Infection with Epstein-Barr virus (EBV) exerts substantially immunomodulating activities in vitro and in vivo. In this context, EBV-induced chemokine production and the influence of EBV on this highly redundant system of inflammatory proteins have hardly been investigated. This study analyzed the production of interleukin-8, RANTES, monocyte chemotactic protein–1, and macrophage inflammatory protein–1α (MIP-1α) on EBV infection of peripheral blood mononuclear cells from immune EBV-seropositive (EBV+) and noninfected EBV-seronegative (EBV−) individuals. EBV failed to induce the production of MIP-1α in EBV+ as well as EBV− individuals, whereas the other chemokines studied were readily expressed. Moreover, EBV completely down-regulated lipopolysaccharide (LPS)– and phytohemagglutinin–induced MIP-1α production up to 4 hours after induction. Reverse transcription–polymerase chain reaction (RT-PCR) analysis of EBV- and LPS-stimulated cultures revealed that EBV inhibited MIP-1α production on the transcriptional level. This effect was abolished by addition of antiglycoprotein (gp)350/220, a monoclonal antibody against EBV's major envelope glycoprotein, which mediates binding of the virus to the EBV receptor, CD21. However, recombinant gp350/220 protein alone did not inhibit the LPS-induced MIP-1α production, indicating that infection of the target cell is indispensable for this effect. In summary, we demonstrate a new immunomodulating activity of EBV on the chemokine system that probably helps the virus to evade the host's immune system favoring lifelong infection.
Introduction
Chemokines are important mediators of the immune system with primarily chemotactic properties. By recruiting phagocytes as well as lymphocytes to extranodal sites of inflammation, chemokines connect unspecific with specific compartments of the immune system and therefore play a pivotal role in developing a rapid, focused, and effective immune response (for a review, see Rollins1).
Chemokines are classified as C, CC, CXC, or CX3C chemokines on the basis of the arrangement of 1 or 2 N-terminal cysteine residues.2 CXC and CC chemokines consist of multiple 8- to 10-kd proteins each with overlapping, but to some extent, opposite actions.3 CXC chemokines, such as interleukin-8 (IL-8), almost exclusively act on neutrophils, whereas chemokines of the CC group, such as RANTES (regulated on activation normal T cell expressed and secreted), monocyte chemotactic protein–1 (MCP-1), or macrophage inflammatory protein–1α (MIP-1α) have a broader spectrum of action predominantly attracting monocytes, macrophages, lymphocytes, and natural killer (NK) cells.3CC chemokines, especially MIP-1α, proved to be key elements in the development of antiviral immunity.4 Besides promotion of coxsackievirus B3-induced myocarditis, MIP-1α is indispensable for viral clearance in influenza virus–induced pneumonitis in vivo.5 Moreover, MIP-1α is critical for protection against murine cytomegalovirus infection in vivo through recruitment of NK cells and accompanied interferon gamma (IFN-γ) production.6 Together with MIP-1β and RANTES, MIP-1α was identified as one of the major human immunodeficiency virus (HIV)–suppressive factors secreted by CD8+ T cells.7 Further actions of MIP-1α comprise the modulation of macrophage functions, such as stimulation of tumor necrosis factor α (TNF-α) production,8 the preferential chemotaxis of CD8+ T cells4 as well as the inhibition of hematopoietic stem cell proliferation.1 Another indication of its important role in antiviral defense derives from Kaposi sarcoma–associated herpesvirus (KSHV). KSHV integrated into its genome homologues of human MIP-1α that are assumed to counteract some functions of the human homologue.9,10 An MIP-1α homologue encoded by other human herpesviruses, including Epstein-Barr virus (EBV), has not been described yet. However, the rhesus macaque rhadinovirus, which seems to be the homologue to KSHV, as well as human molluscum contagiosum poxvirus also encode for viral CC chemokines, the latter one acting as an antagonist on human chemokine receptor CCR8.11 12
Epstein-Barr virus is a human herpesvirus with a selective tropism for B lymphocytes and is associated with certain human malignancies.13 Lifelong EBV infection is established in the long-living compartment of resting memory B cells.14Reactivation is presumably achieved by infected B cells, which intermittently recirculate to secondary lymphoid tissues.14 The cellular and subcellular events of EBV reactivation are only poorly understood. However, suppression of local immunity by immunomodulating strategies, such as expression of viral IL-10,15 down-regulation of TNF-α production,16 or inhibition of IFN-α secretion through open reading frame BARF1 product,17 are helpful bystander mechanisms of the virus to evade the host's cellular immunity.
Little is known about the influence of EBV on the chemokine system. Multiple chemokines are likely to be involved in the course of infectious mononucleosis (IM) causing the typical polymorphous cellular infiltrate.13 18 In this study, we define the chemokine production of noninfected EBV-seronegative (EBV−) and immune EBV-seropositive (EBV+) individuals in response to in vitro EBV infection. T cells were removed in certain experiments to determine the influence of memory T cells on selective chemokine production.
Materials and methods
Preparation of infectious B95-8 virus
Infectious EB virions were obtained as described elsewhere in detail.19 In brief, the marmoset cell line B95-8 was cultivated for 10 days in a humidified atmosphere containing 5% CO2 without changing the culture medium. Cells and medium were then frozen at −20°C for 2 hours to release intracellular virus particles. Afterward, cell suspension was filtrated twice through a sterile 0.45-μm filter, aliquoted in 1.5-mL portions and stored at −80°C. Virus preparation was repeatedly shown to be free of mycoplasma contamination. Using quantitative real-time polymerase chain reaction (PCR),20 it was determined that the preparation contained approximately 3.5 × 105 EBV genome equivalents (geq)/mL.
Cell preparation and EBV serology
Peripheral blood mononuclear cells (PBMCs) from buffy coats or from heparinized whole blood of healthy volunteers were isolated by standard density centrifugation (Ficoll Separation Solution; Biochrom, Berlin, Germany). After 2 wash steps in phosphate-buffered saline (PBS, pH 7.2) cells were resuspended in RPMI 1640 medium (Biochrom) supplemented with l-glutamine 2 mM, penicillin 100 U/mL, streptomycin 100 μg/mL (Gibco BRL, Karlsruhe, Germany), and 10% heat-inactivated fetal calf serum (Gibco BRL). T-cell depletion of PBMCs from 19 EBV+ and 11 EBV− blood donors followed exactly the same protocol as described before.21The EBV serostatus was determined using enzyme-linked immunosorbent assay (ELISA) for IgG antibodies against Epstein-Barr nuclear antigen 1 (EBNA-1) and for IgG and IgM antibodies against EBV early antigen (Biotest, Dreieich, Germany),22 as already done previously.19 21
Culture conditions
Cells were stimulated in 24-well flat bottom plates (Nunc, Roskilde, Denmark) at a concentration of 2 × 106cells/mL. Total volume was 1 mL/well. Cells were cultured in a humidified atmosphere containing 5% CO2 for 48 hours unless otherwise mentioned. The EBV preparation was used in a 1:5 dilution (ie, 200 μL for each stimulation, corresponding to a concentration of 7 × 104 geq/mL). Lipopolysaccharide (LPS) derived from Escherichia coli (Sigma, Deisenkirchen, Germany) was used at a final concentration of 10 ng/mL. Phytohemagglutinin (PHA; Sigma) was used at a concentration of 1 ng/mL. Anti-EBNA1 (EBNA.OT1x) and neutralizing anti-gp350/220 (EBV.OT6) antibodies (kindly provided by J. Middeldorp, Amsterdam, The Netherlands) were used at a concentration of 100 μg/mL.23,24 J. Middeldorp also provided truncated gp350/220 protein that lacks the transmembrane domain. It was expressed in Chinese hamster ovary cells and purified by immunoaffinity and anion-exchange chromatography as described elsewhere in detail.24 The C-terminal membrane anchor is necessary for virus budding and capsulation through the cell membrane but has no binding activity for the EBV receptor, CD21.25 The N-terminal part of the EBNA1 protein, p72 (provided by W. Hinderer, Biotest), originally served as a capture antigen for anti-EBNA1 ELISA.22 It was used as a control antigen in our experiments. Both the recombinant gp350/220 and the p72 protein were used at a concentration of 100 μg/mL.
Chemokine determination
Commercially available ELISA systems were used for chemokine quantification. ELISA performance followed exactly the manufacturer's recommendations. Test kits for MIP-1α and RANTES were purchased from R & D Systems (Minneapolis, MN), for MCP-1 from Biosource International (Camarillo, CA), and for IL-8 from Bender Medsystems (Vienna, Austria).
RNA extraction and complementary DNA synthesis
Total RNA was extracted from 2 × 106PBMCs using the RNeasy Mini Kit (Qiagen, Hilden, Germany). RNA was eluted in 40 μL distilled RNAse free water. For reverse transcription (RT), 18 μL total RNA was incubated with 2 μL random hexamers (Gibco; 50 ng/mL) for 10 minutes at 70°C. Afterward, complementary DNA (cDNA) synthesis was performed in a final volume of 40 μL for 1 hour at 37°C using Superscript II RT (Gibco).
Real-time PCR assay
A quantitative real-time RT-PCR assay was established for MIP-1α messenger RNA (mRNA) detection using the TaqMan technology. Primer and probe for MIP-1α were designed by means of Primer Express software (Applied Biosystems, Weiterstadt, Germany). The underlying sequence for MIP-1α cDNA was obtained from GenBank database, accession number AF043339. Sequences read as follows (5′ 3′): forward primer CAT CAC TTg CTg CTg ACA Cg, reverse primer TgT ggA ATC TgC Cgg gAg, probe VIC-CgA CCg CCT gCT gCT TCA gCT ACA-TAMRA with VIC and TAMRA representing the reporter and quencher dye, respectively. The β-actin gene coamplification was used for relative quantification using a primer/probe set purchased from PE Applied Biosystems (TaqMan β-actin Control Reagents). Master mix for MIP-1α and β-actin determination comprised 25 μL TaqMan Universal Mastermix (Applied Biosystems), 1.5 μL of each forward and reverse primer (10 μM) for MIP-1α or 1.0 μL (3 μM) for β-actin, 1.0 μL of probe (10 μM) for MIP-1α or 1.0 μL (2 μM) for β-actin, and 2 μL cDNA. Distilled water was added for a final volume of 50 μL. RNA without RT was run in parallel with each sample. Cycler conditions were 10 minutes at 95°C followed by 40 cycles consisting of 15 seconds at 95°C and 1 minute at 60°C. Threshold values were calculated as the upper 10-fold SD of the background fluorescence signal measured over all cycles defined by the baseline. The baseline was set manually from cycle 3 to the cycle before the exponential increase of the first PCR kinetics was to be observed. The threshold cycle (Ct), which is inversely proportional to the starting copy numbers26 and is defined as the PCR cycle at which the reporter fluorescence signal exceeds the threshold value of the respective analysis, was used for quantification. MIP-1α expression levels were calculated in relation to β-actin expression and calibrated against values derived from respective unstimulated controls. According to the ABI Prism User Bulletin no. 2, the following term was used to express the relative amounts of mRNA (ΔΔCt calculation):where Ct MIPx as well as CtActinx refer to the Ct values for MIP-1α and β-actin of the respective samples and CtMIPcalibr as well as Ct Actincalibrrefer to the Ct values of the calibrator, that is, the unstimulated control.
Statistical analysis
Values in Table 1 represent individual differences between the respective stimulated and unstimulated samples. Differences are given as mean ± SD. Graphs always show mean ± SD of different numbers of experiments, as indicated in the figure legends. For comparison between the individual groups, the Mann Whitney Utest and, where applicable, the Wilcoxon test for related samples were used. A P < .05 was considered to be significant.
Chemokine production in response to EBV stimulation of PBMCs before and after removal of T cells from EBV− and EBV+ healthy adults
| . | PBMCs . | Without T cells . | ||
|---|---|---|---|---|
| EBV− (n = 11) . | EBV+ (n = 19) . | EBV− (n = 11) . | EBV+ (n = 19) . | |
| IL-8 | 294 ± 297* | 1381 ± 900*† | 316 ± 302‡ | 732 ± 534†‡ |
| RANTES | 423 ± 384* | 461 ± 363† | 165 ± 263* | 210 ± 240† |
| MCP-1 | 339 ± 739* | 781 ± 1026† | 1017 ± 1307* | 1783 ± 1237† |
| MIP-1α | 0.5 ± 1.7 | 0.9 ± 2.7 | 2.8 ± 9.3 | 3.3 ± 5.4 |
| . | PBMCs . | Without T cells . | ||
|---|---|---|---|---|
| EBV− (n = 11) . | EBV+ (n = 19) . | EBV− (n = 11) . | EBV+ (n = 19) . | |
| IL-8 | 294 ± 297* | 1381 ± 900*† | 316 ± 302‡ | 732 ± 534†‡ |
| RANTES | 423 ± 384* | 461 ± 363† | 165 ± 263* | 210 ± 240† |
| MCP-1 | 339 ± 739* | 781 ± 1026† | 1017 ± 1307* | 1783 ± 1237† |
| MIP-1α | 0.5 ± 1.7 | 0.9 ± 2.7 | 2.8 ± 9.3 | 3.3 ± 5.4 |
Values represent mean values ± SD of differences between the respective stimulated and unstimulated cultures and are given in pg/mL.
The different symbols per row (*, †, ‡) identify for each chemokine respective groups for which P < .05.
Results
Chemokine induction by EBV
The PBMC cultures of 11 EBV− as well as of 19 EBV+ healthy adults were investigated for EBV-induced chemokine secretion following a 48-hour infection. Synthesis of IL-8 as a representative CXC chemokine was examined as well as of RANTES, MCP-1, and MIP-1α as main CC chemokines involved in early inflammation. To eliminate the probable influence of memory T cells on selective chemokine production, cultures were analyzed before and after T-cell depletion. Table 1 shows induced production of IL-8 after stimulation with EBV, which was significantly higher in EBV+ than in EBV− individuals. This difference between EBV+ and EBV− donors was also seen after T-cell depletion. Because removal of T cells significantly decreased IL-8 secretion in EBV+adults, it is likely that memory T cells were the main source of IL-8 in this setting. Production of RANTES before and after T-cell depletion exhibited no significant differences between EBV+and EBV− donors. The significantly lower production of RANTES after T-cell depletion indicates that T lymphocytes were the main producers of RANTES on EBV induction. MCP-1 production showed an inverse secretion pattern seen for RANTES with the highest amounts of MCP-1 found after the removal of T cells. This is in accordance with monocytes being a major source of MCP-1 in the peripheral blood. In contrast to these chemokines, MIP-1α secretion of EBV+ and EBV− individuals remained undetectable before as well as after T-cell depletion (Table1).
Inhibition of MIP-1α expression
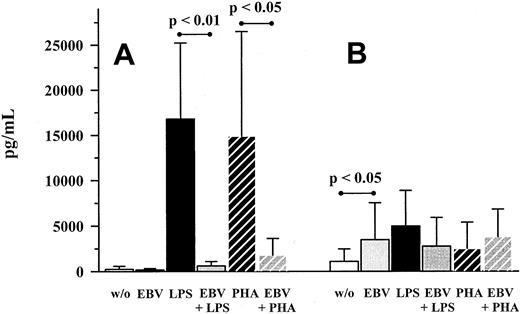
The lack of MIP-1α production became more apparent in relation to LPS- and PHA-induction as depicted in Figure1. LPS and PHA induced MIP-1α and MCP-1 production after 48 hours of stimulation. On the other hand, EBV only induced MCP-1 production at a level comparable to LPS- or PHA-stimulated amounts, whereas MIP-1α levels remained unchanged. When EBV-stimulated cultures were coincubated with LPS or PHA, production of MIP-1α was almost completely down-regulated (Figure 1).
Production of MIP-1α and MCP-1 on EBV infection and LPS or PHA costimulation.
PBMC cultures were incubated with EBV (strain B95-8, diluted 1:5) for 48 hours. LPS (10 ng/mL) or PHA (1 μg/mL), respectively, were also used as stimulants, either alone or in combination with EBV. MIP-1α (A) and MCP-1 (B) were quantified in culture supernatants by means of ELISA. Shown are mean ± SD of 8 individuals. P values refer to the figure bars combined by the dotted ends of the given lines.
Production of MIP-1α and MCP-1 on EBV infection and LPS or PHA costimulation.
PBMC cultures were incubated with EBV (strain B95-8, diluted 1:5) for 48 hours. LPS (10 ng/mL) or PHA (1 μg/mL), respectively, were also used as stimulants, either alone or in combination with EBV. MIP-1α (A) and MCP-1 (B) were quantified in culture supernatants by means of ELISA. Shown are mean ± SD of 8 individuals. P values refer to the figure bars combined by the dotted ends of the given lines.
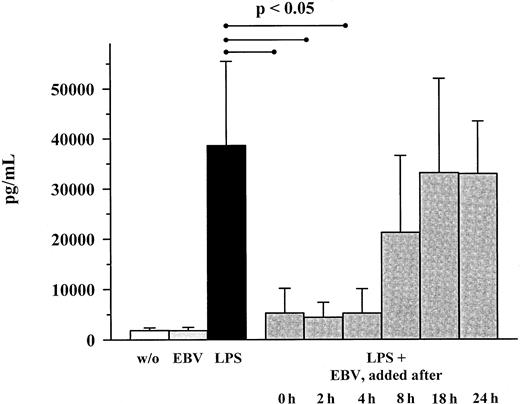
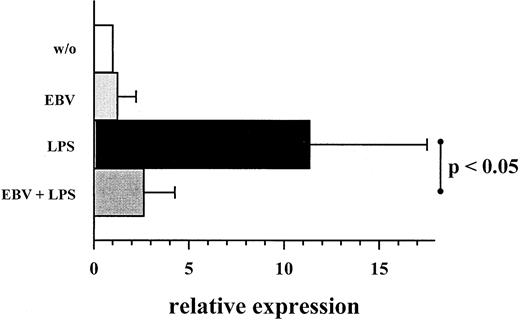
Addition of EBV up to 4 hours after the onset of LPS stimulation still inhibited MIP-1α secretion (Figure 2). However, addition of EBV at a later time point had only a marginal effect on MIP-1α secretion. Transcriptional analysis by real-time quantitative TaqMan PCR revealed an endogenous expression of MIP-1α mRNA in unstimulated PBMC controls, which was significantly increased by LPS stimulation. Incubation with EBV reduced the amount of LPS-induced MIP-1α mRNA approximately to levels observed for unstimulated cells (Figure 3).
Kinetics of MIP-α production in LPS- and EBV-costimulated PBMC cultures.
Cultures were stimulated with LPS (10 ng/mL); EBV (B95-8; 1:5 dilution) was added at the indicated time points. Shown are mean ± SD of 3 individuals. P values refer to the figure bars combined by the dotted ends of the given lines.
Kinetics of MIP-α production in LPS- and EBV-costimulated PBMC cultures.
Cultures were stimulated with LPS (10 ng/mL); EBV (B95-8; 1:5 dilution) was added at the indicated time points. Shown are mean ± SD of 3 individuals. P values refer to the figure bars combined by the dotted ends of the given lines.
Expression of MIP-1α mRNA on EBV infection and LPS costimulation.
Same experiment settings were used as described in Figure 1. RNA detection and quantification was performed using RT and real-time TaqMan PCR technology. Quantification of MIP-1α mRNA was based on expression relative to the coamplified amount of β-actin mRNA. Values are expressed in arbitrary units and represent multiplicities of the respective unstimulated controls. Shown are mean ± SD of 7 individuals. P values refer to the figure bars combined by the dotted ends of the given line.
Expression of MIP-1α mRNA on EBV infection and LPS costimulation.
Same experiment settings were used as described in Figure 1. RNA detection and quantification was performed using RT and real-time TaqMan PCR technology. Quantification of MIP-1α mRNA was based on expression relative to the coamplified amount of β-actin mRNA. Values are expressed in arbitrary units and represent multiplicities of the respective unstimulated controls. Shown are mean ± SD of 7 individuals. P values refer to the figure bars combined by the dotted ends of the given line.
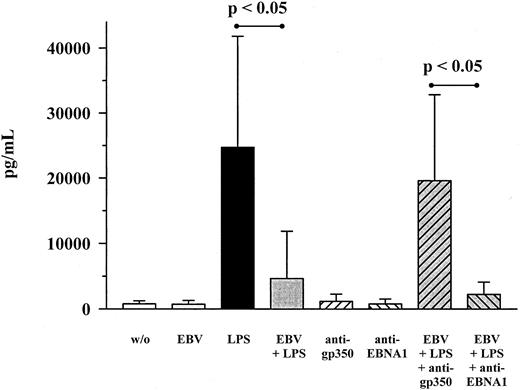
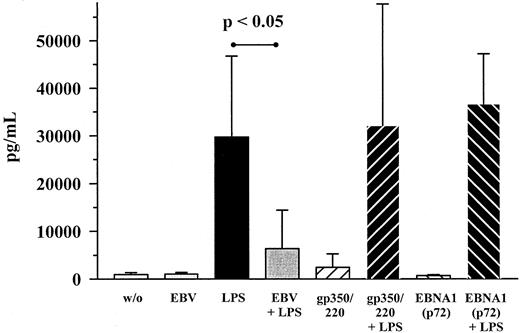
To further investigate the mechanism of MIP-1α inhibition by EBV, monoclonal antibodies against EBV's major envelope glycoprotein gp350/220 as well as against the EBNA1 antigen were used. With respect to anti-gp350/220 antibodies, cocultivation together with LPS- and EBV-treated cultures led to an almost complete reversal of EBV's inhibiting effect on MIP-1α production (Figure4). Anti-EBNA1 antibodies, on the other hand, had no effect. When the truncated form of recombinant gp350/220 protein was used instead of whole EB virions, LPS-induced MIP-1α expression was not inhibited (Figure 5). This also held true for the N-terminal part of the EBNA1 protein, p72.
Influence of gp350/220 antibody on the production of MIP-1α on EBV infection and LPS costimulation.
PBMC cultures were costimulated with EBV (diluted 1:5) and LPS (10 ng/mL) for 48 hours, either in the presence of neutralizing anti-gp350/220 antibodies (100 μg/mL) or not. As a control, anti-EBNA1 antibodies (100 μg/mL) were used in the same way. Shown are mean ± SD of 7 individuals. P values refer to the figure bars combined by the dotted ends of the given lines.
Influence of gp350/220 antibody on the production of MIP-1α on EBV infection and LPS costimulation.
PBMC cultures were costimulated with EBV (diluted 1:5) and LPS (10 ng/mL) for 48 hours, either in the presence of neutralizing anti-gp350/220 antibodies (100 μg/mL) or not. As a control, anti-EBNA1 antibodies (100 μg/mL) were used in the same way. Shown are mean ± SD of 7 individuals. P values refer to the figure bars combined by the dotted ends of the given lines.
Recombinant gp350/220 protein does not inhibit LPS-induced MIP-1α production.
Costimulation of PBMC cultures with EBV (1:5 dilution) and LPS (10 ng/mL) was compared to cultures stimulated with LPS and a recombinant gp350/220 protein lacking the C-terminal membrane anchor (100 μg/mL). Stimulation with the N-terminal EBNA1 protein, p72 (100 μg/mL), was performed as a control experiment. MIP-1α concentrations were again determined after 48 hours of stimulation. Shown are mean ± SD of 5 individuals. P values refer to the figure bars combined by the dotted ends of the given line.
Recombinant gp350/220 protein does not inhibit LPS-induced MIP-1α production.
Costimulation of PBMC cultures with EBV (1:5 dilution) and LPS (10 ng/mL) was compared to cultures stimulated with LPS and a recombinant gp350/220 protein lacking the C-terminal membrane anchor (100 μg/mL). Stimulation with the N-terminal EBNA1 protein, p72 (100 μg/mL), was performed as a control experiment. MIP-1α concentrations were again determined after 48 hours of stimulation. Shown are mean ± SD of 5 individuals. P values refer to the figure bars combined by the dotted ends of the given line.
Discussion
Primary infection with EBV is tightly controlled in the immunocompetent host mainly due to a large expanding compartment of EBV-specific CD8+ cytotoxic T lymphocytes.27Tonsils affected by IM are, in contrast, characterized by a more polymorphous cellular infiltrate indicating the efforts of the immune system to get the infection under control.13,18 Chemokines are likely to play a key role in initiating and maintaining this mixed cellularity consisting of granulocytes, lymphocytes, macrophages, and NK cells. Indeed, mRNA transcripts for MIP-1α, RANTES, IFN-γ–inducible protein-10, and monokine induced by IFN-γ were detected in EBV-infected tonsils of patients with IM.28Furthermore, tissue specimens of patients with morbus Hodgkin,29,30 posttransplantation lymphoproliferative disorders (PTLDs),28,31 nasopharynx carcinoma (NPC),32 or EBV-induced hemophagocytic syndrome (HPS)33 showed expression of different CC and CXC chemokines partially including IL-8, MCP-1, MIP-1α, and RANTES. However, only for NPC and HPS was the production of protein described. Interestingly, a selective expression of CC chemokines has been observed for influenza virus infection in vitro, probably explaining the mononuclear infiltrate in influenza virus–induced airway inflammation.34
Apart from these studies, the chemokine system to date has hardly been investigated in the context of EBV infection. Interestingly, most data on chemokine expression and EBV infection derive from in vivo studies, whereas in vitro studies are pending. This is the first study that systematically addressed the question of chemokine induction by EBV. We show significant secretion of IL-8, as well as of RANTES and MCP-1, on EBV challenge. Differences between EBV+ and EBV− individuals were likely due to the presence of primed T cells, because removal of T cells abrogated any differences between those 2 populations. As expected, IL-8 and RANTES were predominantly of T-cell origin, whereas MCP-1 was primarily derived from monocytes.1
Because the production of CC as well as of CXC chemokines was readily observed after stimulation with EBV, it came to us as a surprise that MIP-1α secretion was not found. This situation is comparable to the cytokine system, in which EBV is able to induce several proinflammatory cytokines, apart from TNF-α.35Similar to the down-regulation of TNF-α production by EBV,16 we showed in this study that EBV also inhibits LPS- and PHA-induced MIP-1α expression of human PBMCs. Because MIP-1α is a strong inductor of TNF-α in human monocytes,8 it seems possible that TNF-α inhibition by EBV is mediated through blockage of MIP-1α production. The effect of EBV on LPS-induced MIP-1α synthesis was time dependent and only effective up to 4 hours after addition of LPS. The quantitative analysis of mRNA production proved that EBV inhibits MIP-1α expression on the transcriptional level. The results we found in PBMCs are inconsistent with findings for neutrophils, which were reported to express and secrete MIP-1α on EBV stimulation.36 However, the neutrophils used in the study by McColl and colleagues36 may have been prestimulated by hypotonic lysis of erythrocytes. Like others, we were not able to confirm any MIP-1α expression in pure neutrophil preparations after challenge with Newcastle disease, Sendai, or EB viruses37 (also our unpublished observations, May 2001).
Recent studies have already supposed a direct influence of gp350/220 on cytokine production of monocytes and neutrophils.35 36Therefore, we examined the role of gp350/220 on MIP-1α production more in detail. Addition of anti-gp350/220 antibodies to PBMC cultures completely abolished the inhibiting effect of EBV on MIP-1α production, suggesting that this effect was specific for EBV. Recombinant gp350/220 protein alone was insufficient to block the LPS-induced MIP-1α expression. These results exclude a direct interaction of gp350/220 with its target cell alone being responsible for inhibition of MIP-1α expression, but emphasize that infection is necessary to achieve inhibition of MIP-1α production.
A possible reason EBV blocked MIP-1α expression is that EBV can also infect human monocytes, as was shown recently.38 In that study, infection of monocytes was permissive for replication and resulted in a significantly reduced phagocytic activity. Because monocytes are the main producer cells of MIP-1α,1inhibition of its production by EBV, therefore, might occur in monocytes. This becomes more evident when looking at B lymphocytes, the actual targets for EBV. B-cell infection and establishment of latency by EBV seems to be contradictory to the inhibition of MIP-1α production in that latently infected tumor cells from T/NK cell lymphoma and Burkitt lymphoma,31 or EBV-transformed B-cell lines39 express MIP-1α mRNA, although protein secretion has not been proven for these cell types. Nevertheless, it is tempting to speculate that for EBV a comparable situation emerged as it did for HIV-1. In fact it was shown that the HIV-1–encoded tatprotein down-regulated the expression of MIP-1α in activated T cells by overexpression of a transcription factor crucial for MIP-1α expression in monocytes, termed MIP-1α nuclear protein 1 (MNP-1).40 41 Whether changes in levels of MNP-1 or MNP-2 contribute to the effect of EBV on MIP-1α expression has to be elucidated.
In summary, we demonstrated that EBV down-regulates MIP-1α expression of human PBMCs, like EBV does for TNF-α as well.16 Our experiments defined another intriguing immunomodulatory activity of EBV, which probably is of advantage for the virus to persist in the host for life.
We are indebted to the help of Jaap Middeldorp, Amsterdam, Netherlands, who provided the monoclonal antibodies against EBNA1 and gp350/220 proteins as well as the recombinant gp350/220 protein. We would further like to thank Walter Hinderer, formerly of Biotest, Dreieich, Germany, who helped us with the recombinant p72 protein of EBNA1. We kindly appreciate the excellent laboratory performance of Annette Willemsen and Maren Behrensen as well as the helpful comments of Luis Guembes Hidalgo, Edmonton, Alberta, Canada in preparing the manuscript.
Supported in part by grant 800/B3 from the 2000 National Government Research Found of the University of Lübeck. The ABI PRISM 7700 Sequence Detection System used in this study was sponsored by “Lübeck-Hilfe für Krebskranke Kinder,” “Schleswig-Holstein Krebshilfe,” and “Possehl-Stiftung.”
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Wolfram J. Jabs, First Department of Medicine, Division of Nephrology, University of Lübeck School of Medicine, Ratzeburger Allee 160, 23538 Lübeck, Germany; e-mail:wjabs@gmx.de.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal