Abstract
Multiple myeloma (MM) is characterized by the accumulation of malignant plasma cells in the bone marrow caused primarily by failure of normal homeostatic mechanisms to prevent the expansion of postgerminal center plasma cells. We have examined the molecular mechanisms that promote the survival of MM cells and have identified a key role for myeloid cell factor–1 (Mcl-1), an antiapoptotic member of the Bcl-2 family. These experiments were initiated by the observation that MM cells were exquisitely sensitive to culture in the presence of actinomycin D: caspase activation occurred within 3 hours of treatment and cells were not protected by interleukin-6, the main MM cell growth and survival factor. Actinomycin D–induced apoptosis was blocked by proteasome inhibitors, suggesting that a labile protein was required for MM cell survival. Further analysis demonstrated that Mcl-1 was likely to be the labile factor governing MM cell survival. Mcl-1 protein levels decreased rapidly after culture in the presence of actinomycin D in concordance with effector caspase activation, but addition of proteasome inhibitors reversed the loss of Mcl-1 and maintained cell viability. The levels of other antiapoptotic proteins, including Bcl-2 and members of the inhibitors-of-apoptosis family, were unaffected by these interventions. Furthermore, Mcl-1 antisense oligonucleotides caused a rapid down-regulation of Mcl-1 protein levels and the coincident induction of apoptosis, whereas overexpression of Mcl-1 delayed actinomycin D–induced apoptosis with kinetics that correlated with expression levels of Mcl-1. These data indicate that Mcl-1 expression is required for the survival of MM cells and may represent an important target for future therapeutics.
Introduction
Multiple myeloma (MM) is a B-cell malignancy characterized by the accumulation of plasma cells with a low proliferation index and an extended life span.1,2 Despite improvements in event-free and long-term survival resulting from autologous stem cell transplantation, MM remains an incurable disease in the majority of patients.3,4 In the initial phases of this disease, the malignant plasma cells are dependent on the supportive environment of the bone marrow, which is at least partly responsible for the up-regulation of antiapoptotic factors in the tumor cells.5,6 Translocations and aneuploidy are detected in the earliest stage of the disease (monoclonal gammopathy of undetermined significance),7-9 and karyotypic abnormalities accumulate as the disease progresses.2,10 In the later stages of disease, genetic lesions result in the activation of oncogenes that promote cell growth (Ras) and the loss of tumor suppressors (p16Ink4a, p53), and the disease becomes bone marrow stroma–independent.2 The molecular basis for the poor prognosis conferred by other genetic lesions, especially deletions of chromosome 13q, remains to be elucidated.11
Interleukin-6 (IL-6) is important for the growth and survival12-14 of myeloma cells and increases resistance to a variety of therapeutic interventions.15-17 Bone marrow (BM) stromal cells provide IL-6 in a paracrine fashion, leading to activation of the Ras/mitogen-activated protein kinase (MAPK) and Jak/signal transducers and activators of transcription (STAT) pathways.2,18,19 The Ras/MAPK pathway is considered more important for MM cell proliferation20; however, it is likely that downstream effectors of Ras also play a role in cell survival.21 In some experimental systems, STAT3 has been shown to activate genes that promote MM cell survival, including Bcl-XL and myeloid cell factor–1(Mcl-1).22,23 Constitutive expression of STAT3 has been demonstrated in freshly isolated MM cells and may confer an important survival advantage in vivo.22
Expression of antiapoptotic members of the Bcl-2 family appears to be an important factor for the clinical resistance of MM. Five antiapoptotic Bcl-2 family members (Bcl-2, Bcl-XL, A1, Mcl-1, and Bcl-w) have been identified in mammalian cells.24-26 Bcl-2 has been found to be highly expressed in malignant plasma cells.27-29 Bcl-XL expression was detected more frequently in patients at relapse and was shown to correlate with resistance to chemotherapy.30,31 Mcl-1 is distinct from Bcl-2 and Bcl-XL in that it lacks a BH4 domain and is a large protein (40 kd as compared with 26 kd for Bcl-2) that encodes additional domains with Pro-Glu-Ser-Thr–like sequences (PEST) that could account for its short half-life (although recent data question the importance of the putative Pro-Glu-Ser-Thr sequences in regulating Mcl-1 turnover).32-34 Mcl-1 levels in MM cells were reported to be induced by IL-6 in a STAT3-dependent mechanism.23 The roles of A1 and Bcl-W in MM remain to be determined. Thus, MM cells express at least 3 antiapoptotic Bcl-2 family members, and much remains to be learned about the relative importance of each for survival under different conditions, such as growth in the presence or absence of IL-6 or in response to antineoplastic agents.
The studies presented in this manuscript implicate Mcl-1 as a critical survival factor for MM cells. This hypothesis was initiated by the observation that inhibition of transcription in MM cells led to a rapid activation of apoptosis pathways that could be prevented by the addition of proteasome inhibitors to the cultures. These findings suggested the presence of a labile protein in MM cells whose turnover was mediated by proteasomal degradation and whose continued expression in these cells was required for their survival. Molecular analyses indicated that Mcl-1 could be the labile survival factor targeted by actinomycin D, and further experiments demonstrated that inhibition of Mcl-1 expression by means of antisense oligonucleotides led to the rapid induction of apoptosis. Overexpression of Mcl-1 in MM cells delayed the activation of caspases in a manner consistent with the hypothesis that a threshold level of Mcl-1 is required for MM cell survival. These data demonstrate that Mcl-1 expression is critical for the survival of MM cells by preventing the activation of endogenous apoptotic pathways, and Mcl-1 may therefore play an important role in the clinical behavior of these malignant cells.
Materials and methods
Cell lines
The MM cell lines 8226, ARP1, and ANBL-6 were kindly provided by Dr Guido Tricot (Arkansas Cancer Research Center, Little Rock, AR) and Dr Diane Jelinek (Mayo Clinic, Rochester, MN). Cells were cultured in RPMI-1640 medium supplemented with 50 IU/mL penicillin G, 50 μg/mL streptomycin, 2 mM L-glutamine, 10% fetal bovine serum (FBS) (all from Life Technologies, Gaithersburg, MD), and 1 ng/mL recombinant human IL-6 (R&D Systems, Minneapolis, MN). In experiments performed in our laboratory and by others, these cell lines have demonstrated distinct patterns of resistance and cell-cycle checkpoint arrest in response to antineoplastic agents. The 8226 cell line encodes mutated K-Ras (G12A), is resistant to dexamethasone, and grows in the absence of IL-6. ANBL-6 encodes wild-type Ras and requires IL-6 for its growth and survival. ARP1 grows more slowly in the absence of IL-6; like the other lines, ARP1 has no functional p53 pathway. In some experiments, the proteasome inhibitors MG115 or MG132 (Calbiochem, La Jolla, CA) were added 30 minutes before addition of actinomycin D. The flavopiridol was obtained from the Developmental Therapeutics Program, Division of Cancer Treatment and Diagnosis, National Cancer Institute, National Institutes of Health (Bethesda, MD).
Terminal deoxynucleotidyl transferase-mediated deoxyuridine 5-triphosphate–biotin end labeling assays and Hoechst staining
Terminal deoxynucleotidyl transferase-mediated deoxyuridine 5-triphosphate–biotin end labeling (TUNEL) assays were performed by means of the ApopTag Plus Fluorescein in Situ Apoptosis Detection Kit according to the manufacturer's instructions (Intergen, Purchase, NY). Slides were washed in phosphate-buffered saline (PBS), counterstained with 1 μM Hoechst 33342 (Sigma Chemical, St Louis, MO), and mounted with the use of Vectashield Mounting Medium (Vector Laboratories, Burlingame, CA). Fluorescent micrographs were taken by means of a Zeiss (Thornwood, NY) Axiovert 10 microscope and digitally recorded with Oncor (Gaithersburg, MD) Imaging System Software.
Caspase assays
Cells were pelleted and lysed in 50 mM Hepes (pH 7.9), 100 mM NaCl, 10 mM EDTA, 1% Triton X-100, 4 mM NaPPi, 10 mM NaF, 2 mM NaVO4, 1 mM phenylmethyl sulfonyl fluoride (PMSF), 2 μg/mL aprotinin, and 2 μg/mL leupeptin. Lysates were clarified by centrifugation for 10 minutes at 12 000 rpm and 4°C, and extracts were stored at −80°C. Protein concentrations were determined by means of the BCA protein assay (Pierce, Rockford, IL). Caspase assays were performed in 96-well plates. Each well contained 200 μL caspase assay buffer (20 mM Hepes [pH 7.5], 10% glycerol, 2 mM dithiothreitol [DTT]) to which 10 μg cell extract and 10 μM Ac-DEVD-AMC fluorogenic substrate (PharMingen, San Diego, CA) were added. The reactions were incubated at 37°C for 2 hours. Measurement of the AMC cleavage product was performed by means of a CytoFluor II Microplate Fluorescence Reader (PerSeptive Biosystems, Bedford, MA) with an excitation wavelength of 360 nm and an emission wavelength of 460 nm. Experiments were performed in triplicate, and results were expressed as the mean ± SD.
Antibodies for Western blotting
The following antibodies (Abs) were used: anti–Mcl-1 (rabbit polyclonal Ab S-19; band size 40 kd) from Santa Cruz Biotechnology (Santa Cruz, CA); anti–Mcl-1 mouse monoclonal Ab, clone RC31 (NeoMarkers, Fremont, CA); antihuman Bcl-2 (26 kd) mouse monoclonal Ab clone 124 and anti-Bax (23 kd) rabbit polyclonal Ab (both from Upstate Biotechnology, Lake Placid, NY); anti–Bcl-XL (26 kd) rabbit polyclonal Ab (Transduction Laboratories, Lexington, KY); anti–Bag-1 (50-, 46-, 33-kd isoforms) monoclonal Abs (3.9F1E11 plus 3.10G3E2) (NeoMarkers); anti-XIAP (57 kd) mouse monoclonal Ab clone 48 (BD Biosciences, San Jose, CA); anti-HA monoclonal Ab HA-11 (BAbCO, Richmond, CA); anti-actin (42 kd) polyclonal Ab I-19 (Santa Cruz Biotechnology).
Western blot analysis
Cell lysates were prepared in modified RIPA buffer (50 mM Tris-HCl [pH 7.4], 1% NP-40, 0.25% Na-deoxycholate, 150 mM NaCl, 1 mM EDTA, 1 mM NaF, 1 mM Na3VO4, 1 mM PMSF, 5 μg/mL aprotinin, and 5 μg/mL leupeptin). Protein extracts (50 μg) were size-fractionated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to Immobilon-P membranes (Millipore, Bedford, MA). After transfer, filters were blocked in PBS/3% nonfat dry milk (PBS-MLK) and then incubated overnight at 4°C in PBS-MLK containing 1 μg/mL primary antibody. The filters were washed 3 times in PBS with 0.05% Tween 20 (PBST) and were incubated for 90 minutes at room temperature with secondary antibody (horseradish peroxidase conjugated) diluted 1:5000 in PBS-MLK. Filters were washed 3 times in PBST and rinsed with water, and the signal was detected by electrogenerated chemiluminescence (ECL) (Amersham Life Science, Arlington Heights, IL). Levels of Mcl-1 protein were subjected to quantitative analysis by means of a Personal Densitometer SI (Molecular Dynamics, Sunnyvale, CA) and ImageQuant software (Molecular Dynamics).
Antisense and inverted oligonucleotides
Chimeric phosphodiester/phosphorothioate oligodeoxynucleotides (ODNs) were synthesized by Midland Certified Reagent (Midland, TX). The sequence for the Mcl-1 antisense (AS) ODN was selected on the basis of previously reported data35 and corresponds to nucleotides 981 through 1000 of the Mcl-1 coding region. The ODNs contain a 5′-fluorescein moiety attached by phosphoramidite cyano-ethyl chemistry; this allows noninvasive determination of the efficiency of delivery into cells. To protect the ODN against nuclease-mediated degradation, 5 linkages at each end are phosphorothioate, with the central 9 linkages being phosphodiester. The sequence of the AS ODN is 5′ F-C/A/T/C/C/C-A-G-C-C-T-C-T-T-T/G/T/T/T/A 3′, where F represents fluorescein, virgules represent phosphorothioate linkages, and hyphens represent phosphodiester bonds. The sequence of the inverted ODN that is used as a negative control is 5′ F-A/T/T/T/G/T-T-T-C-T-C-C-G-A-C/C/C/T/A/C 3′.
Reversible cell permeabilization
In order to efficiently deliver ODN into MM cells, the streptolysin-O (SLO) (Sigma) technique was used to reversibly permeabilize cells as previously reported.35 SLO was dissolved in Mg2+/Ca2+–free PBS at 1000 U/mL followed by the addition of DTT to 20 mM and incubation at 37°C for 2 hours. Aliquots of the activated SLO were stored at −20°C until needed. Cells were washed and resuspended at 5 × 106cells per 200 μL in serum-free RPMI 1640. SLO was added at 12 U/106 cells in the presence or absence of 20 μM ODN and incubated at 37°C for 10 minutes. These conditions were established in preliminary experiments as being optimal for efficient ODN introduction into 8226 cells with minimal toxicity. Cells were resealed by the addition of 1 mL complete medium (RPMI/10% fetal bovine serum [FBS]) and incubation at 37°C for 30 minutes and were then transferred to flasks containing 9 mL complete medium. At the times indicated in the text, cells were removed for caspase assays, Western blots, and cytospins.
Lentivirus vector construction and virus production
The self-inactivating lentivirus vector pHR′-CMVgfpSIN was obtained from Dr Stephen Russell (Mayo Clinic) with permission from Dr Didier Trono (University of Geneva Medical School, Switzerland).36 The pHR′–Mcl-1 vector was derived by replacing the BamHI-XhoI DNA fragment encoding enhanced green fluorescent protein (EGFP) with a BamHI-SalI fragment encoding Mcl-1 (excised from pGEM–Mcl-1; kindly provided by Dr Hsin-Fang Yang-Yen, National Taiwan University Medical School, Taipei). The pHR′-HA–Mcl-1 vector was generated by introducing a BamHI-SalI fragment encoding the HA–Mcl-1 complementary DNA, which was generated by polymerase chain reaction using pGEM–Mcl-1 as template for the following primers: 5′-GGAGGATCCGGCAATGTACCCATACGATGTGCCAGACTATGCGTTTGGCCTCAAAAGAAA-3′, which encodes a BamHI site, and 5′-GAGGTCGACTCTATCTTATTAGATATGCCA-3′, which encodes a SalI site.
Lentiviruses pseudotyped with the VSV-G envelope protein were generated by calcium phosphate cotransfection of 293T cells (1.5 × 106 cells per 60-mm dish) with the following plasmids: 1.25 μg pMD.G (envelope plasmid), 3.75 μg packaging plasmid pCMVΔR8.91, and 3.75 μg vector plasmid.37 The precipitate was added to the cultures with gentle swirling. After 18 hours, cells were washed twice with serum-free Dulbecco modified Eagle medium (DMEM), and fresh DMEM/10% FBS was added. The viral supernatants were collected 48 hours later and either used immediately or frozen on dry ice and stored at −80°C.
Lentivirus infection of MM cells
The 8226 cells were cultured in 24-well plates with 1 × 105 cells per well in a volume of 0.5 mL RPMI/10% FBS with polybrene (8 μg/mL). We added 500 μL virus supernatant, and cells were infected for 24 hours at 37°C. Infected cells were expanded in complete medium plus IL-6 (1 ng/mL) without drug selection.
Isolation of CD138+ myeloma cells
Mononuclear cells were isolated by Ficoll-Hypaque centrifugation, washed extensively, and incubated with anti-CD138–coated microbeads as per the manufacturer's instructions (Miltenyi Biotec, Auburn, CA). CD138+ cells were purified by magnetic separation, washed, and then cultured under conditions described in the text. Purified cells were more than 95% positive for clonal kappa or lambda light chain.
Results
Evidence for a labile survival factor in MM
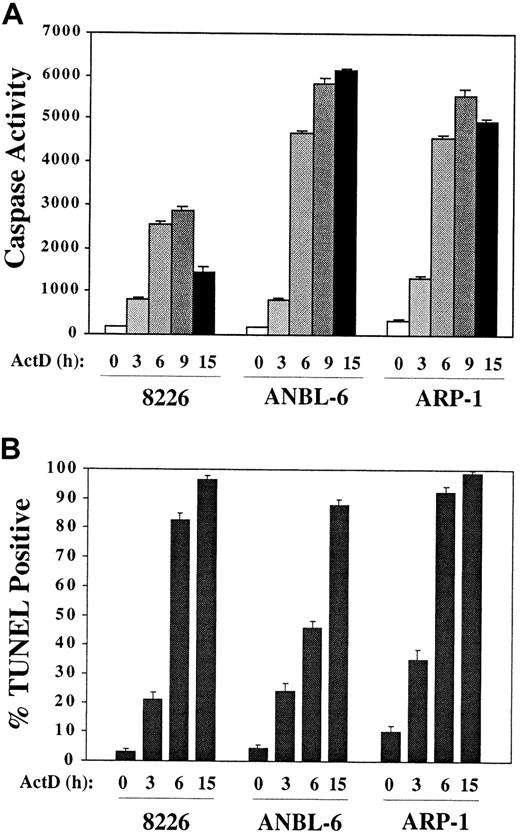
These studies began with the hypothesis that inhibiting transcription in IL-6–treated MM cell lines would block induction of survival pathways and render them more sensitive to treatment with other antineoplastic agents. We cultured 8226, ANBL-6, and ARP1 in complete medium plus IL-6 with or without the addition of actinomycin D. Surprisingly, caspase activity was induced within 3 hours in actinomycin D–containing cultures and reached high levels by 6 hours (Figure 1A). TUNEL positivity followed similar kinetics (Figure 1B). The IC50 for actinomycin D (ie, the dose that induced 50% apoptosis at 15 hours) was 0.1 μM. This very rapid induction of apoptosis was not observed with other chemotherapeutic agents commonly used to treat MM, including melphalan (10 μM), dexamethasone (1 μM), and doxorubicin (1 μM). Therefore, the mechanism of action of actinomycin D was examined in more detail.
Rapid induction of apoptosis by culture of MM cells in the presence of actinomycin D.
MM cell lines were cultured in RPMI/10% FBS plus IL-6 (1 ng/mL) and actinomycin D (1 μg/mL) for the times indicated. (A) Aliquots of cells were removed and protein extracts assayed for caspase activity by means of the fluorogenic substrate DEVD-AMC. (B) Cytospin preparations were prepared at the times indicated, and TUNEL assays were performed.
Rapid induction of apoptosis by culture of MM cells in the presence of actinomycin D.
MM cell lines were cultured in RPMI/10% FBS plus IL-6 (1 ng/mL) and actinomycin D (1 μg/mL) for the times indicated. (A) Aliquots of cells were removed and protein extracts assayed for caspase activity by means of the fluorogenic substrate DEVD-AMC. (B) Cytospin preparations were prepared at the times indicated, and TUNEL assays were performed.
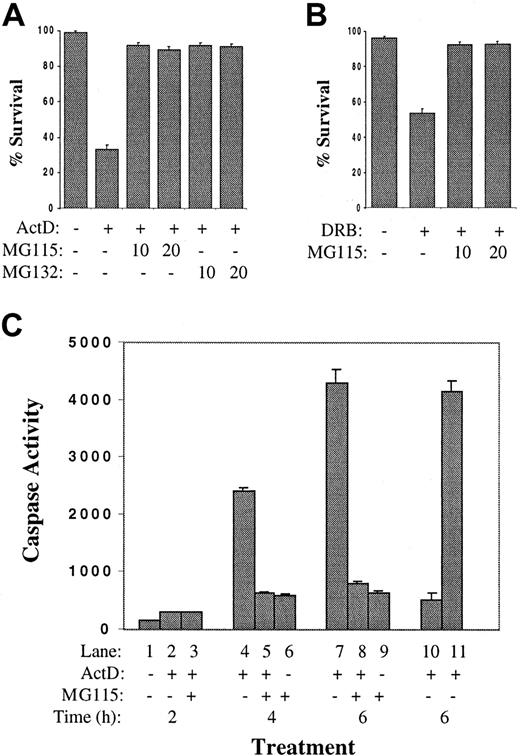
A hypothesis was formulated that MM cells express a short-lived survival factor and that inhibition of messenger RNA (mRNA) synthesis with actinomycin D would result in its degradation, promoting apoptosis of MM cells. Since the turnover of most short-lived proteins is mediated by the ubiquitin-proteasome pathway, experiments were designed to determine if proteasome inhibitors would protect actinomycin D–treated MM cells from the rapid induction of apoptosis. As shown in Figure 2A, pretreatment of 8226 cells with either of 2 proteasome inhibitors (MG115 or MG132) inhibited apoptosis after 6 hours of culture in actinomycin D. The 8226 cells were then cultured with a second inhibitor of RNA polymerase II, 5,6-dichloro-1-β-D-ribofuranosylbenzimidazole (DRB), a ribonucleoside analog that inhibits transcription by preventing the phosphorylation of the RNA polymerase II carboxy-terminal domain.38,39 In contrast to actinomycin D, DRB does not inhibit RNA polymerases I or III and does not alter the physical integrity of DNA.40Culture of 8226 cells in DRB resulted in approximately 50% apoptosis by 6 hours (Figure 2B); as with actinomycin D, this was blocked by MG115. These data were confirmed by caspase assays (Figure 2C). Significant effector caspase activity was induced by actinomycin D at 4 hours and 6 hours (lanes 4,7). This was inhibited by the addition of MG115 (lanes 5,8). Control experiments (lanes 10,11) demonstrate that the caspase activity detected in the protein extracts of apoptotic 8226 cells (from lane 7) is inhibited by the addition of the caspase inhibitor zVAD-fmk to the assay (lane 10), but not by the addition of MG115 (lane 11). Therefore, MG115 is not a caspase inhibitor per se, but prevents caspase activation in actinomycin D–treated MM cells indirectly through its inhibition of proteasome activity. These data support the hypothesis that MM cells encode a protein survival factor with a short half-life and that inhibition of its synthesis with subsequent rapid turnover is the apoptosis-inducing signal mediated by actinomycin D.
Proteasome inhibitors block actinomycin D–induced apoptosis of MM cell lines.
(A) The 8226 cells were cultured in complete medium plus IL-6 with or without the addition of actinomycin D (1 μg/mL). Indicated cultures were preincubated for 30 minutes with proteasome inhibitor MG115 or MG132 at 10 or 20 μM. After 6 hours, TUNEL assays were performed and the survival percentage was determined. (B) Cultures were prepared as in panel A, except that DRB (0.1 mM) was added instead of actinomycin D. (C) Actinomycin D–induced caspase activation is prevented by proteasome inhibitors. The 8226 cells were cultured in complete medium plus IL-6 with the addition of the proteasome inhibitor MG115 (10 μM) and /or actinomycin D, as indicated. Cells were cultured for 2 hours (lanes 1-3), 4 hours (lanes 4-6), or 6 hours (lanes 7-9), and caspase 3 activity was determined. In lane 10, an aliquot of protein extract from lane 7 was incubated with zVAD-fmk (25 μM final) immediately prior to caspase assay; caspase activity was inhibited as expected. In lane 11, MG115 (10 μM) was added to lane 7 extracts just prior to caspase assay to demonstrate that MG115 is not a direct inhibitor of caspase activity.
Proteasome inhibitors block actinomycin D–induced apoptosis of MM cell lines.
(A) The 8226 cells were cultured in complete medium plus IL-6 with or without the addition of actinomycin D (1 μg/mL). Indicated cultures were preincubated for 30 minutes with proteasome inhibitor MG115 or MG132 at 10 or 20 μM. After 6 hours, TUNEL assays were performed and the survival percentage was determined. (B) Cultures were prepared as in panel A, except that DRB (0.1 mM) was added instead of actinomycin D. (C) Actinomycin D–induced caspase activation is prevented by proteasome inhibitors. The 8226 cells were cultured in complete medium plus IL-6 with the addition of the proteasome inhibitor MG115 (10 μM) and /or actinomycin D, as indicated. Cells were cultured for 2 hours (lanes 1-3), 4 hours (lanes 4-6), or 6 hours (lanes 7-9), and caspase 3 activity was determined. In lane 10, an aliquot of protein extract from lane 7 was incubated with zVAD-fmk (25 μM final) immediately prior to caspase assay; caspase activity was inhibited as expected. In lane 11, MG115 (10 μM) was added to lane 7 extracts just prior to caspase assay to demonstrate that MG115 is not a direct inhibitor of caspase activity.
Identity of a putative labile survival factor
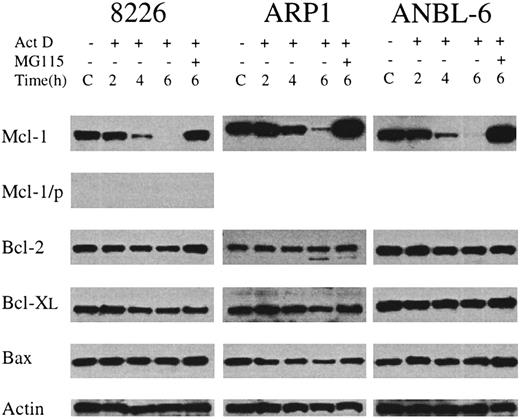
We endeavored to determine the identity of the survival factor by looking for antiapoptotic proteins whose levels of expression rapidly decline in the presence of actinomycin D, but are maintained by the addition of proteasome inhibitors to the cultures. To this end, we assessed the levels of Bcl-2, Bcl-XL, and Mcl-1 in MM cells cultured with IL-6 alone (control) or with actinomycin D (1 μg/mL) for 2 hours, 4 hours, and 6 hours. Additional cultures were preincubated with MG115 before adding actinomycin D. As shown in Figure3, there were no significant changes in the levels of prosurvival family members Bcl-2 and Bcl-XL. Levels of death-inducer Bax were also unchanged. However, the level of Mcl-1 decreased dramatically during culture in the presence of actinomycin D. The decline was especially rapid for 8226 and ANBL-6, with 2% and 3% of control expression at 4 hours, respectively; for all 3 cell lines, the levels of Mcl-1 remaining after 6 hours of culture in actinomycin D were less than 1% of the untreated control. Pretreatment with the proteasome inhibitor MG115 completely blocked actinomycin D–induced Mcl-1 degradation and resulted in Mcl-1 levels that were greater than control. This probably occurred owing to the rapid inhibition of proteasome activity by MG115 before actinomycin D could block new synthesis of Mcl-1. The turnover of Mcl-1 protein occurs with kinetics similar to that of the inhibition of synthesis and turnover of the Mcl-1 mRNA, although levels of the mRNA were not rescued by MG115 (data not shown). Taken together, these data indicate that culture of myeloma cells in the presence of actinomycin D results in rapid degradation of the Mcl-1 mRNA and protein and that protein levels are rescued by proteasome inhibitors, which also block actinomycin D–induced apoptosis.
Changes in the level of expression of Bcl-2 family members during treatment with actinomycin D.
MM cell lines were cultured in IL-6–containing medium without (C) or with the addition of actinomycin D (1 μg/mL) for 2 to 6 hours. In one culture from each cell line, MG115 (10 μM) was added 30 minutes before actinomycin D, and then cells were incubated for an additional 6 hours. At the indicated times, protein extracts were prepared, and equal amounts of total protein were subjected to Western blot analysis. The target of the primary antibody used in each case is indicated along the left margin. In the lane marked Mcl-1/p, the immunizing peptide was preincubated with the anti–Mcl-1 antibody prior to addition to the Western blot filter. Actin levels are included as a control for equal protein loading.
Changes in the level of expression of Bcl-2 family members during treatment with actinomycin D.
MM cell lines were cultured in IL-6–containing medium without (C) or with the addition of actinomycin D (1 μg/mL) for 2 to 6 hours. In one culture from each cell line, MG115 (10 μM) was added 30 minutes before actinomycin D, and then cells were incubated for an additional 6 hours. At the indicated times, protein extracts were prepared, and equal amounts of total protein were subjected to Western blot analysis. The target of the primary antibody used in each case is indicated along the left margin. In the lane marked Mcl-1/p, the immunizing peptide was preincubated with the anti–Mcl-1 antibody prior to addition to the Western blot filter. Actin levels are included as a control for equal protein loading.
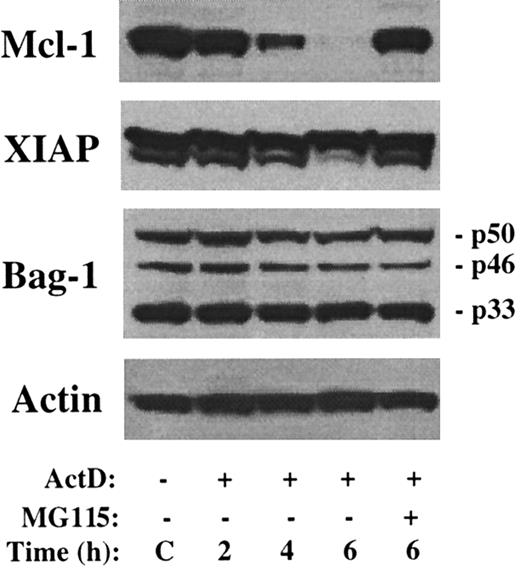
Downregulation of Mcl-1 was also recently implicated as a potential inducer of apoptosis in flavopiridol-treated CLL cells.41In these experiments, levels of the antiapoptotic proteins XIAP and Bag-1 were rapidly reduced by culture in flavopiridol, leading to speculation that these proteins also exhibited short half-lives. Therefore, the turnover of these proteins was examined following culture in the presence of actinomycin D. As shown in Figure4, there is minimal change in the level of XIAP or any of the 3 isoforms of Bag-1 during a 6-hour treatment with actinomycin D, whereas Mcl-1 levels rapidly decrease. Therefore, decreased levels of XIAP or Bag-1 do not account for the rapid induction of apoptosis by actinomycin D.
No change in level of XIAP or Bag-1 by actinomycin D.
The 8226 cells were cultured in the presence of actinomycin D and/or MG115 for the times indicated. Western blot analysis was performed with antibodies as indicated along the left margin. The sizes of the 3 Bag-1 variants are noted.
No change in level of XIAP or Bag-1 by actinomycin D.
The 8226 cells were cultured in the presence of actinomycin D and/or MG115 for the times indicated. Western blot analysis was performed with antibodies as indicated along the left margin. The sizes of the 3 Bag-1 variants are noted.
Inhibition of Mcl-1 by AS ODN
If inhibition of Mcl-1 synthesis is the critical mechanism leading to apoptosis of actinomycin D–treated cells, then downregulation of Mcl-1 levels by more specific mechanisms would be expected to induce apoptosis independent of other cellular insults. To test this hypothesis, an Mcl-1 AS ODN was introduced into 8226 cells according to the same methodology that had been shown to be highly effective in analysis of Mcl-1 function during phorbolmyristate acetate–induced differentiation of U937 cells.35 The Mcl-1 AS ODN used was a 20-mer containing 5 phosphorothioate-linked nucleotides at each end, with 9 central phosphodiester bonds. In addition, the ODN was 5′-tagged with fluorescein to allow simple determination of the efficacy of cellular uptake. Cells were transiently permeabilized with SLO, the ODN was introduced, and then cells were resealed by the addition of serum. Controls included 8226 cells treated with SLO only, and those treated with SLO plus a fluorescein-tagged ODN with an inverted nucleotide sequence (Inv ODN). In all experiments, permeabilization with or without addition of ODN resulted in less than 10% acute cell death, and more than 95% of cells were flourescein positive, indicating efficient uptake of the ODN. Representative data from a series of experiments are shown in Figure 5.
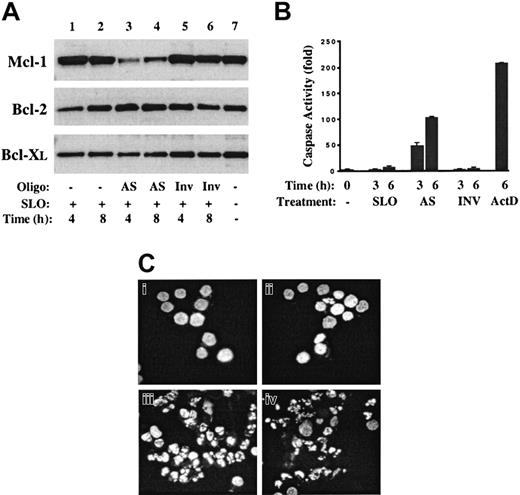
Mcl-1 AS ODNs inhibit Mcl-1 protein levels and induce apoptosis of MM cells.
Mcl-1 AS ODN or inverted control ODN (Inv) was introduced into 8226 cells by means of streptolysin-O (SLO). (A) At 4 and 8 hours after transfection, cell lysates were prepared, and Western blot analysis was performed with the use of antibodies directed against Mcl-1, Bcl-2, or Bcl-XL. Lane 7 contains extracts from 8226 cells grown under standard conditions (complete medium plus IL-6). (B) At 3 or 6 hours after the introduction of AS or Inv ODN, cells were harvested and cell lysates analyzed for effector caspase activity. Extracts in the lane marked ActD were from cells cultured for 6 hours in the presence of actinomycin D (1 μg/mL). (C) The 8226 cells were treated with SLO alone (Ci), SLO plus Inv ODN (Cii), SLO plus AS ODN (Ciii), or actinomycin D (1 μg/mL) (Civ), and cultured for an additional 6 hours. Cytospin preparations were fixed and then stained with Hoechst 33342. Magnification, × 400.
Mcl-1 AS ODNs inhibit Mcl-1 protein levels and induce apoptosis of MM cells.
Mcl-1 AS ODN or inverted control ODN (Inv) was introduced into 8226 cells by means of streptolysin-O (SLO). (A) At 4 and 8 hours after transfection, cell lysates were prepared, and Western blot analysis was performed with the use of antibodies directed against Mcl-1, Bcl-2, or Bcl-XL. Lane 7 contains extracts from 8226 cells grown under standard conditions (complete medium plus IL-6). (B) At 3 or 6 hours after the introduction of AS or Inv ODN, cells were harvested and cell lysates analyzed for effector caspase activity. Extracts in the lane marked ActD were from cells cultured for 6 hours in the presence of actinomycin D (1 μg/mL). (C) The 8226 cells were treated with SLO alone (Ci), SLO plus Inv ODN (Cii), SLO plus AS ODN (Ciii), or actinomycin D (1 μg/mL) (Civ), and cultured for an additional 6 hours. Cytospin preparations were fixed and then stained with Hoechst 33342. Magnification, × 400.
Introduction of Mcl-1 AS ODN resulted in a significant decrease in the levels of Mcl-1 protein at 4 and 8 hours (Figure 5A, lanes 3-4) corresponding to 10- and 7-fold reductions, respectively. In contrast, treatment with SLO alone (lanes 1-2) or SLO plus Inv ODN (lanes 5-6) had no effect. The specificity of the AS treatment is demonstrated by the lack of effect on the levels of Bcl-2 and Bcl-XL. To determine the effect of Mcl-1 AS treatment on apoptosis pathways, cell extracts were prepared from cells at 3 and 6 hours after treatment and caspase assays were performed. Introduction of Mcl-1 AS ODN resulted in a 50- to 100-fold induction of effector caspase activity at these times, while controls (SLO alone and Inv ODN) were negative. The induction of apoptosis by Mcl-1 AS ODN was confirmed by staining treated cells with Hoechst 33342 to determine the integrity of nuclear structure. As shown in Figure 6C, whereas SLO- and Inv ODN–treated cells demonstrate a normal chromatin pattern, cells treated with AS ODN for 6 hours have condensed chromatin and fragmented nuclei characteristic of apoptosis. An identical pattern is observed following 6 hours of culture in actinomycin D (Figure 6D). Taken together, these data indicate that introduction of Mcl-1 AS ODN into 8226 cells results in rapid inhibition of the synthesis of Mcl-1 with its subsequent degradation and that this alone is sufficient to induce apoptosis.
Overexpression of Mcl-1 confers resistance to actinomycin D.
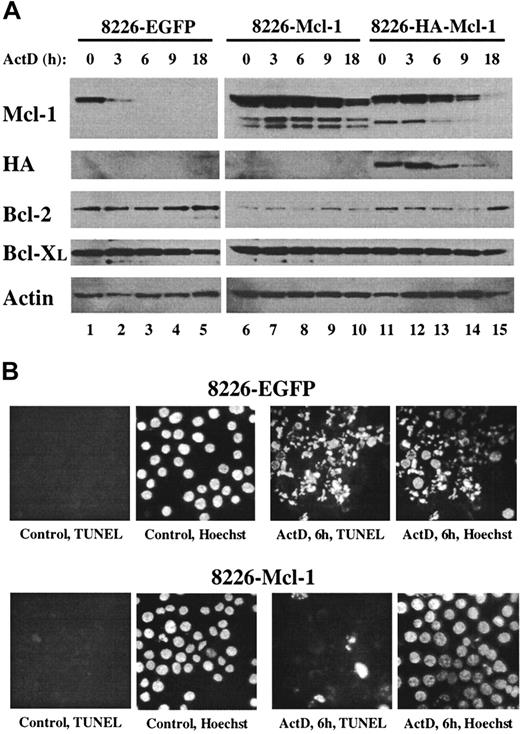
(A) Pools of 8226 cells were expanded after infection with recombinant lentiviruses expressing EGFP, Mcl-1, or HA–Mcl-1. Cells were cultured for the indicated times in medium containing actinomycin D, and protein extracts were subjected to Western blot analysis by means of antibodies with specificities as indicated along the left margin. (B) The 8226–EGFP or 8226–Mcl-1 pools were cultured in normal growth medium (control) or in medium containing actinomycin D for 6 hours. Cytospin preparations were assayed in situ by TUNEL and counterstained with Hoechst 33342. Identical fields are shown for each TUNEL/Hoechst pair (magnification, × 400).
Overexpression of Mcl-1 confers resistance to actinomycin D.
(A) Pools of 8226 cells were expanded after infection with recombinant lentiviruses expressing EGFP, Mcl-1, or HA–Mcl-1. Cells were cultured for the indicated times in medium containing actinomycin D, and protein extracts were subjected to Western blot analysis by means of antibodies with specificities as indicated along the left margin. (B) The 8226–EGFP or 8226–Mcl-1 pools were cultured in normal growth medium (control) or in medium containing actinomycin D for 6 hours. Cytospin preparations were assayed in situ by TUNEL and counterstained with Hoechst 33342. Identical fields are shown for each TUNEL/Hoechst pair (magnification, × 400).
Overexpression of Mcl-1 protects MM cells from actinomycin D–induced apoptosis
The preceding data suggested that MM cells must maintain a threshold level of Mcl-1 protein to prevent the spontaneous induction of apoptosis. This hypothesis predicts that MM cells engineered to overexpress Mcl-1 would tolerate culture in the presence of actinomycin D for longer periods before the induction of apoptosis. In Mcl-1–overexpressing cells, the time required for the proteasome-mediated degradation of Mcl-1 down to subthreshold levels would be expected to take longer and would therefore delay caspase activation. To test this, an Mcl-1 recombinant lentivirus was used to overexpress Mcl-1 or HA-tagged Mcl-1 in 8226 cells. As a control for the effects of virus infection, a lentivirus expressing EGFP was used. Pools of infected 8226 cells were generated, and Mcl-1 protein levels were determined under normal growth conditions and after culture in the presence of actinomycin D. The levels of Mcl-1 expression were significantly greater in 8226–Mcl-1 and 8226–HA–Mcl-1 pools as compared with 8226-EGFP cells (Figure 6A, untreated cells in lanes 1,6,11). The lower-molecular-weight bands observed in the lanes from Mcl-1–transfected 8226 cells may represent degradation products or alternatively spliced forms of Mcl-1. The levels of endogenous Mcl-1 decreased rapidly in the 8226-EGFP cells during culture in the presence of actinomycin D (lanes 2-5). In contrast, high levels of Mcl-1 were maintained in both Mcl-1–infected pools, with a decrease noted only after 9 and 18 hours of culture in actinomycin D. The higher level of recombinant Mcl-1 expression in the 8226–Mcl-1 pool as compared with the HA–Mcl-1 pool probably reflects a higher infection efficiency in the former. The expression of the HA-tagged Mcl-1 was confirmed by its slower migration with the use of anti–Mcl-1 antibody and by direct anti-HA analysis. As had been observed in previous cultures of actinomycin D–treated MM cells, the levels of Bcl-2, Bcl-XL, and actin did not change significantly during this experiment.
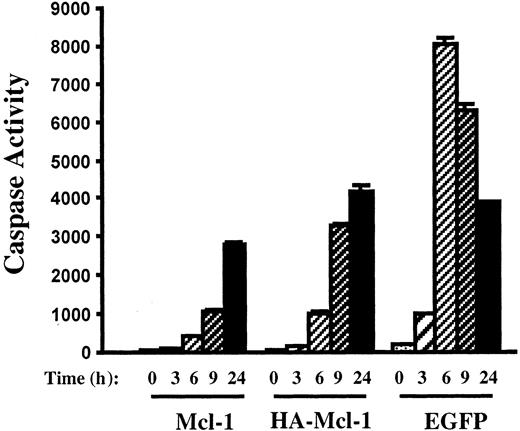
The effect of overexpression of Mcl-1 on actinomycin D–induced apoptosis was determined. After 6 hours of culture in actinomycin D, 8226-EGFP cells were nearly all apoptotic as determined by Hoechst staining and TUNEL assays (Figure 6B). In contrast, 8226–Mcl-1 cells had few TUNEL-positive cells, and nuclear stucture was intact. Caspase induction by actinomycin D also correlated inversely with the level of Mcl-1 expression (Figure 7). The 8226-EGFP cells demonstrated caspase activity beginning 3 hours after culture in actinomycin D, with high levels observed at 6 hours. In the 8226–Mcl-1 cells, caspase activity was minimally increased at 6 hours and continued to rise at 9 and 24 hours. HA–Mcl-1 cells exhibited a similar slow rise in caspase activation, but the delay was less pronounced than with 8226–Mcl-1 cells. Together, these data indicate that Mcl-1 overexpression significantly delays the induction of actinomycin D–induced apoptosis with kinetics that are consistent with a threshold level of Mcl-1 being critical for MM cell survival.
Inhibition of caspase activation by Mcl-1 overexpression.
Pools of 8226 cells were expanded after infection with lentiviruses encoding Mcl-1, HA–Mcl-1, or EGFP. After culture in the presence of actinomycin D for the indicated times, cell extracts were prepared and caspase activity was determined.
Inhibition of caspase activation by Mcl-1 overexpression.
Pools of 8226 cells were expanded after infection with lentiviruses encoding Mcl-1, HA–Mcl-1, or EGFP. After culture in the presence of actinomycin D for the indicated times, cell extracts were prepared and caspase activity was determined.
Downregulation of Mcl-1 in freshly isolated MM cells
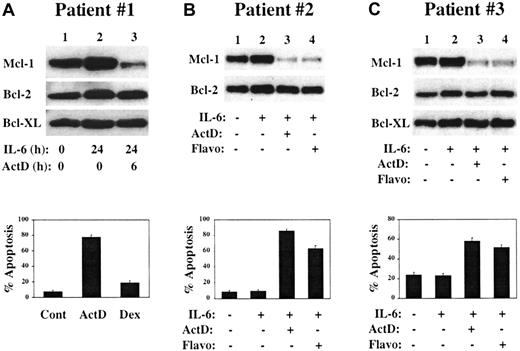
To determine the importance of Mcl-1 in freshly isolated MM cells, CD138+ MM cells were purified from the BM of patients with relapsed MM. For patient no. 1, cells were cultured with or without IL-6 for 24 hours, and actinomycin D was added to one culture for the final 6 hours of the culture period. As shown in Figure8A, Mcl-1 was highly expressed in the absence of IL-6 and its level appeared to increase after culture in IL-6 (lane 1 may be relatively underloaded, making this interpretation difficult). Addition of actinomycin D resulted in a rapid and significant decline in Mcl-1 level (lane 3), with no changes observed in levels of Bcl-2 or Bcl-XL. Cells from cultures grown in the presence of IL-6 for 24 hours followed by addition of actinomycin D or dexamethasone for 6 hours were subjected to TUNEL assay. As shown in the lower part of Figure 8A, significant apoptosis occurs after culture in actinomycin D but not in dexamethasone. Of note, this patient was treated in a Phase 1 study of actinomycin D given at 15 μg/kg/d intravenously for 5 consecutive days and achieved a partial clinical response with a decrease in M-component from 1.1 g/dL to barely detectable and a reduction of BM plasmablasts from 35% to 7%. Further data from this clinical trial will be needed to determine if these laboratory and clinical correlations will be more generalized.
Response of freshly isolated CD138+ cells to culture in actinomycin D or flavopiridol.
CD138+ cells were isolated from the BM of patients with relapsed MM. (A) Cells were cultured for the indicated times in the presence or absence of IL-6 (1 ng/mL) or actinomycin D (1 μg/mL), and protein extracts were subjected to Western blot analysis by means of antibodies with the specificities shown along the left margin. In the lower part of panel A, aliquots of CD138+ cells were cultured for 24 hours in medium containing IL-6 with the addition of actinomycin D (1 μg/mL) or dexamethasone (1 μM) for the final 6 hours. Cytospin preparations were analyzed by TUNEL with Hoechst 33342 counterstain. (B) CD138+ MM cells were cultured for 20 hours in the presence or absence of IL-6 (1 ng/mL) as indicated. Actinomycin D (1 μg/mL) or flavopiridol (200 nM) was added to the indicated cultures for the entire 20-hour incubation period, after which some cells were harvested for Western blot analysis while others were used in TUNEL assays (lower portion of the panel). (C) CD138+ BM cells from a third MM patient were treated as described in panel B, except that the cells were cultured for a 6-hour period.
Response of freshly isolated CD138+ cells to culture in actinomycin D or flavopiridol.
CD138+ cells were isolated from the BM of patients with relapsed MM. (A) Cells were cultured for the indicated times in the presence or absence of IL-6 (1 ng/mL) or actinomycin D (1 μg/mL), and protein extracts were subjected to Western blot analysis by means of antibodies with the specificities shown along the left margin. In the lower part of panel A, aliquots of CD138+ cells were cultured for 24 hours in medium containing IL-6 with the addition of actinomycin D (1 μg/mL) or dexamethasone (1 μM) for the final 6 hours. Cytospin preparations were analyzed by TUNEL with Hoechst 33342 counterstain. (B) CD138+ MM cells were cultured for 20 hours in the presence or absence of IL-6 (1 ng/mL) as indicated. Actinomycin D (1 μg/mL) or flavopiridol (200 nM) was added to the indicated cultures for the entire 20-hour incubation period, after which some cells were harvested for Western blot analysis while others were used in TUNEL assays (lower portion of the panel). (C) CD138+ BM cells from a third MM patient were treated as described in panel B, except that the cells were cultured for a 6-hour period.
CD138+ MM cells from patient no. 2 were cultured with or without IL-6 for 20 hours (Figure 8B). Actinomycin D or flavopiridol was added to IL-6–containing cultures for the entire 20-hour culture period. Flavopiridol is a cyclin-dependent kinase (CDK) inhibitor that has been shown to induce apoptosis and/or cell cycle arrest of a variety of tumor cell types and that is currently in Phase 2 clinical trials.42,43 Recent data suggest that by inhibiting CDK7/cyclin H and/or CDK9/cyclin T kinase activity, flavopiridol prevents the phosphorylation of the carboxy-terminal domain of RNA polymerase II, thus blocking transcription elongation.44-47 Flavopiridol may thus act in a manner analagous to DRB and, like actinomycin D, can broadly inhibit transcription of cellular mRNAs.48 Data using MM cell lines support this model (I.G., B.Z., and R.G.F., manuscript in preparation). When MM cells from patient no. 2 were cultured in actinomycin D or flavopiridol, a dramatic decrease in Mcl-1 levels was noted, along with the induction of apoptosis (lower part of Figure 8B). Similarly, when CD138+ MM cells from patient no. 3 were cultured under these conditions for 6 hours, both transcription inhibitors induced a rapid decline in Mcl-1 levels with induction of apoptosis of the MM cells (Figure 8C). These data are representative of our data with primary patient cultures and indicate that as with MM cell lines, freshly isolated MM cells require the expression of Mcl-1 for their survival.
Discussion
Understanding the molecular pathways that regulate tumor cell survival under normal growth conditions and in response to therapeutic interventions is critical for the design of targeted therapies for specific neoplasms.49 The experiments presented in this manuscript led to the hypothesis that Mcl-1 expression was critical for the survival of MM cells. Turnover of the Mcl-1 protein after brief periods of culture in actinomycin D (or other inhibitors of RNA polymerase II, such as DRB and flavopiridol), and inhibition of Mcl-1 degradation by proteasome inhibitors, correlated precisely with activation or inhibition of apoptosis in all MM cell lines examined, including multiple clinical isolates. Although other antiapoptotic proteins were shown not to be involved in actinomycin D–induced apoptosis, the nonspecific mechanism of action of transcription inhibitors required further experimental proof for the direct role of Mcl-1. AS ODNs were used to reduce Mcl-1 levels, and these experiments confirmed that specific targeting of Mcl-1 led to rapid caspase activation and nuclear destruction. Others have demonstrated that AS inhibition of Mcl-1expression during phorbol ester–induced myeloid differentiation of U937 cells resulted in apoptosis, and suggested that a threshold level of Mcl-1 expression was required to maintain viability.35 This model differs from ours in that U937 cells appear to require Mcl-1 expression during a brief period of differentiation during which they are susceptible to apoptosis. Previous data from this group showed that cultured neutrophils undergo spontaneous apoptosis accompanied by loss of Mcl-1 and that interventions to support neutrophil survival also maintained Mcl-1 levels.50 Our data are more similar to these latter findings in that expression of Mcl-1 appears to be required for survival of MM cells under normal growth conditions; inhibition of Mcl-1 with the use of AS ODN was sufficient to induce death in the absence of other noxious insults to the cell. The increased resistance to actinomycin D conferred by lentivirus-mediated Mcl-1 overexpression is consistent with the model in which a critical threshold level of Mcl-1 is required for viability of MM cells. The increased levels of Mcl-1 set the rheostat at a higher level with the result that after inhibition of Mcl-1 synthesis, the time for Mcl-1 degradation to the apoptotic threshold is increased by about 6 hours. The correlation between the levels of overexpression of Mcl-1 or HA–Mcl-1 with the degree of resistance to actinomycin D fits nicely with our model. It should be noted that others had previously demonstrated a requirement for macromolecular synthesis for the survival of hematopoietic tumor cells and had predicted the existence of a short-lived survival factor that inhibited constitutive apoptosis pathways in these cells.51 52 However, the identity of this factor was unknown.
The survival of all somatic cells requires the continuous input of intrinsic and extrinsic signals to suppress apoptosis.53Specific cell types have unique requirements for survival that depend on local environment, growth factors, and cell-cell and cell–extracellular matrix interactions. For MM, the bone marrow provides this specialized trophic microenvironment through direct contacts of MM with stromal cells and the production of a variety of cytokines, interleukins, and angiogenic factors.2,5,54-56We speculate, that in addition to the extrinsic signals that regulate MM growth and survival, Mcl-1 plays a critical cell-autonomous role in promoting survival. The extent to which extrinsic signals regulate Mcl-1 in MM remains unclear. It has been reported57 that IL-6 up-regulates Mcl-1; however, preliminary data show that in many cases, freshly isolated CD138+ MM cells express high levels of Mcl-1 levels that are maintained in an IL-6–independent manner for up to 24 hours of culture (data not shown). Analysis of Mcl-1 expression at different stages of differentiation in the LN showed that high levels were observed in the germinal center (where Bcl-2 levels were low), whereas long-lived mantle-zone B cells express Bcl-2 but not Mcl-1.58 Interestingly, the survival of peripheral blood B cells has been shown to correlate with Mcl-1 levels.59Since normal plasma cells undergo expansion and differentiation in the germinal center, it is of interest to consider whether they maintain expression of Mcl-1 during their finite life spans in the BM and whether downregulation of Mcl-1 regulates plasma cell death. We speculate that continuous expression of Mcl-1 after the exit of MM cells from the LN germinal center may play a central role in the survival and clonal expansion of these slowly proliferative cells. Critical issues remain to be resolved, including whether Mcl-1 is constitutively expressed in MM, the role the BM microenvironment plays in regulating its expression, and the role Mcl-1 plays in the life cycle of normal plasma cells.
Mcl-1 appears to have both overlapping and unique properties compared with other prosurvival Bcl-2 family members, and it remains unclear how similar the antiapoptotic functions of Mcl-1 are to those of Bcl-2 and Bcl-XL. The functions performed at the mitochondrial membrane may be the same for each, thereby suggesting that it is the sum of antiapoptotic activity that is important.24-26 Or perhaps Mcl-1 does have unique activities that cannot be replaced by other, related factors. Mcl-1 localizes to the mitochondrion as well as other intracellular membranes,60 and preliminary evidence suggests that it can regulate ion channels.61 Furthermore, Mcl-1 can associate with proapoptotic Bcl-2 family members (eg, Bax), although some data suggest that the pattern of association is distinct.62-64 Mcl-1 differs from Bcl-2 and Bcl-XL in structure,34 in the short half-life of its mRNA and protein,34,65 in the regulation of its promoter,23,66-68 and perhaps in its attenuated ability to protect cells from a variety of chemotherapeutic insults.1,64,69 Genetic manipulations in mice again demonstrate similarities and differences.70-72 Of special importance is the recent finding that Mcl-1 transgenic mice develop a spectrum of low- and high-grade lymphomas after a long latency period (longer than 1 year), suggesting the potential role of Mcl-1 as an oncogene in B-cell neoplasms.73
The genetic lesions implicated in tumor initiation and progression also burden tumor cells with physiologic alterations that create a heavy “apoptotic load.”53 Such derangements consist of the drive to proliferate aberrantly,74 genomic instability,75 loss of attachment to extracellular matrix,76 growth under conditions of hypoxia or growth factor deprivation,77 and others.78 To suppress these apoptotic signals below a critical threshold, tumor cells use multiple survival mechanisms: mutations in pathways that regulate p53 function,79 increased expression of Bcl-2 or IAP family members,80 loss of parts of the core apoptosis apparatus,81 and dysregulated signal transduction pathways.82,83 Given this tenuous balance between apoptosis and survival, cancer cells may be particularly susceptible to loss of a single survival mechanism. The data in this paper suggest that MM cells follow this paradigm. Thus, MM cell lines as well as de novo MM cells express multiple antiapoptotic proteins,2often do not encode functional p53,84 and frequently contain a dysregulated Akt pathway.85 86 Despite these protective mechanisms, our data demonstrate that inhibition of Mcl-1 alone induces rapid activation of apoptosis, even with continuous expression of other antiapoptotic proteins. The notion that a short-lived protein is critical for myeloma cell survival suggests that Mcl-1 may be an attractive therapeutic target.
We thank Dr Hsin-Fang Yang-Yen for providing the Mcl-1 complementary DNA and Drs Didier Trono and Stephen Russell for providing the lentivirus vector and packaging plasmids.
Supported by a Translation Research Grant from the Leukemia and Lymphoma Society of America (R.G.F.).
B.Z. and I.G. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Robert G. Fenton, University of Maryland Greenebaum Cancer Center, Bressler Research Building, 655 W Baltimore St, Rm 7-023, Baltimore, MD 21201; e-mail: rfent001@umaryland.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal