Mutations of c-KIT causing spontaneous activation of the KIT receptor kinase are associated with sporadic adult human mastocytosis (SAHM) and with human gastrointestinal stromal tumors. We have classified KIT-activating mutations as either “enzymatic site” type (EST) mutations, affecting the structure of the catalytic portion of the kinase, or as “regulatory” type (RT) mutations, affecting regulation of an otherwise normal catalytic site. Using COS cells expressing wild-type or mutant KIT, 2 compounds, STI571 and SU9529, inhibited wild-type and RT mutant KIT at 0.1 to 1 μM but did not significantly inhibit the Asp816Val EST mutant associated with SAHM, even at 10 μM. Using 2 subclones of the HMC1 mast cell line, which both express KIT with an identical RT mutation but which differ in that one also expresses the Asp816Val EST mutation, both compounds inhibited the RT mutant KIT, thereby suppressing proliferation and producing apoptosis in the RT mutant-only cell line. Neither compound suppressed activation of Asp816Val EST mutant KIT, and neither produced apoptosis or significantly suppressed proliferation of the cell line expressing the Asp816Val mutation. These studies suggest that currently available KIT inhibitors may be useful in treating neoplastic cells expressing KIT activated by its natural ligand or by RT activating mutations such as gastrointestinal stromal tumors but that neither compound is likely to be effective against SAHM. Furthermore, these results help establish a general paradigm whereby classification of mutations affecting oncogenic enzymes as RT or EST may be useful in predicting tumor sensitivity or resistance to inhibitory drugs.
Introduction
The c-KIT protooncogene encodes the KIT protein,1 the tyrosine kinase receptor for stem cell factor (SCF).2 KIT is essential for normal development of mast cells in humans and other mammals.3 Adult-type human mastocytosis is characterized by mutations in c-KIT codon 816, which cause constitutive activation of the KIT kinase.4-7 A number of mast cell lines and canine mast cell tumors also express activating c-KITmutations,4,8,9 and small molecules that inhibit mutant activated KIT effectively kill these cell lines.10
In our previous work, we have classified KIT-activating mutations into 2 major groups.11 One group, exemplified by codon 816 mutations found in human mastocytosis, causes residue substitutions in the activation loop lying at the entrance to the enzymatic pocket.9 Because these mutations affect the structure of the enzymatic pocket, we have called them “enzymatic pocket” or “enzymatic site” type (EST) mutations. The second group, exemplified by mutations causing residue substitutions, or in-frame deletions or insertions in the intracellular juxtamembrane region found in canine mast cell tumors and in human gastrointestinal stromal tumors,9,12 affects the regulation of the activity of an otherwise normal enzymatic site.13 We have termed these “regulatory” type (RT) mutations. Importantly, different KIT inhibitors vary in their ability to inhibit these different types of mutant KIT and in their ability to inhibit similar types of mutations occurring in different species.10 Because of these differences, we have proposed that human mastocytosis be classified according to the presence or absence of specific activating mutations and that the ability of a potential therapeutic agent to inhibit the specific activating mutation expressed by a patient's neoplastic mast cells be demonstrated prior to initiation of a therapeutic trial.14 15
In this paper, we extend to human mast cells and human KIT our original observations on different classes of activating KIT mutations and their tendency to be inhibited by different pharmacologic agents. Furthermore, we show that not only does the class of mutations affect the sensitivity of activated KIT to inhibition but that specific amino acid substitutions in the same codon may impart differential sensitivity to inhibitors. Because it is relatively easy to exclude an EST mutation by sequencing short stretches of tumor DNA and because many clinically available enzyme inhibitors are developed to inhibit the wild-type enzyme, the paradigm we are proposing that separates out mutations that activate by changing the primary structure of the enzymatic site may be useful in understanding mechanisms of drug resistance of other enzymes besides KIT and may help guide therapy in a number of neoplastic diseases.
Materials and methods
Cell lines
For these studies we used 2 subclones of the human mast cell leukemia line HMC1.16 One subclone, HMC1.1, expresses a valine to glycine substitution in codon 560 (Val560Gly) of the KIT intracellular juxtamembrane region. The other, HMC1.2, expresses the Val560Gly mutation and a second substitution, aspartate to valine in codon 816 (Asp816Val) in the kinase enzymatic site. These 2 mutations were originally found together in the HMC1 cell line and were shown to cause SCF-independent constitutive activation of KIT by Furitsu et al.4 For study of specific KIT mutations, COS cells were used for transient expression of human wild-type and mutated c-KIT complementary DNAs, as described previously.13
Inhibitors
KIT phosphorylation assay
Cells were serum-starved overnight, incubated with or without inhibitors for 1 hour and with or without SCF (200 μg/mL, 10 minutes), followed by immunoprecipitation of cell lysates with anti-KIT antibodies (generously provided by Dr Keith Langley of Amgen, Thousand Oaks, CA), fractionation of proteins by sodium dodecyl sulfate–polyacrylamide gel electrophoresis, and immunoblotting with antiphophotyrosine antibody (Upstate Biotechnology, Lake Placid, NY). Blots were stripped and reprobed with anti-KIT antibody as described previously.13
Cell growth and apoptosis assay
Cells cultured in the presence or absence of inhibitors were counted daily in a hemocytometer using trypan blue exclusion. Proliferation assay was repeated at least 3 times. Apoptosis was examined using DNA fragmentation assay. Briefly, cells were grown in the presence or absence of inhibitors, genomic DNA was isolated and separated by agarose gel electrophoresis, and DNA fragments were visualized under UV light by ethidium bromide staining.
Results
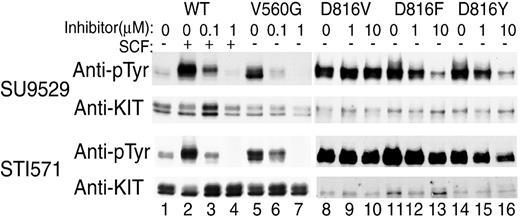
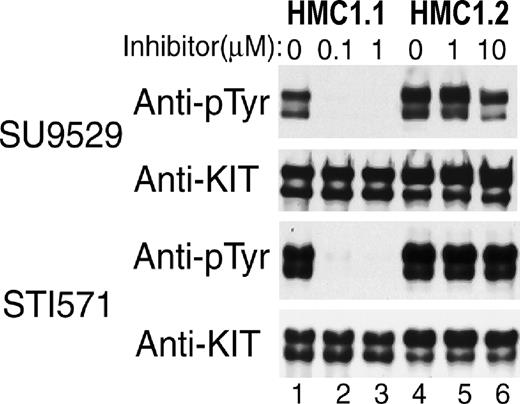
Both STI571 and SU9529 prevent the phosphorylation of wild-type KIT induced by its natural ligand SCF and inhibit SCF-independent constitutive phosphorylation of KIT caused by the Val560Gly juxtamembrane RT activating mutation at 0.1 to 1 μM (Figure1). In contrast, both inhibitors fail to inhibit SCF-independent constitutive phosphorylation of KIT containing the Asp816Val EST mutation associated with adult human mastocytosis even at 10 μM. Similarly, both drugs inhibit spontaneous KIT phosphorylation in the HMC1.1 subclone (Figure2, lanes 1-3), which expresses the Val560Gly activating mutation, but at 10 μM they fail to inhibit spontaneous phosphorylation of KIT in the HMC1.2 subclone, which expresses both the Val560Gly and Asp816Val activating mutations (Figure2, lanes 4-6). As would be predicted if the activating mutations caused the proliferation of the mast cells and were necessary for their survival, both inhibitors induce apoptosis of the HMC1.1 cells, causing the death of this line, but fail to kill the HMC1.2 cells (Figures3 and4).
Differential sensitivity of RT, EST, and differently substituted codon 816 mutant KIT to KIT inhibitors.
Antiphosphotyrosine blots of immunoprecipitated KIT expressed in COS cells show a low level of spontaneous phosphorylation of wild-type (WT) KIT (lane 1), which increases in response to SCF stimulation (lane 2). Both inhibitors at 0.1 to 1 μM prevent this ligand-induced phosphorylation (lanes 3-4). RT mutant KIT with Val560Gly substitution shows a high level of SCF-independent phosphorylation (lane 5), and the phosphorylation is inhibited by both inhibitors at physiologically achievable (0.1-1 μM) concentrations (lanes 6-7). The EST Asp816Val mutation commonly found in human mastocytosis also shows high spontaneous phosphorylation (lane 8) but is resistant to inhibition by either KIT inhibitor at 1 to 10 μM (lanes 9-10). Substitution of phenylalanine or tyrosine for aspartate 816, rarely found in human mastocytosis, also results in high spontaneous phosphorylation (lanes 11, 14), but these 2 mutant variants respond to the inhibitors at 1 to 10 μM (lanes 12-13, 15-16) unlike Asp816Val KIT. However, they are still an order of magnitude less sensitive than are RT mutant or wild-type KIT and are not valid therapeutic targets with currently available drugs.
Differential sensitivity of RT, EST, and differently substituted codon 816 mutant KIT to KIT inhibitors.
Antiphosphotyrosine blots of immunoprecipitated KIT expressed in COS cells show a low level of spontaneous phosphorylation of wild-type (WT) KIT (lane 1), which increases in response to SCF stimulation (lane 2). Both inhibitors at 0.1 to 1 μM prevent this ligand-induced phosphorylation (lanes 3-4). RT mutant KIT with Val560Gly substitution shows a high level of SCF-independent phosphorylation (lane 5), and the phosphorylation is inhibited by both inhibitors at physiologically achievable (0.1-1 μM) concentrations (lanes 6-7). The EST Asp816Val mutation commonly found in human mastocytosis also shows high spontaneous phosphorylation (lane 8) but is resistant to inhibition by either KIT inhibitor at 1 to 10 μM (lanes 9-10). Substitution of phenylalanine or tyrosine for aspartate 816, rarely found in human mastocytosis, also results in high spontaneous phosphorylation (lanes 11, 14), but these 2 mutant variants respond to the inhibitors at 1 to 10 μM (lanes 12-13, 15-16) unlike Asp816Val KIT. However, they are still an order of magnitude less sensitive than are RT mutant or wild-type KIT and are not valid therapeutic targets with currently available drugs.
Differential sensitivity of Val560Gly and Asp816Val mutant KIT in HMC1 subclones to KIT inhibitors.
Antiphosphotyrosine blots of immunoprecipitated KIT expressed in 2 HMC1 subclones show that spontaneous phosphorylation of KIT containing only the juxtamembrane RT Val560Gly mutation in the HMC1.1 clone is susceptible to inhibition by both inhibitors at 0.1 to 1 μM (lanes 1-3). In contrast, spontaneous phosphorylation of KIT with both the Val560Gly and the EST Asp816Val mutation in the HMC1.2 clone is resistant to both inhibitors at 1 to 10 μM (lanes 4-6).
Differential sensitivity of Val560Gly and Asp816Val mutant KIT in HMC1 subclones to KIT inhibitors.
Antiphosphotyrosine blots of immunoprecipitated KIT expressed in 2 HMC1 subclones show that spontaneous phosphorylation of KIT containing only the juxtamembrane RT Val560Gly mutation in the HMC1.1 clone is susceptible to inhibition by both inhibitors at 0.1 to 1 μM (lanes 1-3). In contrast, spontaneous phosphorylation of KIT with both the Val560Gly and the EST Asp816Val mutation in the HMC1.2 clone is resistant to both inhibitors at 1 to 10 μM (lanes 4-6).
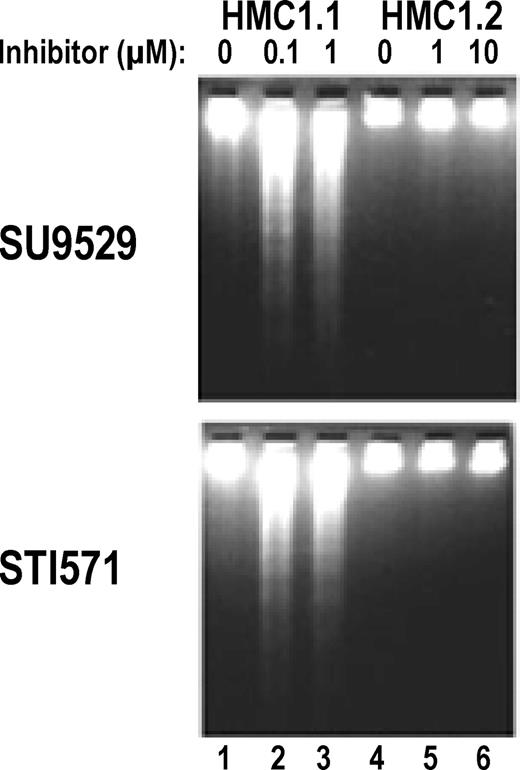
Induction of apoptosis in HMC1.1 but not in HMC1.2 cells by KIT inhibitors.
DNA fragmentation assay shows that the HMC1.1 clone expressing only the Val560Gly juxtamembrane RT mutation undergoes apoptosis, as indicated by formation of DNA “ladders” in the presence of the inhibitors at 0.1 to 1 μM (lanes 2-3). In contrast, the HMC1.2 clone expressing both the Val560Gly mutation and the Asp816Val EST mutation does not show any significant DNA “ladder” in the presence of the inhibitors at 1 to 10 μM (lanes 5-6).
Induction of apoptosis in HMC1.1 but not in HMC1.2 cells by KIT inhibitors.
DNA fragmentation assay shows that the HMC1.1 clone expressing only the Val560Gly juxtamembrane RT mutation undergoes apoptosis, as indicated by formation of DNA “ladders” in the presence of the inhibitors at 0.1 to 1 μM (lanes 2-3). In contrast, the HMC1.2 clone expressing both the Val560Gly mutation and the Asp816Val EST mutation does not show any significant DNA “ladder” in the presence of the inhibitors at 1 to 10 μM (lanes 5-6).
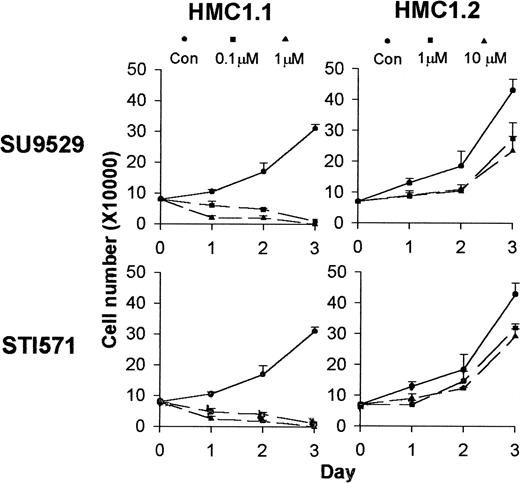
Differential effects of KIT inhibitors on the growth of HMC1.1 and HMC1.2 cells.
Cell proliferation assay shows that incubation of HMC1.1 cells with inhibitors at 0.1 to 1 μM for a 3-day period kills the cells, while treatment of the HMC1.2 cells with inhibitors at 1 to 10 μM only slightly inhibits growth of the cells.
Differential effects of KIT inhibitors on the growth of HMC1.1 and HMC1.2 cells.
Cell proliferation assay shows that incubation of HMC1.1 cells with inhibitors at 0.1 to 1 μM for a 3-day period kills the cells, while treatment of the HMC1.2 cells with inhibitors at 1 to 10 μM only slightly inhibits growth of the cells.
Two rare cases of human mastocytosis have been described in which other amino acids besides valine are substituted for Asp816 in the enzymatic site of the KIT kinase.7 These 2 variants, involving substitution of either tyrosine or phenylalanine, also cause SCF-independent constitutive phosphorylation of KIT. Interestingly, these 2 mutants are partially inhibited by the KIT inhibitors at 1 to 10 μM (Figure 1, lanes 11-16), concentrations that are totally ineffective against the most common Asp816Val-substituted mutant (Figure 1, lanes 8-10). Unfortunately, both of these variant EST mutant KITs are an order of magnitude less sensitive than the wild-type or RT mutant KIT, and neither of these inhibitors appear to have a high enough therapeutic index to be valid candidates for inhibiting the mutant kinases in a clinical trial. Nevertheless, these data do show that KIT kinases with different residue substitutions in codon 816 of the enzymatic site may show differences in susceptibility to specific pharmacologic inhibitors.
Discussion
The data presented here show unequivocally that different classes of activating KIT mutations respond differentially to KIT inhibitors. These data extend our previous studies of nonhuman mammalian KIT-activating mutations to the actual mutations found in various forms of human disease and support our proposals for classification of mutations as RT or EST mutations and for classification of human disease according to the type of the mutations expressed in specific tumors.10,11,14 15 Our current results also highlight the need to identify specific variants of mutant KIT expressed by individual patients when one contemplates rational therapy.
It appears likely that activating mutations affecting other enzymes may also be classified as regulatory or enzymatic site in type and that this paradigm may prove to be generally useful in predicting drug resistance and guiding therapy. A recent study by Gorre et al19 supports this prediction. In the study, the authors report that the development of resistance to STI571 in 6 of 9 patients with BCR-ABL–positive chronic myeloid leukemia was associated with acquisition of mutations that directly affected the active site of the enzyme, and resistance in the other 3 patients was associated with BCR-ABL gene amplification. We would classify the former as EST mutations and the gene amplification (as well as the original translocation forming the BCR-ABL fusion gene) as RT mutations or events. Gorre et al suggest that the patients with gene amplification might be susceptible to treatment with higher doses of STI571 but that those with active site mutations may require treatment with a different drug, a speculation with which we agree. Although their study may be viewed as retrospective in nature, the association of resistance in a previously sensitive tumor with the acquisition of an EST mutation supports the general clinical utility of the paradigm we are proposing.
The incidental finding that different amino acid substitutions in codon 816 of the enzymatic site give rise to differential sensitivity to drugs is not unexpected, but it represents the first documentation with human mutant activated KIT to support individualization of therapy based on the response of specific mutant proteins to specific drugs. Accordingly, our data suggest that despite the previously reported ability of the KIT inhibitor STI571 to kill an HMC1 line at 0.1 μM,18 currently available KIT inhibitors may be ineffective in treating human adult-type mastocytosis. On the other hand, neoplastic processes characterized by RT KIT-activating mutations, such as gastrointestinal stromal tumors, should be susceptible to inhibition by a relatively wide variety of inhibitors, including those that inhibit wild-type KIT. It has been our experience that different RT mutations, in a given species, show similar sensitivities regardless of the specific amino acid substitution10 (additional data not shown). This observation supports the concept that the enzymatic site in our proposed RT mutants does not differ significantly from the enzymatic site of wild-type KIT. It follows that a drug that is a “good fit” for the wild-type enzymatic site and is capable of sterically blocking the enzymatic reaction would be likely to be also effective against a RT mutant but would not necessarily be effective against an EST mutant. This concept may aid in identifying potentially clinically useful drugs.
The data presented also shed a unique perspective on the cause of mast cell neoplasms. The fact that the HMC1.1 and 1.2 cell lines are only known to differ by the presence or absence of the Asp816Val mutation makes them an ideal model for determining the role of KIT activation in the factor-independent growth and survival of these cells. The key observation is the fact that both drugs inhibit the RT activating KIT mutation, and both drugs are capable of killing the HMC1.1 cell line, which expresses only that mutation. However, neither of these drugs are capable of inhibiting the EST mutation found in the HMC1.2 clone, and neither are capable of inhibiting the growth and survival of that cell line. Together, these observations show that the ability to kill neoplastic mast cells expressing activated KIT is associated with the ability to inhibit the mutated, activated KIT rather than to inhibit some other unknown kinases. Thus, this finding strongly supports the hypothesis that it is the mutated, activated KIT molecule itself that is the cause of adult-type mastocytosis.
Supported by grants to the research of B.J.L. from the Leukemia Foundation of America, Translational Research Award; National Institutes of Health, RO1AR43356; and SUGEN, Sponsored Research Award.
This work was included in a presentation by C.A. et al at the 42nd Annual Meeting of the American Society of Hematology, December 1-5, 2000, San Fransicso, CA.
S.D. and G.M. are employed by a company or a competitor of a company whose product was studied in the present work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
B. Jack Longley, Dept of Dermatology, Section of Dermatopathology, College of Physicians and Surgeons, Columbia University, 630 West 168th St, New York, NY 10032; e-mail:jl691@columbia.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal