectingBCL11A; another case studied by FICTION and a cHL with cytogenetic 2p change investigated by fluorescence in situ hybridization showed signal patterns suggesting breakpoints in the region spanned by the REL probe. These data indicate thatREL rather than BCL11A may be the target of the 2p13 alterations in cHL.
Introduction
The chromosomal alterations underlying Hodgkin lymphoma (HL) remain largely unknown.1-4Cytogenetic banding studies suffer from the low proportion and genetic complexity of the mononuclear large Hodgkin (H) cells and multinucleated Reed-Sternberg (RS) cells, which constitute the neoplastic compartment in HL.5-8 Combined immunophenotypic and interphase cytogenetic studies by means of the FICTION (fluorescence immunophenotyping and interphasecytogenetics as a tool for theinvestigation of neoplasms) techniques have shown numerical chromosome abnormalities to be present in all cases of HL and to be limited to the HRS cells.9 Recently, comparative genomic hybridization (CGH) with DNA from microdissected HRS cells revealed recurrent gains in 2p, 9p, and 12q in the most prevalent HL category, that is, classical HL (cHL).10 Gains in 2p were observed in more than 50% of cHL with a minimal region of gain containing 2 closely linked candidate oncogenes, BCL11A and REL (see Joos et al,10 and the accompanying article by Joos et al,26 page 1381). REL encodes a nuclear factor (NF)–κB transcription factor and is frequently amplified in various B-lineage neoplasias.11-16BCL11A was identified from B-cell lymphoma–associated t(2;14)(p13;q32) translocations and codes for a zinc-finger transcription factor. BCL11A is overexpressed in B-cell lymphomas with t(2;14) and HL cell lines.17
Here we investigated 46 primary cHLs for alterations of RELand BCL11A loci by interphase cytogenetics. Our results confirm gains in 2p including high-level amplifications to be highly recurrent in cHL and suggest REL rather thanBCL11A to be a target of these alterations.
Study design
Patient material
Samples from 73 patients were included in the study. Fresh tumor material was submitted to the Institute of Human Genetics of the University Hospital Kiel as part of the diagnostic workup. Based on the availability of cytospin slides of sufficient quality for the specific FICTION analyses,9,18 46 cases (63%) were eligible for the study including 44 cases of cHL (Table1) and 2 samples from benign lymphoid hyperplasias. Central histopathologic review (by M.L.H.) could be performed in 24 cHLs and confirmed the histologic diagnosis of cHL in all these cases. In addition, 2 cHLs (cHL45 and cHL46) with a karyotype containing structural alterations in 2p were selected for fluorescent in situ hybridization (FISH). The karyotype of the Hodgkin sarcoma cHL46 was reported previously (case 11 in Schlegelberger et al4).
Results of the FICTION analyses for the REL andBCL11A loci in 44 cHLs
| . | Age at diagnosis, y . | Sex . | Estimated proportion of HRS cells . | HRS cells evaluated . | Copies of D2Z1median (range) . | Copies of REL median (range) . | Scores* median (range) . | REL probe split . | RELstatus . | BCL11A status . | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| REL-D2Z1 . | REL/D2Z1 . | ||||||||||
| 1 | 25 | M | 1/100 | 17 | 4 (4-6) | 30 (10-30) | 26 (5-26) | 7.5 (2-7.5) | No | A | A |
| 2 | 21 | F | 1/50 | 24 | 5 (4-6) | ∼ 30 | 25 (24-26) | 6 (5-7.5) | No | A | A |
| 3 | 46 | M | 1/200 | 30 | 2 (2-4) | 8.5 (7-12) | 6.5 (2-10) | 3.8 (1.5-6) | No | A | A |
| 4 | 29 | F | 1/100 | 35 | 3 (3-6) | 11 (9-30) | 8 (5-24) | 3.7 (2.7-5) | No | A | A |
| 5 | 41 | M | 1/500 | 18 | 3 (3-4) | 10 (10-25) | 7 (1-21) | 3.3 (1.3-6.3) | No | A | A |
| 6 | 68 | F | 1/500 | 15 | 3 (2-9) | 10 (6-30) | 7 (2-21) | 3 (1.5-4.4) | No | A | A |
| 7 | 32 | F | 1/1000 | 7 | 4 (2-5) | 10 (7-30) | 6 (4-25) | 3 (2.5-6) | No | A | A |
| 8 | 28 | M | 1/1000 | 7 | 3 | 9 (7-12) | 6 (4-9) | 3 (2.3-4) | No | A | B |
| 9 | 21 | M | 1/500 | 6 | 4 | 10 (8-14) | 6 (4-10) | 2.5 (2-3.5) | No | G | G |
| 10 | 43 | F | 1/1000 | 7 | 4 (3-6) | 8 (7-10) | 4 (3-4) | 1.8 (1.7-2) | No | G | G |
| 11 | 23 | F | 1/100 | 9 | 4 (3-8) | 8 (4-17) | 4 (0-9) | 1.7 (1-2.7) | No | G | G |
| 12 | 27 | F | 1/50 | 18 | 3 (3-6) | 4 (4-7) | 1 (1-4) | 1.3 (1.3-2.3) | No | B | B |
| 13 | 36 | F | 1/1000 | 16 | 3 (3-8) | 4 (3-9) | 1 (0-2) | 1.3 (1-1.7) | No | B | B |
| 14 | 59 | M | 1/100 | 21 | 4 (3-4) | 4 (3-6) | 1 (0-2) | 1.2 (1-1.5) | No | B | B |
| 15 | 24 | F | 1/5000 | 6 | 4 | 4.5 (4-5) | 0.5 (0-1) | 1.1 (1-1.3) | No | B | B |
| 16 | 21 | F | 1/50 | 20 | 4 (4-6) | 4 (4-6) | 0 (0-2) | 1 (1-1.5) | No | B | B |
| 17 | 27 | M | 1/200 | 10 | 5.5 (3-6) | 5.5 (3-6) | 0 (0-1) | 1 (1-1.3) | Yes | R | B |
| 18 | 28 | M | 1/200 | 15 | 2 (2-3) | 2 (2-4) | 0 (0-1) | 1 (1-1.3) | No | B | B |
| 19 | 30 | M | 1/100 | 11 | 4 (3-6) | 4 (3-6) | 0 (0-1) | 1 (1-1.3) | No | B | B |
| 20 | 70 | F | 1/200 | 10 | 5 (4-6) | 5 (4-6) | 0 (0-1) | 1 (1-1.2) | No | B | B |
| 21 | 36 | M | 1/500 | 9 | 4 (4-5) | 5 (4-5) | 0 (0-1) | 1 (1-1.2) | No | B | B |
| 22 | 56 | M | 1/100 | 9 | 5 (5-6) | 5 (5-6) | 0 (0-1) | 1 (1-1.2) | No | B | B |
| 23 | 49 | M | 1/500 | 9 | 4 (3-6) | 4 (3-6) | 0 | 1 | No | B | B |
| 24 | 26 | M | 1/200 | 18 | 4 (3-6) | 4 (3-6) | 0 | 1 | No | B | B |
| 25 | 19 | M | 1/500 | 5 | 3 (3-4) | 3 (3-4) | 0 | 1 | No | B | B |
| 26 | 8 | M | 1/200 | 14 | 2 (2-4) | 2 (2-4) | 0 | 1 | No | B | B |
| 27 | 40 | M | 1/100 | 13 | 2 | 2 | 0 | 1 | No | B | B |
| 28 | 46 | M | 1/100 | 19 | 4 (3-6) | 4 (3-6) | 0 [(-1)-2] | 1 (0.8-1.7) | No | B | B |
| 29 | 20 | M | 1/100 | 14 | 4.5 (4-7) | 4.5 (4-7) | 0 [(-1)-1] | 1 (0.8-1.2) | No | B | B |
| 30 | 42 | F | 1/200 | 11 | 3 (2-5) | 3 (3-5) | 0 [(-1)-1] | 1 (0.7-1.5) | No | B | B |
| 31 | 41 | M | 1/100 | 12 | 4.5 (4-6) | 4 (4-6) | −0.5 [(-1)-0] | 0.9 (0.7-1) | No | B | B |
| 32 | 8 | M | 1/200 | 10 | ND | 9 (4-10) | NA | NA | No | NA | NA |
| 33 | 35 | M | 1/1500 | 14 | ND | 8 (4-10) | NA | NA | No | NA | NA |
| 34 | 34 | M | 1/1500 | 7 | ND | 6 (6-10) | NA | NA | No | NA | NA |
| 35 | 25 | F | 1/1000 | 6 | ND | 6 (6-10) | NA | NA | No | NA | NA |
| 36 | 15 | F | 1/500 | 7 | ND | 5 (4-10) | NA | NA | No | NA | NA |
| 37 | 20 | M | 1/100 | 14 | ND | 5 (5-10) | NA | NA | No | NA | NA |
| 38 | 66 | F | 1/100 | 20 | ND | 4.5 (3-8) | NA | NA | No | NA | NA |
| 39 | 57 | M | 1/200 | 20 | ND | 4 (4-10) | NA | NA | No | NA | NA |
| 40 | 66 | M | 1/300 | 7 | ND | 4 (3-5) | NA | NA | No | NA | NA |
| 41 | 23 | F | 1/100 | 26 | ND | 3.5 (3-7) | NA | NA | No | NA | NA |
| 42 | 23 | M | 1/1000 | 11 | ND | 2 (2-3) | NA | NA | No | NA | NA |
| 43 | 53 | M | 1/1000 | 10 | ND | 2 (2-3) | NA | NA | No | NA | NA |
| 44 | 28 | M | 1/200 | 26 | ND | 2 (1-3) | NA | NA | No | NA | NA |
| . | Age at diagnosis, y . | Sex . | Estimated proportion of HRS cells . | HRS cells evaluated . | Copies of D2Z1median (range) . | Copies of REL median (range) . | Scores* median (range) . | REL probe split . | RELstatus . | BCL11A status . | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| REL-D2Z1 . | REL/D2Z1 . | ||||||||||
| 1 | 25 | M | 1/100 | 17 | 4 (4-6) | 30 (10-30) | 26 (5-26) | 7.5 (2-7.5) | No | A | A |
| 2 | 21 | F | 1/50 | 24 | 5 (4-6) | ∼ 30 | 25 (24-26) | 6 (5-7.5) | No | A | A |
| 3 | 46 | M | 1/200 | 30 | 2 (2-4) | 8.5 (7-12) | 6.5 (2-10) | 3.8 (1.5-6) | No | A | A |
| 4 | 29 | F | 1/100 | 35 | 3 (3-6) | 11 (9-30) | 8 (5-24) | 3.7 (2.7-5) | No | A | A |
| 5 | 41 | M | 1/500 | 18 | 3 (3-4) | 10 (10-25) | 7 (1-21) | 3.3 (1.3-6.3) | No | A | A |
| 6 | 68 | F | 1/500 | 15 | 3 (2-9) | 10 (6-30) | 7 (2-21) | 3 (1.5-4.4) | No | A | A |
| 7 | 32 | F | 1/1000 | 7 | 4 (2-5) | 10 (7-30) | 6 (4-25) | 3 (2.5-6) | No | A | A |
| 8 | 28 | M | 1/1000 | 7 | 3 | 9 (7-12) | 6 (4-9) | 3 (2.3-4) | No | A | B |
| 9 | 21 | M | 1/500 | 6 | 4 | 10 (8-14) | 6 (4-10) | 2.5 (2-3.5) | No | G | G |
| 10 | 43 | F | 1/1000 | 7 | 4 (3-6) | 8 (7-10) | 4 (3-4) | 1.8 (1.7-2) | No | G | G |
| 11 | 23 | F | 1/100 | 9 | 4 (3-8) | 8 (4-17) | 4 (0-9) | 1.7 (1-2.7) | No | G | G |
| 12 | 27 | F | 1/50 | 18 | 3 (3-6) | 4 (4-7) | 1 (1-4) | 1.3 (1.3-2.3) | No | B | B |
| 13 | 36 | F | 1/1000 | 16 | 3 (3-8) | 4 (3-9) | 1 (0-2) | 1.3 (1-1.7) | No | B | B |
| 14 | 59 | M | 1/100 | 21 | 4 (3-4) | 4 (3-6) | 1 (0-2) | 1.2 (1-1.5) | No | B | B |
| 15 | 24 | F | 1/5000 | 6 | 4 | 4.5 (4-5) | 0.5 (0-1) | 1.1 (1-1.3) | No | B | B |
| 16 | 21 | F | 1/50 | 20 | 4 (4-6) | 4 (4-6) | 0 (0-2) | 1 (1-1.5) | No | B | B |
| 17 | 27 | M | 1/200 | 10 | 5.5 (3-6) | 5.5 (3-6) | 0 (0-1) | 1 (1-1.3) | Yes | R | B |
| 18 | 28 | M | 1/200 | 15 | 2 (2-3) | 2 (2-4) | 0 (0-1) | 1 (1-1.3) | No | B | B |
| 19 | 30 | M | 1/100 | 11 | 4 (3-6) | 4 (3-6) | 0 (0-1) | 1 (1-1.3) | No | B | B |
| 20 | 70 | F | 1/200 | 10 | 5 (4-6) | 5 (4-6) | 0 (0-1) | 1 (1-1.2) | No | B | B |
| 21 | 36 | M | 1/500 | 9 | 4 (4-5) | 5 (4-5) | 0 (0-1) | 1 (1-1.2) | No | B | B |
| 22 | 56 | M | 1/100 | 9 | 5 (5-6) | 5 (5-6) | 0 (0-1) | 1 (1-1.2) | No | B | B |
| 23 | 49 | M | 1/500 | 9 | 4 (3-6) | 4 (3-6) | 0 | 1 | No | B | B |
| 24 | 26 | M | 1/200 | 18 | 4 (3-6) | 4 (3-6) | 0 | 1 | No | B | B |
| 25 | 19 | M | 1/500 | 5 | 3 (3-4) | 3 (3-4) | 0 | 1 | No | B | B |
| 26 | 8 | M | 1/200 | 14 | 2 (2-4) | 2 (2-4) | 0 | 1 | No | B | B |
| 27 | 40 | M | 1/100 | 13 | 2 | 2 | 0 | 1 | No | B | B |
| 28 | 46 | M | 1/100 | 19 | 4 (3-6) | 4 (3-6) | 0 [(-1)-2] | 1 (0.8-1.7) | No | B | B |
| 29 | 20 | M | 1/100 | 14 | 4.5 (4-7) | 4.5 (4-7) | 0 [(-1)-1] | 1 (0.8-1.2) | No | B | B |
| 30 | 42 | F | 1/200 | 11 | 3 (2-5) | 3 (3-5) | 0 [(-1)-1] | 1 (0.7-1.5) | No | B | B |
| 31 | 41 | M | 1/100 | 12 | 4.5 (4-6) | 4 (4-6) | −0.5 [(-1)-0] | 0.9 (0.7-1) | No | B | B |
| 32 | 8 | M | 1/200 | 10 | ND | 9 (4-10) | NA | NA | No | NA | NA |
| 33 | 35 | M | 1/1500 | 14 | ND | 8 (4-10) | NA | NA | No | NA | NA |
| 34 | 34 | M | 1/1500 | 7 | ND | 6 (6-10) | NA | NA | No | NA | NA |
| 35 | 25 | F | 1/1000 | 6 | ND | 6 (6-10) | NA | NA | No | NA | NA |
| 36 | 15 | F | 1/500 | 7 | ND | 5 (4-10) | NA | NA | No | NA | NA |
| 37 | 20 | M | 1/100 | 14 | ND | 5 (5-10) | NA | NA | No | NA | NA |
| 38 | 66 | F | 1/100 | 20 | ND | 4.5 (3-8) | NA | NA | No | NA | NA |
| 39 | 57 | M | 1/200 | 20 | ND | 4 (4-10) | NA | NA | No | NA | NA |
| 40 | 66 | M | 1/300 | 7 | ND | 4 (3-5) | NA | NA | No | NA | NA |
| 41 | 23 | F | 1/100 | 26 | ND | 3.5 (3-7) | NA | NA | No | NA | NA |
| 42 | 23 | M | 1/1000 | 11 | ND | 2 (2-3) | NA | NA | No | NA | NA |
| 43 | 53 | M | 1/1000 | 10 | ND | 2 (2-3) | NA | NA | No | NA | NA |
| 44 | 28 | M | 1/200 | 26 | ND | 2 (1-3) | NA | NA | No | NA | NA |
A indicates amplified; G, gain; B, balanced (for definitions see text); R, rearranged; ND, not determined; NA, not applicable; M, male; and F, female.
REL-D2Z1 is number of signals for theREL probe minus number of signals for the CEP2 probe;REL/D2Z1 is number of signals for the REL probe divided by number of signals for the CEP2 probe.
FISH and combined immunophenotyping and interphase cytogenetics (FICTION)
Pooled BAC clones RP11-440P5 and RP11-158I21 labeled with Spectrum Orange (SO, Vysis, Downers Grove, IL) were used as probes for the BCL11A locus; pooled BAC clones RP11-373L24 and RP11-498O5 or Alu-polymerase chain reaction (PCR) products from CEPH-YAC clones 747H5 and 927G9 labeled with Spectrum Green (SG; Vysis) served as probes for the REL locus.16,17 The chromosome 2 centromeric probe was obtained from Vysis (CEP2, locusD2Z1, SO). For FICTION, indirect immunophenotyping using a monoclonal antibody to CD30 (Dako, Hamburg, Germany) and aminomethyl-coumarin (AMCA)–labeled secondary antibodies (rabbit antimouse, goat antirabbit, and donkey antigoat; Jackson-Immunoresearch/Dianova, Hamburg, Germany) was performed as described.9,18 After fixation, 1.5 μL hybridization solution containing 200 ng of each labeled DNA (or 0.5-1 μL CEP2) and 5 μL Cot1-DNA (Gibco/Life-Technologies, Eggenstein, Germany) in 10 μL 2 times standard sodium citrate (SSC), 50% formamide, and 10% dextran sulfate were applied. Following denaturation at 70°C for 12 minutes and incubation overnight at 37°C, slides were washed in 0.1 times SSC 3 times for 5 minutes at 60°C and mounted in antifade solution. Slides were analyzed by use of a Zeiss Axioskop-2 fluorescence microscope (Göttingen, Germany) equipped with appropriate filter sets (AHF, Tübingen, Germany) and documented using the ISIS imaging system (Metasystems, Altlussheim, Germany). A median of 13 (range, 5-35) HRS cells identified by virtue of the CD30 positivity and typical morphology were evaluated per slide. FISH analyses were done similarly on cells left from chromosome analysis as detailed elsewhere.18
Results and discussion
FICTION of cHL and lymphoid hyperplasias was performed blindly without knowledge of the histopathologic diagnosis. In the 2 patients with reactive lymphoid hyperplasias, CD30+ cells were detected but recorded to lack HRS morphology. There was no evidence for copy number changes or structural alterations in 2p in these cells as well as in 50 CD30− cells scored for each assay as internal controls in the cHL cases.
By FICTION applying REL-YAC and BCL11A-BAC pools, the median numbers of BCL11A and RELcopies in the CD30+ HRS cells ranged from 2 to approximately 30 in the 44 cHLs (Table 1). Median 2p13 copy numbers of more than 2 or 4 were detected in 38 (86%) and 24 (55%) samples, respectively, confirming supernumerary copies of 2p to be common in cHL. Even considering that most HRS cells are triploid or tetraploid1-4,9 more than half (55%) of the cHLs seem to have gains including amplifications in 2p13, which is in line with the CGH analyses detecting 2p gains in 6 of 12 HLs.10
To determine the chromosome 2 copy number of the tumor cells and to relate the number of REL to the number of centromere 2 copies, FICTION was performed combining D2Z1 with theREL-BAC pool probe, which produces more distinct signals than the REL-YAC pool probe allowing a more accurate signal counting. In 27 of the 31 cases (87%) that could be analyzed with the CD30/D2Z1/REL-BAC assay, the median centromere 2 copy number exceeded the diploid range (Figure1A,B). To adjust the REL locus for the centromere 2 copy number, 2 different scores,REL-D2Z1 andREL/D2Z1, were calculated and for each case median and range of both scores were determined. TheREL-D2Z1 score gives the absolute number of additional signals for the REL locus, theREL/D2Z1 score indicates the relative amplification of REL compared to the number of copies of the chromosome 2 as estimator of ploidy. Eleven of the 31 cHL cases (35%) displayed a REL/D2Z1 score of more than 1.5 indicating gains of 2p, with 8 of them (26%) having aREL/D2Z1 score of at least 3 suggesting high-level amplifications (Table 1). Thus, the high-level amplifications were detected more frequently in the present FICTION study (8 of 31) than in the published CGH series (0 of 12), which could indicate that the genomic size of some amplicons might be too small for detection by CGH.10
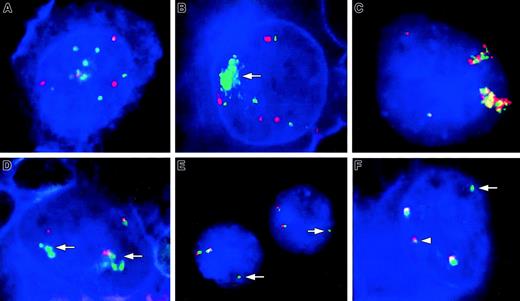
Involvement of theREL and BCL11A loci in cHL detected by FISH and FICTION.
(A,B) FICTION analyses using probes for REL (green signals) and CEP2 (red signals) showing multiple copies of REL in cHL11 (A) and a high-level amplification of REL (arrow) in cHL1 (B). The tumor cells of both cases contain 4 copies of CEP2. (C-F) FISH (C,E) and FICTION (D,F) analyses with probes for REL(green signals) and BCL11A (red signals) loci. (C) FISH in case cHL45 with del(2)(p14p23) and add(2)(p?)x2 identified a massive copy gain of both REL and BCL11A loci in HRS nuclei most likely due to 2 chromosomes containing a 2p amplicon. Two colocalized signals for each probe indicate 2 additional intactREL/BCL11A loci. (D) Amplification of the RELlocus (arrows) without coamplification of BCL11A locus in cHL8. (E,F) Signal patterns indicating a translocation breakpoint within the region spanned by the REL probe or, less likely, between the REL and BCL11A loci. (E) FISH in case cHL46 with t(2;22)(p16;q12) and der(9)t(2;9)(p16;p22) revealed 2 colocalized signals for the REL and BCL11A loci and an additional isolated signal for the REL locus (arrow). (F) FICTION in cHL17 showing 2 intact BCL11A/REL fusions and one split of the REL probe, displayed as one isolated green (arrow) and one red/tiny green fusion signal (arrowhead).
Involvement of theREL and BCL11A loci in cHL detected by FISH and FICTION.
(A,B) FICTION analyses using probes for REL (green signals) and CEP2 (red signals) showing multiple copies of REL in cHL11 (A) and a high-level amplification of REL (arrow) in cHL1 (B). The tumor cells of both cases contain 4 copies of CEP2. (C-F) FISH (C,E) and FICTION (D,F) analyses with probes for REL(green signals) and BCL11A (red signals) loci. (C) FISH in case cHL45 with del(2)(p14p23) and add(2)(p?)x2 identified a massive copy gain of both REL and BCL11A loci in HRS nuclei most likely due to 2 chromosomes containing a 2p amplicon. Two colocalized signals for each probe indicate 2 additional intactREL/BCL11A loci. (D) Amplification of the RELlocus (arrows) without coamplification of BCL11A locus in cHL8. (E,F) Signal patterns indicating a translocation breakpoint within the region spanned by the REL probe or, less likely, between the REL and BCL11A loci. (E) FISH in case cHL46 with t(2;22)(p16;q12) and der(9)t(2;9)(p16;p22) revealed 2 colocalized signals for the REL and BCL11A loci and an additional isolated signal for the REL locus (arrow). (F) FICTION in cHL17 showing 2 intact BCL11A/REL fusions and one split of the REL probe, displayed as one isolated green (arrow) and one red/tiny green fusion signal (arrowhead).
Compared to CGH, the resolution of FISH and FICTION is significantly higher and allows a comparative analysis of both candidate oncogenes REL and BCL11A. In 42 of the 44 cHL cases studied by FICTION, the copy numbers of BCL11Aand REL were in complete agreement suggesting that most structural changes in 2p cHL led to gains of both loci. This holds also true for cHL45 with a hyperploid karyotype containing complex changes including 3 aberrant chromosomes, 2 described as del(2)(p14p23) and add(2)(p?)x2, in which FISH analyses revealed one (3 of 20) or 2 (17 of 20) high-level amplifications of both BCL11A andREL in the large HRS nuclei (Figure 1C).
In contrast, FICTION in cHL8 displayed an amplification of theREL but not of the BCL11A locus (Figure 1D). Moreover, cHL17 presented several colocalizations of the RELand BCL11A probes and one additional signal of theREL probe with one of the REL signals colocalizing with a BCL11A signal showing significantly decreased intensity. Similarly, FISH in cHL46 described recently to contain a t(2;22)(p16;q12) and a der(9)t(2;9)(p16;p22) by cytogenetic analysis4 revealed 34 of 100 nuclei with one isolated REL signal in addition to 2 fusions of oneBCL11A and REL signal each. The signal patterns in the latter 2 cases suggest a breakpoint within the region covered by the REL probe (Figure 1E,F). Thus, in a total of 3 cHLs investigated herein signal patterns indicating selective alteration of the REL locus without obvious affection of BCL11Awere observed. This suggests that REL or a closely linked gene other than BCL11A might be a target of the 2p changes in cHL.
REL plays essential roles in regulation of proliferation, maturation, differentiation, and apoptosis of B-lymphocytes exerting many of its functions via interaction with other members of the NF-κB family.19-21 Considering that constitutive NF-κB activation represents a common feature of HRS cells, it is tempting to speculate that genomic alterations of the REL gene, like mutations of the NFKBIA and NKFBIE genes, represent a means of NF-κB activation in cHL.22-25
The authors thank Claudia Becher, Reina Zühlke-Jenisch, and Dorit Schuster for their excellent technical assistance and the pathologists of the German Lymphoma Consultation Centers for providing histopathologic diagnoses.
Supported by the Interdisziplinäres Zentrum für Klinische Krebsforschung (IZKF) Kiel. J.I.M.-S. is a scholar of the Gobierno de Navarra.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Reiner Siebert, Institute of Human Genetics, University Hospital Kiel, Schwanenweg 24, 24105 Kiel, Germany; e-mail:rsiebert@medgen.uni-kiel.de.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal