Cell surface antigen CD109 is a glycosylphosphatidylinositol (GPI)–linked glycoprotein of approximately 170 kd found on a subset of hematopoietic stem and progenitor cells and on activated platelets and T cells. Although it has been suggested that T-cell CD109 may play a role in antibody-inducing T-helper function and it is known that platelet CD109 carries the Gov alloantigen system, the role of CD109 in hematopoietic cells remains largely unknown. As a first step toward elucidating the function of CD109, we have isolated and characterized a human CD109 cDNA from KG1a and endothelial cells. The isolated cDNA comprises a 4335 bp open-reading frame encoding a 1445 amino acid (aa) protein of approximately 162 kd that contains a 21 aa N-terminal leader peptide, 17 potential N-linked glycosylation sites, and a C-terminal GPI anchor cleavage–addition site. We report that CD109 is a novel member of the α2 macroglobulin (α2M)/C3, C4, C5 family of thioester-containing proteins, and we demonstrate that native CD109 does indeed contain an intact thioester. Analysis of the CD109 aa sequence suggests that CD109 is likely activated by proteolytic cleavage and thereby becomes capable of thioester-mediated covalent binding to adjacent molecules or cells. In addition, the predicted chemical reactivity of the activated CD109 thioester is complementlike rather than resembling that of α2M proteins. Thus, not only is CD109 potentially capable of covalent binding to carbohydrate and protein targets, but the t½of its activated thioester is likely extremely short, indicating that CD109 action is highly restricted spatially to the site of its activation.
Introduction
To identify new surface antigens expressed by primitive hematopoietic stem and progenitor cells, we raised a series of monoclonal antibodies (mAbs) against the primitive CD34+acute myeloid leukemia cell line, KG1a.1-3 Four of these mAbs—8A3, 7D1, 8A1, and 7C5—recognized a novel glycoprotein of approximately 170 kd that was expressed in a restricted pattern in the hematopoietic compartment and by endothelial cells.4Subsequently found to be identified by a number of additional mAbs, this antigen was designated CDw109 in 19935 and CD109 in 1996.6
Antibodies to CD109 recognize monomeric polypeptides of about 170 kd and 150 kd in lysates of KG1a cells, T-cell lines, and activated T lymphoblasts, endothelial cells, and activated platelets.3,7-11 Peptide mapping and amino acid (aa) analysis indicate that the 150-kd form is likely derived proteolytically from the 170-kd form.3,10 An additional band of about 120 kd that is occasionally observed arises through calcium-dependent proteolysis of the larger forms.3,10CD109 contains several N-linked endoglycosidase H-sensitive hybrid-type glycans but no O-linked glycans.3,10 Consistent with this finding, ABH blood group antigens have recently been shown to be carried by platelet CD109.12 KG1a CD109 is susceptible to cleavage with phosphatidylinositol-specific phospholipase C (PI-PLC), indicating that CD109 is bound to the cell membrane by a GPI anchor.5,6,9-11 On some cell types, a proportion of surface CD109 is resistant to PI-PLC but is sensitive to GPI-phospholipase D, indicating that some of the CD109 GPI anchors are acylated on inositol.10,13 Although the biologic relevance of such anchor heterogeneity is unknown, it suggests that discrete populations of CD109 may have distinct lipid solubility characteristics and, therefore, may partition differentially into distinct membrane-lipid microdomains. Indeed, T-cell CD109 is considerably more soluble in nonionic detergents than are several other GPI-anchored proteins,11 and it does not appear to colocalize with membrane-associated src-family kinases after T-cell activation.11
The expression of CD109 has been studied in greatest detail in hematopoietic cells.3,14,15 As defined by mAb binding, CD109 appears initially on a subset (3%-35%) of fetal and adult CD34+ bone marrow (BM) mononuclear cells.14,15Notably, almost all myelo-erythroid and MK progenitors in fetal and adult BM are found in this CD34+CD109+ subset and not in the CD34+/CD109−fraction.14 Moreover, CD109 expression is strongest on cells expressing the highest levels of CD34, suggesting that the most primitive candidate hematopoietic stem cells (HSCs) may also be contained within the CD109+ subset. Consistent with this notion, we have shown that the CD34+CD109+fraction contains cells that are CD38lo, Thy-1 (CD90)+, and AC133+ and that are able to efflux the mitochondrial dye rhodamine 123 (Rho123), characteristics associated with the most primitive HSCs.14 Indeed, the CD34+CD109+ bone marrow fraction contains all assayable cobblestone area–forming cell activity (known to correlate with long-term in vivo repopulating ability), with the CD109+Rho123lo subset of CD34+cells containing cobblestone area–forming cells at frequencies of approximately 1 in 10 cells plated.14 Thus, it is likely that CD109 marks primitive HSCs. As hematopoietic differentiation proceeds, however, CD109 expression (as determined by mAb binding) becomes progressively restricted—initially to myeloid precursors, and then to megakaryoblastic progenitors—and finally becomes undetectable on resting, mature blood cells.
Curiously, CD109 reappears as an activation antigen on platelets and T cells.3,7-9 After activation with a variety of agonists, platelets become CD109+, expressing approximately 2000 ± 400 mAb 8A3 binding sites per cell.3 The Gov platelet alloantigens, which have been implicated in posttransfusion purpura, neonatal alloimmune thrombocytopenia, and refractoriness to platelet transfusion, have recently been shown to correspond to noncarbohydrate platelet CD109 epitopes.3,12,13 Previously undefined,10,16,17 the immunogenicity of the Gov antigens is now known to be similar to that of the HPA-5 antigens and is exceeded only by that of the HPA-1 antigens.18
CD109 is also expressed by activated T cells. After T-cell activation in vitro, CD109 becomes detectable on CD4+ and CD8+ T cells by day 1, peaks by day 6, and thereafter decreases in the absence of additional interleukin-2.3,7-9 The precise function of CD109 on hematopoietic precursors and on activated platelets and T cells is unknown. However, the observation that T-cell–dependent antibody production is abrogated in vitro by the CD109 mAb LDA1suggests that CD109 may play a role in B-cell–T-cell interactions.7
As a first step toward elucidating the function of CD109, we have isolated and characterized a human CD109 cDNA. We report that CD109 is a novel member of the α2 macroglobulin (α2M)/C3, C4, C5 family of thioester-containing proteins and demonstrate that native CD109 contains an intact thioester.
Materials and methods
Cell culture
KG1a12 and Chinese hamster ovary (CHO) cells were grown in RPMI 1640 and F12(Ham) (Life Technologies, Burlington, ON, Canada), respectively, each supplemented with 10% heat-inactivated fetal calf serum (Hyclone, Logan, UT), 2 mM L-glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin (Life Technologies). Cells were obtained from the ATCC and were maintained at 37°C in 5% CO2.
Antibodies
CD109 mAbs 8A3 and 7D1 were raised against KG1a cells as described previously,3 1B38 and LDA17 were gifts of Irwin Bernstein (Fred Hutchinson Cancer Research Center, Seattle, WA) and Nicole Suciu-Foca (Columbia University, New York, NY), respectively, and TEA 2/16 was from PharMingen (San Diego, CA). CD71 mAb D5119 (gift of Hans-Joachim Gross, University of Würzburg, Germany) was used as an immunoprecipitation control.
Immuno-affinity purification and partial amino acid sequencing of CD109
KG1a cells (1 × 1010) were washed in ice-cold phosphate-buffered saline (PBS) (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4), pH 7.4, supplemented with 0.2 mM EDTA, were resuspended in 300 mL ice-cold lysis buffer (0.01 M Tris-HCl, pH 8.1, 0.15 M NaCl, and 0.5% Nonidet P-40 [NP40]) supplemented with 2 mM phenylmethylsulfonyl fluoride, 1 mg/mL bovine serum albumin, and 2 mM EDTA, and were lysed on ice for 20 minutes with stirring.20,21 After centrifugation at 100 000g for 30 minutes, the lysate was brought to 0.5 M NaCl, and CD109 antigen was purified by cross-linked immuno-affinity chromatography. Briefly, 10 mg of CD109 mAb 8A3 was coupled to 1 mL Protein A–Sepharose beads (Amersham Pharmacia Biotech, Baie d'Urfe, QC, Canada) using dimethyl pimelimidate (Pierce, Rockford, IL). After loading the lysate, the column was washed sequentially with lysis buffer containing 0.5 M NaCl and 0.1% sodium dodecyl sulfate (SDS), respectively. Bound CD109 was eluted with 0.05 M diethylamine (pH 11.5) containing 0.5% deoxycholate (DOC) adjusted to pH 8.1 with 0.1 N HCl,21,22 and its purity was assessed by SDS–polyacrylamide gel electrophoresis (PAGE) and silver staining.23
Purified CD109 was fractionated further by preparative 7.5% SDS-PAGE, and the 170-kd Coomassie-stained band was excised and digested overnight with endoproteinase Lys-C or Asp-N (Roche, Laval, QC, Canada). Resultant peptides were extracted and separated by tandem ion exchange and reverse-phase chromatography and were sequenced with an Applied Biosystems Procise sequencer.24
Labeling of cells and immunoprecipitation of CD109
KG1a cells were labeled with 125I-sodium iodide20 or were biotinylated with Sulfo-NHS-LC-LC-Biotin (Pierce). Lysates of labeled cells were then clarified by centrifugation at 14 000g for 5 minutes, brought to 0.5 M NaCl, and used for immunoprecipitation.20 21 Immune complexes were collected on Protein A–Sepharose beads or on rabbit anti–mouse immunoglobulin-coated Protein A–Sepharose beads and were washed sequentially with lysis buffer containing 0.5 M NaCl and 0.5% DOC–0.1% SDS, respectively.
Methylamine treatment of immunoprecipitates
Immunoprecipitates were washed further with 0.2 M HEPES, pH 8, 0.1% NP40, were resuspended in 1 mL fresh 0.4 mM methylamine (Sigma, Oakville, ON, Canada) in HEPES–NP40 for 30 minutes at 37°C, and were washed once with HEPES–NP40 and once in lysis buffer containing 0.1% SDS–0.5% DOC. Immunoprecipitates were then dissociated in SDS–gel sample buffer and were analyzed by SDS-PAGE.
Analysis of immune complexes and Western blotting
Radiolabeled immune complexes were analyzed by SDS-PAGE–autoradiography, and biotinylated complexes were analyzed by SDS-PAGE–Western blotting, using streptavidin-conjugated horseradish peroxidase–coupled chemiluminescence (Pierce). In some experiments, CD109 was detected with mAb 1B3, which detects a denaturation-resistant epitope of CD109 (D.R.S., A.C.S., unpublished observation, January 2000), followed by either iodinated Protein A22 or horseradish peroxidase–conjugated goat anti–mouse immunoglobulin-coupled chemiluminescence (Pierce).
cDNA library screening
A 4 kb EcoRI–XhoI fragment of rat ESTR4712325 was radiolabeled with α-32P–dCTP using the random primer synthesis method26 and was used to screen a λ phage Uni-ZAP human umbilical vein endothelial cell (HUVEC) cDNA library (Stratagene, La Jolla, CA).27Plaque-purified clones were rescued into pBS SK+ by in vivo excision and were amplified, and inserts were evaluated by restriction endonuclease digestion and DNA sequence analysis. A KG1a cDNA library28 constructed in λ ZAP Express (gift of Robert Hawley) was screened as above using a clone H6 (Figure1A) probe. Positive plaques were purified, rescued into pBK-CMV by in vivo excision, and analyzed as above.
HUVEC and KG1a cDNA library-derived CD109 cDNAs and predicted protein.
(A) Aligned restriction maps of the largest HUVEC (H6, H7)– and KG1a (K1)–derived CD109-specific cDNA fragments and the positions of the corresponding coding and untranslated regions are shown. K1 and deduced K1-H7 cDNAs contain identical ORFs but divergent 3′UTRs. White box, UTR; gray box, coding sequence; An, poly(A) tail; Sm,Sma I; E, EcoRI; Sa, SacI; B, BamHI; H,HindIII; Xb, XbaI; P, PstI. (B) CD109 is a novel GPI-linked member of the α2M family of thioester-containing proteins. The translated K1 sequence (Figure 1) predicts a 1445-aa protein of about 162 kd bearing a cleavable 21-aa N-terminal leader peptide and a C-terminal consensus GPI anchor cleavage–addition signal, with cleavage predicted to occur after aa 1420. CD109 shares the overall domain structure of the α2M family, containing a thioester signature sequence (aa 918-924) approximately two thirds of the way along the molecule, a thioester reactivity-defining hexapeptide (aa 1030-1035) ending in VIH about 100 aa further downstream and a putative bait region (approximately aa 651-683) lying roughly in the middle of the protein.
HUVEC and KG1a cDNA library-derived CD109 cDNAs and predicted protein.
(A) Aligned restriction maps of the largest HUVEC (H6, H7)– and KG1a (K1)–derived CD109-specific cDNA fragments and the positions of the corresponding coding and untranslated regions are shown. K1 and deduced K1-H7 cDNAs contain identical ORFs but divergent 3′UTRs. White box, UTR; gray box, coding sequence; An, poly(A) tail; Sm,Sma I; E, EcoRI; Sa, SacI; B, BamHI; H,HindIII; Xb, XbaI; P, PstI. (B) CD109 is a novel GPI-linked member of the α2M family of thioester-containing proteins. The translated K1 sequence (Figure 1) predicts a 1445-aa protein of about 162 kd bearing a cleavable 21-aa N-terminal leader peptide and a C-terminal consensus GPI anchor cleavage–addition signal, with cleavage predicted to occur after aa 1420. CD109 shares the overall domain structure of the α2M family, containing a thioester signature sequence (aa 918-924) approximately two thirds of the way along the molecule, a thioester reactivity-defining hexapeptide (aa 1030-1035) ending in VIH about 100 aa further downstream and a putative bait region (approximately aa 651-683) lying roughly in the middle of the protein.
Northern blot hybridization analysis
KG1a cell total RNA was extracted using TRIzol (Life Technologies).29 Twenty micrograms total RNA was then analyzed by Northern blot analysis30 using a 635-bp K1/K1-H7 common probe generated by polymerase chain reaction (PCR) using the oligonucleotide primers K1(3225) 5′-3225GGAGTCTGAATTCAGTAGAGG3245-3′ and K1(5n) 5′-3859CAGCAACATCTAAATCAAAGGC3838-3′ with the K1 cDNA as template and a 732-bp K1-H7 specific probe generated by PCR using the primers K7(4670) 5′-4670CCTAGATTCTTAAGCATTATTAC4692-3′ and K7(U1n) 5′- 5401CAGCAAGCATCAGATGTC5384-3′ with the H7 cDNA fragment (Figure 1A) as template (numbering as in Figure2). RNA loading and integrity were assessed using a 736-bp human hypoxanthine phosphoribosyltransferase probe generated by PCR using the primers F98 5′-GCCCTGGCGTCGTGATTAGTGATG-3′ and R384 5′-AAGCAGATAGCCACAGAACTAGAAC-3′. To determine the tissue expression of CD109, a human multiple-tissue RNA membrane (Human RNA Master Blot; Clontech, Palo Alto, CA) containing poly(A) RNAs from 50 tissues was probed with a radiolabeled 725 bp (nt 3085-3809)BamHI–XbaI K1 cDNA fragment (Figure 1A; numbering as in Figure 2).
CD109 cDNA and predicted protein sequences.
The K1 and K1-H7 CD109 cDNAs and the corresponding 1445 aa protein sequence are shown (GenBank accession number AF410459). Nucleotides are numbered relative to the translation initiation codon, with the corresponding aa numbering shown in parentheses. K1 and K1-H7 3′UTRs, and the positions of the corresponding poly(A) tails [(a)n)] are shown. Peptides identified by immuno-affinity purification–microsequencing of CD109 are singly underlined. Potential sites of N-linked glycosylation, the thioester signature sequence (aa residues 918-924), and the corresponding downstream thioester reactivity-defining hexapeptide motif (aa residues 1030-1035) are marked by open boxes. Additionally, the amino-terminal leader peptide (double underline), the bait region (dotted underline), the translation stop (*), and the GPI anchor cleavage–addition site (open triangle) are shown.
CD109 cDNA and predicted protein sequences.
The K1 and K1-H7 CD109 cDNAs and the corresponding 1445 aa protein sequence are shown (GenBank accession number AF410459). Nucleotides are numbered relative to the translation initiation codon, with the corresponding aa numbering shown in parentheses. K1 and K1-H7 3′UTRs, and the positions of the corresponding poly(A) tails [(a)n)] are shown. Peptides identified by immuno-affinity purification–microsequencing of CD109 are singly underlined. Potential sites of N-linked glycosylation, the thioester signature sequence (aa residues 918-924), and the corresponding downstream thioester reactivity-defining hexapeptide motif (aa residues 1030-1035) are marked by open boxes. Additionally, the amino-terminal leader peptide (double underline), the bait region (dotted underline), the translation stop (*), and the GPI anchor cleavage–addition site (open triangle) are shown.
Reverse transcription–polymerase chain reaction
RNA isolated as above was treated with RQ1 RNAse-free DNAse (Promega, Madison, WI), sequential phenol-chloroform and chloroform extraction, and ethanol precipitation, and cDNA was prepared with SuperScript I reverse transcriptase (Life Technologies).29CD109 K1-H7–specific transcripts were detected using the oligonucleotide primer pair H6(7-1) 5′-4201CTGTCCTCCTGTGACCTT4218-3′ and H7(U2) 5′-4841ATCTACTGAGACCACTGG4824-3′ (numbering as in Figure 2) using 50 μL hot-start PCR reactions.29
In vitro transcription/translation
The KG1a-derived CD109 K1 clone in pBK-CMV was digested withNotI–SalI to liberate a cDNA fragment containing the entire open-reading frame (ORF), including the translation initiation codon but without the 3′ untranslated region (UTR), which was then inserted into NotI/SalI–digested pBS II KS(−) such that the CD109 cDNA was placed downstream of the T7 promoter. The pBS KS II T7/K1 construct (1.2 μg) was transcribed and translated in vitro using the T7/T3 TNT Coupled Reticulocyte Lysate System (Promega) and 35S-methionine (Amersham Pharmacia Biotech) according to the manufacturer's protocol. Five microliters reaction mix was then added to 195 μL lysis buffer supplemented with 50 μg/mL aminoethylbenzenesulfonylfluoride, 1 μg/mL antipain, 1 μg/mL leupeptin, 1 μg/mL pepstatin, and 1 μg/mL aprotinin (ICN, Montreal, QC, Canada). Then, 1.5 μg of one of the CD109 mAbs 1B3, 8A3, or LDA1 or of the control CD71 mAb D51 was added, and the resultant mix was incubated on ice for 1 hour.
Protein A–Sepharose beads (5 μL packed beads per immunoprecipitate) were preincubated for 20 minutes on ice with unlabeled KG1a lysate (4 × 107 cells/mL). Beads were washed twice in cold lysis buffer, mixed with the TNT immune complexes for 45 minutes at 4°C, and were washed again in cold lysis buffer. Immune complexes were analyzed by SDS-PAGE.
Expression of CD109 cDNA in Chinese hamster ovary cells
The CD109 K1 cDNA ORF was excised from pBK-CMV as above and was inserted downstream of the CMV promoter (but upstream of the IRES sequence) into EcoRV–Notl cut plRES-EYFP (Clontech) to yield pK1/YFP. Restriction enzyme analysis and DNA sequencing were used to verify the orientation of the insert.
CHO cells were seeded at a density of 1.3 × 106 cells per 10-cm dish. After 24 hours, cells were washed in PBS and were transfected in OPTI-MEM I medium (Life Technologies) with 10 μg pK1/YFP or control pIRES-EYFP and 40 μL Lipofectamine (Life Technologies) per dish, according to the manufacturer's protocol. After 4 hours, cells were washed in PBS and were placed in 5% CO2 at 37°C in F12(Ham) medium supplemented as above. Forty-eight hours thereafter, cells were detached from the plates with citric saline (135 mM KCl, 15 mM sodium citrate), washed twice in PBS–EDTA, incubated with phycoerythrin-conjugated CD109 mAb 8A3, 7D1, or TEA 2/16 for 30 minutes on ice, rinsed twice in PBS–EDTA, and resuspended in 0.5 mL PBS containing 1 μg/mL 7-amino actinomycin D (7-AAD; Sigma). After 10 minutes at room temperature, CD109 expression was determined by flow cytometry through the assessment of mAb binding to gated viable, single, YFP-positive cells.
Results
Immuno-affinity purification and partial amino acid sequencing of CD109
When analyzed by SDS-PAGE and silver staining (not shown), the eluate from the mAb 8A3/Protein A–Sepharose column yielded 2 bands at approximately 170 and 150 kd, characteristic of CD109.3 8-10 Affinity-purified CD109 was then fractionated further by preparative SDS-PAGE, and the larger band was excised and digested with endoproteinase Lys-C or Asp-N. Purification and sequence analysis of the resultant peptide fragments yielded 20 peptide sequences ranging in size from 7 to 20 aa. After overlapping sequences were combined, 17 independent CD109-derived peptide sequences were obtained.
cDNA cloning and analysis
BLAST analysis31,32 using these 17 CD109 peptide sequences identified a rat EST R4712325 that encoded the CD109-specific peptide DPKSNLIQQXLSQQ. EST R47123 was subsequently used to probe a λ phage Uni-ZAP HUVEC cDNA library, yielding 8 independent clones that comprised 2 overlapping groups (Figure 1A). The first consisted of 7 clones that were progressive 5′ truncations of the longest example, clone H6, a 3-kb clone containing a 2.7-kb ORF, followed by a 300-bp 3′ UTR ending with a poly(A) tract. The second, consisting of clone H7 (approximately 2 kb), was contiguous with the H6 series cDNAs but contained a longer 3′ UTR that extended an additional 1132 bp before the appearance of a poly(A) tract. Because clone H6 was not full length, we endeavored unsuccessfully to obtain more 5′ CD109 cDNA sequence by rescreening the HUVEC library with H6 itself. A λ ZAP Express KG1a cDNA library was screened using a clone H6 probe, yielding 9 independent clones. As illustrated in Figure 1A, restriction enzyme analysis and DNA sequencing demonstrated that these clones comprised a series of progressive 5′ deletions of the longest example, clone K1 (approximately 4.7 kb), that encompassed clone H6 in its entirety and that contained about 1.3 kb additional 5′ cDNA sequence.
Nucleotide sequences of the 3 overlapping clones—H6, H7, and K1—were determined in their entirety for both strands (Figures 1, 2). Clone K1 contained a 4335-bp ORF flanked by 112 bp 5′ and 300 bp 3′ UTRs, respectively. The putative translation start, though not comprising an optimal Kozak consensus sequence,33 was preceded by stop codons in all 3 frames. In addition (see below), the first 20 codons downstream of this start were predicted to encode a cleavable signal peptide. The K1 3′ UTR contained a canonical polyadenylation signal, AATAAA, 15 bp upstream of the poly(A) tail. The clone H7 3′ UTR was contiguous with that of clones K1 and H6 but extended an additional 1132 bp in the 5′ direction. Two polyadenylation signals were found 34 and 19 bp, respectively, upstream of the H7 poly(A) tail.
Clone K1 encodes CD109
To determine whether cDNA clone K1 did indeed encode CD109, we initially examined the predicted K1 protein sequence. Notably, the clone K1 ORF was found to contain 15 of the 17 CD109derived peptide sequences described above (Figure 2). A 16th peptide appeared to correspond to an actual, but scrambled, CD109 sequence (not shown). Next, we confirmed that the protein encoded by K1 could be detected by CD109-specific mAbs. When transcribed and translated in vitro (Figure3A), K1 yielded a protein of about 160 kd that was recognized by CD109 mAbs 1B3, 8A3, and LDA1. In addition, expression of clone K1 was able to confer high-level mAb 7D1 binding to transfected CHO cells (Figure 3B). Similar staining was observed with the other CD109 mAbs 8A3, and TEA 2/16, and with Govb antiserum. Notably, this binding was abrogated by the treatment of K1 transfected cells with PI-PLC (not shown). In contrast, staining was not detectable on CHO cells transfected with control vectors expressing K1 antisense or an irrelevant cDNA (not shown).
Clone K1 encodes a protein that is recognized by CD109 mAbs.
(A) By in vitro transcription–translation in the presence of35S-methionine, followed by immunoprecipitation, CD109 clone K1 is shown to encode an approximately 160-kd protein (arrow, lane 1) that is recognized by mAb 1B3, but not by control antibodies, including CD71 mAb D51 (lane 2). Positions of radiolabeled molecular weight markers are shown on the left. Similar results were obtained using CD109 mAbs 8A3 and LDA1 (not shown). (B) Expression of clone K1 confers CD109 mAb binding to CHO cells. Control mock-transfected CHO cells (top row), CHO cells transfected with empty pIRES-EYFP vector (middle row), and CHO cells transfected with CD109 expression vector pK1/YFP (bottom row) were stained with CD109 mAb 7D1-PE and 7-AAD. Viable (7-AAD−) cells were then gated (region R1), and single cells within this gate were identified by light scatter (R2, not shown). YFP-expressing cells within this population (R3, left column) were then analyzed for CD109 expression (R4, right column). Expression of the K1 cDNA confers high-level 7D1 binding to CHO cells. Similar specific staining was observed using phycoerythrin conjugates of CD109 mAbs 8A3 and TEA 2/16 (not shown), and with Govb antiserum.53 In contrast, staining was not detectable on control CHO cells or on CHO cells transfected with pIRES-EYFP–based vectors expressing K1 antisense or an irrelevant cDNA and was abrogated by treatment of K1-transfected cells with PI-PLC (not shown).
Clone K1 encodes a protein that is recognized by CD109 mAbs.
(A) By in vitro transcription–translation in the presence of35S-methionine, followed by immunoprecipitation, CD109 clone K1 is shown to encode an approximately 160-kd protein (arrow, lane 1) that is recognized by mAb 1B3, but not by control antibodies, including CD71 mAb D51 (lane 2). Positions of radiolabeled molecular weight markers are shown on the left. Similar results were obtained using CD109 mAbs 8A3 and LDA1 (not shown). (B) Expression of clone K1 confers CD109 mAb binding to CHO cells. Control mock-transfected CHO cells (top row), CHO cells transfected with empty pIRES-EYFP vector (middle row), and CHO cells transfected with CD109 expression vector pK1/YFP (bottom row) were stained with CD109 mAb 7D1-PE and 7-AAD. Viable (7-AAD−) cells were then gated (region R1), and single cells within this gate were identified by light scatter (R2, not shown). YFP-expressing cells within this population (R3, left column) were then analyzed for CD109 expression (R4, right column). Expression of the K1 cDNA confers high-level 7D1 binding to CHO cells. Similar specific staining was observed using phycoerythrin conjugates of CD109 mAbs 8A3 and TEA 2/16 (not shown), and with Govb antiserum.53 In contrast, staining was not detectable on control CHO cells or on CHO cells transfected with pIRES-EYFP–based vectors expressing K1 antisense or an irrelevant cDNA and was abrogated by treatment of K1-transfected cells with PI-PLC (not shown).
CD109 is a GPI-linked thioester-containing protein
Consistent with the known size of CD109, the translated K1 sequence (Figures 1B, 2) predicts a 1445-aa protein of approximately 162 kd bearing a cleavable 21-aa N-terminal leader peptide34 and containing 17 potential N-linked glycosylation sites. As expected, the presence of a C-terminal hydrophobic tail preceded by a short hydrophilic stretch and a cluster of nonbulky amino acids defines a GPI anchor cleavage–addition site, with cleavage predicted to occur after aa 1420.35-37Notably, the translated K1 sequence (Figures 1B, 2) contains the918PYGCGEQ924 thioester motif, defining CD109 as a member of the α2M/C3, C4, C5 superfamily of thioester-containing protease inhibitor and complement proteins.38,39 Indeed, by blast31,32 analysis, CD109 bears 45% to 50% overall sequence similarity to other vertebrate and invertebrate α2M proteins (and was more distantly related to C3 and C4 complement proteins), with particularly high similarity in the region of the thioester motif (Figure 4A) and in 11 additional α2M family-specific conserved sequence blocks.40,41 The overall structural organization and size of CD109 is typical of α2M inhibitors as well (Figure 2B). The thioester lies approximately two thirds of the way along the 162-kd chain, and a hexapeptide motif (residues 1030-1035) that defines the chemical reactivity of the thioester42,43 is found as expected, 100 aa further downstream (Figures 1, 2B, 4B). In addition to these highly conserved regions, each α2M protein also contains a unique bait region in the middle of the molecule that defines substrate specificity.44 Containing cleavage sites for proteases of all 4 mechanistic classes44 45 and with diverse specificities, the bait region confers promiscuous protease inhibitory activity to α2M proteins. Consistent with this, CD109 also contains a putative bait region (approximate residues 651-683; Figures 1B, 2) that, as expected, is unrelated to the corresponding regions of other family members.
CD109 contains an α2M-like thioester, but its chemical reactivity likely resembles that of complement.
(A) The CD109 thioester most closely resembles that of other α2M proteins. Although CD109 bears 45% to 50% overall sequence similarity with other vertebrate and invertebrate α2M proteins, it shares particularly high similarity in the region of the thioester. An alignment of a 50 aa stretch flanking the CD109 thioester and the corresponding regions of the 18 most closely related proteins is shown. Notably, this group does not contain complement proteins. Numbers refer to the corresponding aa coordinates. MUG, murinoglobulin; α1I3, α1-inhibitor III; α1M, α1-macroglobulin; PZP, pregnancy zone protein; OvoM, ovomacroglobulin. Black shading, aa identity; gray shading, aa similarity; no shading, unrelated. (Carp α2M-1, GenBank accession no. [GB no.] AB026128; Carp α2M-2, GB no. AB026129; Carp α2M-3, GB no. AB026130; C elegans ZK337, GB no. Z82090; Limulus α2M, GB no. D83196; Chicken OvoM, GB no. X78801; Guinea Pig α2M, GB no. D84338; Guinea Pig MUG, GB no.D84339; Human PZP, GB no. X54380; Human α2M, GB no. M11313; Lamprey α2M, GB no. D13567; Mouse MUG2, GB no. M65238; Mouse MUG1, GB no.M65736; Mouse α2M, GB no. NM_007376; Rat α1I3, GB no. J03552; Rat α2M, GB no. J02635; Rat α1M, GB no. M84000; XenopusEndodermin, GB no. L63543.) (B) The CD109 thioester reactivity-defining hexapeptide most resembles that of complement proteins. A 20 aa stretch encompassing the CD109 regulatory hexapeptide is aligned with the corresponding regions of human complement C3 and C4b and those of the 2 other α2M-family proteins—α2M and PZP. Although the α2M hexapeptide ends in an LLN triplet, that of CD109, C3, and C4b ends in VIH. Numbers refer to the corresponding aa coordinates. (Human PZP, GB no. X54380; Human α2M, GB no. M11313; Human C3, GB no. K02765; Human C4B, GB no. K02404.)
CD109 contains an α2M-like thioester, but its chemical reactivity likely resembles that of complement.
(A) The CD109 thioester most closely resembles that of other α2M proteins. Although CD109 bears 45% to 50% overall sequence similarity with other vertebrate and invertebrate α2M proteins, it shares particularly high similarity in the region of the thioester. An alignment of a 50 aa stretch flanking the CD109 thioester and the corresponding regions of the 18 most closely related proteins is shown. Notably, this group does not contain complement proteins. Numbers refer to the corresponding aa coordinates. MUG, murinoglobulin; α1I3, α1-inhibitor III; α1M, α1-macroglobulin; PZP, pregnancy zone protein; OvoM, ovomacroglobulin. Black shading, aa identity; gray shading, aa similarity; no shading, unrelated. (Carp α2M-1, GenBank accession no. [GB no.] AB026128; Carp α2M-2, GB no. AB026129; Carp α2M-3, GB no. AB026130; C elegans ZK337, GB no. Z82090; Limulus α2M, GB no. D83196; Chicken OvoM, GB no. X78801; Guinea Pig α2M, GB no. D84338; Guinea Pig MUG, GB no.D84339; Human PZP, GB no. X54380; Human α2M, GB no. M11313; Lamprey α2M, GB no. D13567; Mouse MUG2, GB no. M65238; Mouse MUG1, GB no.M65736; Mouse α2M, GB no. NM_007376; Rat α1I3, GB no. J03552; Rat α2M, GB no. J02635; Rat α1M, GB no. M84000; XenopusEndodermin, GB no. L63543.) (B) The CD109 thioester reactivity-defining hexapeptide most resembles that of complement proteins. A 20 aa stretch encompassing the CD109 regulatory hexapeptide is aligned with the corresponding regions of human complement C3 and C4b and those of the 2 other α2M-family proteins—α2M and PZP. Although the α2M hexapeptide ends in an LLN triplet, that of CD109, C3, and C4b ends in VIH. Numbers refer to the corresponding aa coordinates. (Human PZP, GB no. X54380; Human α2M, GB no. M11313; Human C3, GB no. K02765; Human C4B, GB no. K02404.)
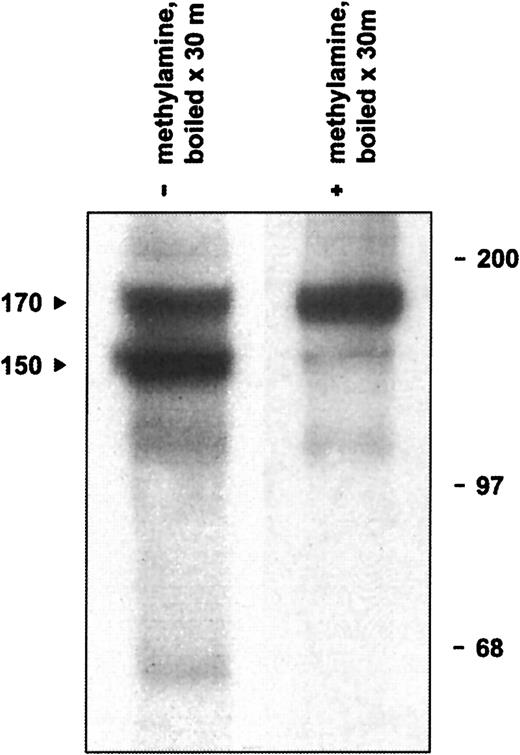
The defining structural feature of the α2M/C3, C4, C5 superfamily is an intrachain thioester bond formed between a cysteinyl side chain sulfhydryl and a glutamine side chain carbonyl in the sequence CGEQ that can be disrupted with small nucleophiles such as methylamine.44,46 In addition, under experimental conditions of heat or chemical denaturation (preparing a sample for SDS-PAGE, for example), both complement and α2M inhibitors may undergo internal nucleophilic attack on the thioester, resulting in autolytic cleavage of the protein.47-49 Although not of physiological significance, this autolytic reaction is useful diagnostically to indicate the presence of an intact thioester bond. Therefore, we determined whether native CD109 could undergo high-temperature autolytic cleavage that could be prevented by pretreatment with methylamine. As illustrated in Figure5, when KG1a-derived mAb 8A3–CD109 immune complexes were treated with 400 mM methylamine before analysis, only a single CD109 band of about 170 kd was subsequently observed. In the absence of methylamine treatment, however, boiling of immune complexes resulted in the appearance of the typical 150-kd form and of an associated 20-kd fragment. In contrast, and consistent with the known inability of standard cell-free systems to support intramolecular thioester formation,50 only a single band was observed when CD109 synthesized in vitro was heat-treated in the absence of methylamine (Figure 3A). Taken together, these observations demonstrate that native CD109 contains an intact thioester. In addition, these data indicate that the 150-kd CD109 band is indeed derived from the 170-kd form as had previously been suggested,3,10 but by autolytic rather than by proteolytic cleavage. The known ability of reducing agents to inhibit the autolytic cleavage of thioester containing proteins such as C348,51,52 thus provides an explanation for the earlier observation that reduction before denaturation decreases the formation of the 150-kd CD109 band.10
Native CD109 contains an intact thioester.
Boiling of radiolabeled KG1a-derived mAb 8A3–CD109 immune complexes results in the appearance of the typical 150- and 170-kd CD109 bands (arrows, lane 1) and of an additional 20-kd band (not shown). If, however, the immune complexes are treated with 400 mM methylamine before boiling, formation of the 150-kd band is greatly inhibited (lane 2). Thus, the 150-kd CD109 band arises by the thioester-mediated autolytic cleavage of the 170-kd form, which is abrogated if the thioester is first disrupted with the small nucleophile methylamine. Positions of molecular weight markers are shown on the right.
Native CD109 contains an intact thioester.
Boiling of radiolabeled KG1a-derived mAb 8A3–CD109 immune complexes results in the appearance of the typical 150- and 170-kd CD109 bands (arrows, lane 1) and of an additional 20-kd band (not shown). If, however, the immune complexes are treated with 400 mM methylamine before boiling, formation of the 150-kd band is greatly inhibited (lane 2). Thus, the 150-kd CD109 band arises by the thioester-mediated autolytic cleavage of the 170-kd form, which is abrogated if the thioester is first disrupted with the small nucleophile methylamine. Positions of molecular weight markers are shown on the right.
Expression pattern of CD109
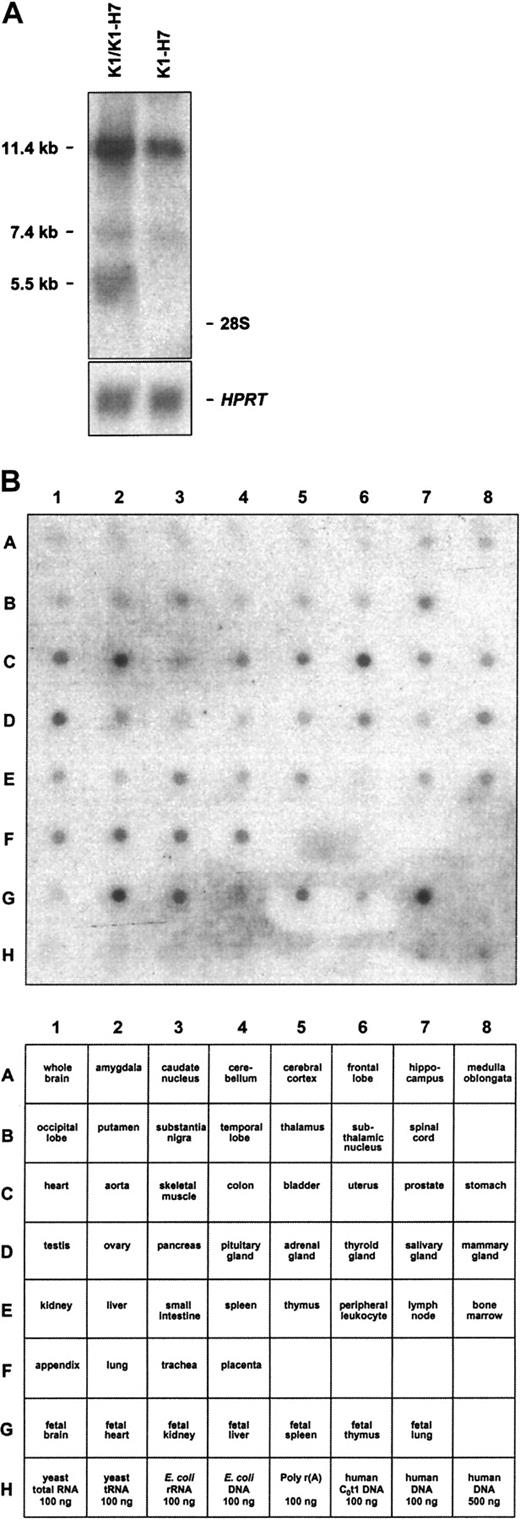
As noted above, 2 distinct CD109 3′ UTRs were isolated by library screening. By RT-PCR analysis, both variants were readily detectable in KG1a and HUVEC RNA (not shown). Consistent with these data, several KG1a CD109 transcripts were also detectable by Northern blot analysis of total RNA (Figure 6A). Notably, a probe expected to recognize both K1 and K1-H7 detected transcripts measuring 5.5 and 7.4 kb, whereas a K1-H7-specific probe detected only the latter. It is likely, therefore, that the 5.5 and 7.4 kb bands correspond to K1 and K1-H7 transcripts, respectively. The relationship of these transcripts to the additional large band detected by both probes is unclear. Northern blot analysis of Jurkat, HeLa, MEG01, and CMK-11-5 cell total RNAs using both K1 and K1-H7 probes yielded similar results (not shown). A cDNA probe expected to recognize both K1 and K1-H7 transcripts was also used to evaluate the tissue range of expression of CD109 with a commercial multiple tissue blot bearing a series of human adult and fetal RNAs. As shown in Figure 6B, CD109 transcripts were detected in a wide range of tissues, with highest levels found in adult uterus, aorta, heart, lung, trachea, and placenta, and in fetal heart, kidney, liver, spleen, and lung. Whether these data indicate true widespread expression or merely reflect endothelial cell expression (or both) is unknown.
Northern blot analysis of CD109-specific transcripts.
(A) KG1a cells contain multiple CD109-specific transcripts. Northern blots of KG1a total RNA probed with K1/K1-H7 common and K1-H7 specific probes are shown. The former probe is expected to detect both K1 and K1-H7 transcripts. Although 3 CD109-specific bands (11.4, 7.4, and 5.5 kb) are revealed by the common probe, the K1-H7–specific probe detects only the 2 larger transcripts. Positions of the 28S and 18S rRNA species are noted on the right. Control hypoxanthine phosphoribosyltransferase hybridizations ensure equal RNA loading. (B) CD109 is expressed widely in adult and fetal tissues. A commercial multiple tissue mRNA blot probed with a CD109 probe expected to detect both K1 and K1-H7 transcripts is shown (top panel). Corresponding RNAs are identified in the bottom panel. CD109 transcripts are detected in a wide range of tissues, with the highest levels found in adult uterus, aorta, heart, lung, trachea, and placenta, and in fetal heart, kidney, liver, spleen, and lung. In all cases, negative control RNAs and DNAs did not yield CD109-specific signals.
Northern blot analysis of CD109-specific transcripts.
(A) KG1a cells contain multiple CD109-specific transcripts. Northern blots of KG1a total RNA probed with K1/K1-H7 common and K1-H7 specific probes are shown. The former probe is expected to detect both K1 and K1-H7 transcripts. Although 3 CD109-specific bands (11.4, 7.4, and 5.5 kb) are revealed by the common probe, the K1-H7–specific probe detects only the 2 larger transcripts. Positions of the 28S and 18S rRNA species are noted on the right. Control hypoxanthine phosphoribosyltransferase hybridizations ensure equal RNA loading. (B) CD109 is expressed widely in adult and fetal tissues. A commercial multiple tissue mRNA blot probed with a CD109 probe expected to detect both K1 and K1-H7 transcripts is shown (top panel). Corresponding RNAs are identified in the bottom panel. CD109 transcripts are detected in a wide range of tissues, with the highest levels found in adult uterus, aorta, heart, lung, trachea, and placenta, and in fetal heart, kidney, liver, spleen, and lung. In all cases, negative control RNAs and DNAs did not yield CD109-specific signals.
Discussion
The restricted pattern of expression of CD109 within hematopoietic cells (CD109 is expressed by a subset of early progenitor and candidate HSCs and by activated platelets and T cells) suggests that it may play a role not only in hematopoiesis but also in cell-mediated immunity and in hemostasis. As a first step toward elucidating the function of CD109, we used an immuno-purification–microsequencing strategy to isolate a cDNA encoding human CD109. Several lines of evidence indicate that the isolated cDNA has been correctly identified. Not only does the clone encode 16 of 17 CD109-derived peptides, but its expression results in the synthesis of a GPI-linked protein that can be detected by multiple CD109-specific mAbs, both in vitro and in vivo. In addition, and confirming its identity, we have recently determined that the Gova and Govb alloantigens are defined by a single nucleotide polymorphism of the cDNA reported here (see accompanying article by Schuh et al,53 page 1692). Nevertheless, the presence of multiple CD109 transcripts by Northern blot analysis and the presence of an additional CD109-related peptide that could not be accounted for by our cDNA raise the possibility that additional CD109 cDNA variants may exist.
The presence of a thioester signature sequence38,39—918PYGCGEQ924—defines CD109 as a member of the α2M/C3, C4, C5 superfamily of thioester-containing proteins. This family comprises 2 general divisions—the α2M-like protease inhibitors and the complement proteins—that are thought to have arisen from a common, ancestral α2M-like molecule.44 By sequence similarity, CD109 is closely related to the α2M inhibitors and more distantly to C3 and C4 proteins. CD109 differs from typical α2M inhibitors in several respects, however. First, though most α2M protease inhibitors exist as oligomers of a 180-kd subunit44 (for example, human α2 macroglobulin occurs in plasma as a 720-kd tetramer), CD109 apparently exists as a monomer.3,5,6 To date, monomeric α2M protease inhibitors have been characterized primarily in rodents,44 although they are believed to exist in other vertebrates as well.54 Second, CD109 is membrane bound through a GPI anchor. Membrane-anchored α2M/C3, C4, and C5 proteins have not been described previously. Third, although various activated human and rodent α2M inhibitors have been shown to interact with 2 cellular receptors—the low-density lipoprotein receptor-related protein–α2M receptor (LRP-α2MR) that mediates the clearance of inhibitor–protease complexes from the circulation55,56 and the α2-macroglobulin signaling receptor (α2MSR) that mediates α2M activation-dependent signals57-61—the carboxyl end of CD109 does not contain the KPTVK motif59 62-67required for receptor binding. And fourth, although CD109 bears much greater overall sequence similarity to α2M proteins than it does to complement, its thioester reactivity-defining hexapeptide (Figure 4B) does not end in LLN as in other α2M proteins; rather, it ends in the 1033VIH1035 triplet characteristic of complement proteins (see below). Overall, therefore, CD109 defines a new member of the α2M family, but one with unusual features.
The defining structural feature of the α2M/C3, C4, C5 superfamily—the intrachain thioester bond—is typically unreactive in the native molecule, except with small nucleophiles such as methylamine. On proteolytic cleavage of the molecule, however—by specific activating enzymes (in the case of the complement proteins) or by a wide range of proteases (in the case of the protease inhibitors)—a conformational change occurs, and the thioester becomes highly reactive toward nucleophiles such that the proteins become covalently bound to nearby macromolecules through ester or amide bonds.44 In the case of complement, this leads to the covalent deposition of C3 and C4 on the target cell and on immune aggregates. In the case of protease inhibitors, covalent binding of the activating protease may similarly occur. By analogy, it is likely that CD109 becomes activated in a similar fashion by proteolytic cleavage and thereby becomes capable of covalent substrate binding.
Curiously, though α2M proteins preferentially bind to substrate molecules by forming ester bonds with hydroxyl groups on carbohydrates or proteins, C3 and C4 generally form amide bonds with proteins.44,68 This differential specificity is determined by the presence or absence of His or Asn in the terminal position of a conserved hexapeptide lying about 100 aa C-terminal to the thioester bond,42,43 (by protein folding, this domain interacts with, and modulates the reactivity of the thioester69). As has recently been elucidated,44,70,71 proteolytically activated Asn-containing molecules undergo direct nucleophilic attack of the thioester carbonyl in an uncatalyzed reaction. In contrast, proteolytic activation of His-containing molecules results in a catalyzed transacylation reaction that involves the initial intramolecular transacylation of the thioester carbonyl to the imidazole ring of His, forming a covalent intramolecular acyl imidazole intermediate. The liberated thioester cysteinyl sulfhydryl then acts as a general base to deprotonate hydroxyl nucleophiles for attack on the acyl imidazole intermediate. The catalyzed (His) reaction thus facilitates transacylation to hydroxyl-containing carbohydrate or protein targets, whereas in the uncatalyzed (Asn) reaction, only primary amine groups of protein targets are sufficiently nucleophilic to attack the thioester bond directly. In addition, thet½ of the reactive thioester is known to be much shorter if His is substituted for Asn, as the intermediate in the catalyzed reaction will react quickly with the most common hydroxyl-containing nucleophile, water.44 70-72 Thus, α2M proteins bearing a carboxyl-terminal His residue can potentially react with both carbohydrate and protein targets, but, by virtue of the short half-life of the reactive thioester, such binding is thought to be tightly restricted spatially to the initial site of activation. Because the CD109 regulatory motif ends in the VIH triplet usually associated with complement, it is likely not only that proteolytically activated CD109 forms ester bonds with hydroxyl groups on carbohydrates or proteins, rather than amide bonds, but also that this reactivity is short-lived and is highly restricted spatially to the site of activation, defining activated CD109 as a locally acting molecule.
To date, though CD109 has been defined primarily as a marker of specific hematopoietic stem and progenitor cell subsets or as potential target antigen in alloimmune platelet destruction, its biologic function has remained obscure. The identification of CD109 as a novel, monomeric, α2M-type inhibitor with complementlike thioester reactivity allows several functional predictions to be made: First, it is likely that CD109 becomes activated by proteolytic cleavage. Although the expression pattern of CD109 immediately suggests a large number of candidate activating proteases that are elaborated during hematopoiesis and during platelet and T-cell activation, the physiologically relevant protease(s) are as yet undefined. Second, it is likely that proteolytic cleavage of CD109 results in a conformational change that leads to the covalent cross-linking of CD109 to adjacent molecules. Such covalent cross-linking activity is essential for the action of C3 and C4 and for the protease inhibitory action of monomeric α2M protease inhibitors. Although it is not yet known whether CD109 can function as a protease inhibitor, by analogy with other monomeric α2M inhibitors, such activity would likely require covalent binding to the activating protease, resulting in the formation of a CD109–protease complex. Third, it is possible that the covalent binding of activated CD109 is not restricted to proteases. Proteolytically activated α2M is able to bind a variety of other proteins and peptides by noncovalent trapping and covalent thioester-mediated binding mechanisms, thereby regulating their plasma stability, transport, and clearance.73-75 Although proteolytically activated monomeric α2M family proteins such as CD109 would not be expected to be able to trap other molecules noncovalently, as are their multimeric counterparts, they likely are capable of covalent binding to other nonprotease substrates. Indeed, in view of the putative complementlike spatially restricted thioester reactivity of activated CD109 and its location as a GPI-linked cell surface molecule, we suggest that CD109 may function as a membrane-bound cross-linking reagent, thereby mediating cell–substrate, cell–matrix, or cell–cell interactions that play roles in hematopoiesis, primary hemostasis, or innate immune responses. Consistent with this notion, the CD109 mAb LDA1 has been reported to abrogate antibody-inducing T-cell helper function during a primary mixed-lymphocyte reaction,7 suggesting that CD109 may play a role in T-cell–antigen-presenting cell or T-cell–B-cell interactions.
The isolation of a cDNA encoding CD109 and the identification of this molecule as a novel membrane-bound member of the α2M/ C3, C4, C5 superfamily of thioester proteins has raised a number of intriguing possibilities regarding its functional role(s) and mechanism of action. We anticipate that biochemical studies using recombinant CD109 and the generation of murine pedigrees carrying specific CD109 mutations will allow these questions to be answered.
We thank Norman Lassam, Ed Conway, David Isenman, and Alex Law for helpful discussions during this work and Willem Ouwehand and David Spaner for reviewing the manuscript.
Supported by grants from the Medical Research Council of Canada and the National Cancer Institute of Canada (A.C.S. and D.R.S.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Andre C. Schuh, Rm 7366, Medical Sciences Bldg, University of Toronto, 1 King's College Circle, Toronto, ON, Canada, M5S 1A8; e-mail: andre.schuh@utoronto.ca.


![Fig. 2. CD109 cDNA and predicted protein sequences. / The K1 and K1-H7 CD109 cDNAs and the corresponding 1445 aa protein sequence are shown (GenBank accession number AF410459). Nucleotides are numbered relative to the translation initiation codon, with the corresponding aa numbering shown in parentheses. K1 and K1-H7 3′UTRs, and the positions of the corresponding poly(A) tails [(a)n)] are shown. Peptides identified by immuno-affinity purification–microsequencing of CD109 are singly underlined. Potential sites of N-linked glycosylation, the thioester signature sequence (aa residues 918-924), and the corresponding downstream thioester reactivity-defining hexapeptide motif (aa residues 1030-1035) are marked by open boxes. Additionally, the amino-terminal leader peptide (double underline), the bait region (dotted underline), the translation stop (*), and the GPI anchor cleavage–addition site (open triangle) are shown.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/99/5/10.1182_blood.v99.5.1683/5/m_h80522225002.jpeg?Expires=1769175008&Signature=pc2mNZYWHXqj2zuoyI~BWQ6~BmpobizAaP2K5PgzXTQnUL-L4tRhf0E-5ikk71u~p9uop-yM1vklJ8A1xq6iTf-cVqB1NIIxZwzIjRlosf5FUztrWdUTcZu~hYxfT~f~OH9EOCgPGBA~CyrGf~oLeNetDSb9nFvTU1ZXYqqaSFhTUL6SUXaf2KD~kx8xeye5XCNmw0e5RGPRNlKlrZqMjLzxmky2d-m8A7-E6dcrFns8XNhKv6DRlM8zcIK56FirrliiHspzApe5JM0lHDl32GnIF9WWJQXbb9QYwAQZUvBNcFAnQ4yH2A7Yzm5elP4uV8QfROlXw3Sxz6ahk6BycQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Fig. 4. CD109 contains an α2M-like thioester, but its chemical reactivity likely resembles that of complement. / (A) The CD109 thioester most closely resembles that of other α2M proteins. Although CD109 bears 45% to 50% overall sequence similarity with other vertebrate and invertebrate α2M proteins, it shares particularly high similarity in the region of the thioester. An alignment of a 50 aa stretch flanking the CD109 thioester and the corresponding regions of the 18 most closely related proteins is shown. Notably, this group does not contain complement proteins. Numbers refer to the corresponding aa coordinates. MUG, murinoglobulin; α1I3, α1-inhibitor III; α1M, α1-macroglobulin; PZP, pregnancy zone protein; OvoM, ovomacroglobulin. Black shading, aa identity; gray shading, aa similarity; no shading, unrelated. (Carp α2M-1, GenBank accession no. [GB no.] AB026128; Carp α2M-2, GB no. AB026129; Carp α2M-3, GB no. AB026130; C elegans ZK337, GB no. Z82090; Limulus α2M, GB no. D83196; Chicken OvoM, GB no. X78801; Guinea Pig α2M, GB no. D84338; Guinea Pig MUG, GB no.D84339; Human PZP, GB no. X54380; Human α2M, GB no. M11313; Lamprey α2M, GB no. D13567; Mouse MUG2, GB no. M65238; Mouse MUG1, GB no.M65736; Mouse α2M, GB no. NM_007376; Rat α1I3, GB no. J03552; Rat α2M, GB no. J02635; Rat α1M, GB no. M84000; XenopusEndodermin, GB no. L63543.) (B) The CD109 thioester reactivity-defining hexapeptide most resembles that of complement proteins. A 20 aa stretch encompassing the CD109 regulatory hexapeptide is aligned with the corresponding regions of human complement C3 and C4b and those of the 2 other α2M-family proteins—α2M and PZP. Although the α2M hexapeptide ends in an LLN triplet, that of CD109, C3, and C4b ends in VIH. Numbers refer to the corresponding aa coordinates. (Human PZP, GB no. X54380; Human α2M, GB no. M11313; Human C3, GB no. K02765; Human C4B, GB no. K02404.)](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/99/5/10.1182_blood.v99.5.1683/5/m_h80522225004.jpeg?Expires=1769175008&Signature=NVoKclc~yFmcgRbhi3SvR8DIc26Pq-DCZxA~YOcvcG~HfFkmqGzlpTxrZdRrwb-~CAgrKS5VebkY8jNThk~7Tnaw-TRa41p5FcLKx4TNScfW-X7GGulb1QDqBOx7vGJcZDCrOhm~mLBx1d9TuAWuLlstT7xnjlvc~2l2nPF2GCwE27wI~wjzSquCMxRXbQxWjVznywz2s7m1-Lz~ozuXBg-79vXdN6mX~3JXNqeloZk-yNgQkCkPE~~PTbQHitAVbHS5ikdZ1LU~yA8BjOKEpesxBItYgf2wDXPTPvtCBORXuafXlybHUVCczYnPmNkdK-L1DkrZB0TiGPhpGn5Oog__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal