Abstract
Therapy-related acute myeloid leukemia (t-AML) in most cases develops after chemotherapy of other malignancies and shows characteristic chromosome aberrations. Two general types of t-AML have previously been identified. One type is observed after therapy with alkylating agents and characteristically presents as therapy-related myelodysplasia with deletions or loss of the long arms of chromosomes 5 and 7 or loss of the whole chromosomes. The other type is observed after therapy with topoisomerase II inhibitors and characteristically presents as overt t-AML with recurrent balanced chromosome aberrations. Recent research suggests that these 2 general types of t-AML can now be subdivided into at least 8 genetic pathways with a different etiology and different biologic characteristics.
Introduction
The genetic events leading to malignant transformation in most human tumors remain more or less concealed. A sequence of specific gene mutations has been identified in a few malignancies such as hereditary colon cancer and has in this disease been related to a stepwise transformation of normal epithelium to dysplastic cells, papillomas and, finally, localized or disseminated cancer.1
Recent research indicates that myelodysplasia (MDS), often progressing to acute myeloid leukemia (AML), may represent a similar although more complicated model for leukemic transformation. Most cases of MDS and AML arise de novo without verified leukemogenic exposures. However, in a subset of 10% to 20% of the patients, the disease arises after previous therapy, most often chemotherapy, of other malignancies. Because the risk of therapy-related MDS (t-MDS) and therapy-related AML (t-AML) after intensive chemotherapy is often increased 100 times or more,2 99 of 100 cases of t-MDS and t-AML observed in this type of patient must be considered as excess cases directly induced by the cytostatic agents.
Approximately 50% to 60% of patients with MDS and AML de novo present chromosome abnormalities of unknown etiology,3,4 and in cases of overt AML there is most often no knowledge about a preleukemic phase of MDS. However, in t-MDS and t-AML, 85% to 90% of the patients show chromosome aberrations generally of the same types as observed in MDS and AML de novo,5-7 and the cytogenetic changes can be related to previous exposure to different chemically well-defined cytostatic agents with a known mechanism of action. In cases of overt t-AML a preleukemic phase of t-MDS, if present, is most often diagnosed at an early stage of the disease because of a regular follow-up with laboratory investigations of patients treated intensively with chemotherapy.
Previous classification of t-MDS and t-AML
For 2 decades, deletions or loss of 5q and 7q or monosomy of these 2 chromosomes have been closely associated with previous therapy with alkylating agents and with presentation of the disease as t-MDS.5-7 The abnormalities of chromosomes 5 and 7 in t-MDS and t-AML have been classified together, and the abnormalities of chromosome 7, which are the most commonly observed, have for this reason sometimes been considered as the primary changes.7
Many years later, another general type of t-AML was observed. Abnormalities of the long arm of chromosome 11 were initially observed in cases of overt t-AML after therapy with the epipodophyllotoxins.8-10 Subsequently, a variety of balanced translocations involving chromosome bands 11q23 or 21q22 in t-MDS and t-AML were shown to be significantly related to previous therapy with the whole group of topoisomerase II inhibitors.11,12 Based on this experience, 2 main groups of t-MDS and t-AML were defined,13 and their different genetic pathways were discussed.7 Much new information has now been obtained for both general types of t-MDS and t-AML, and for this reason we wish to propose a revised model for the genetic pathways of these diseases.
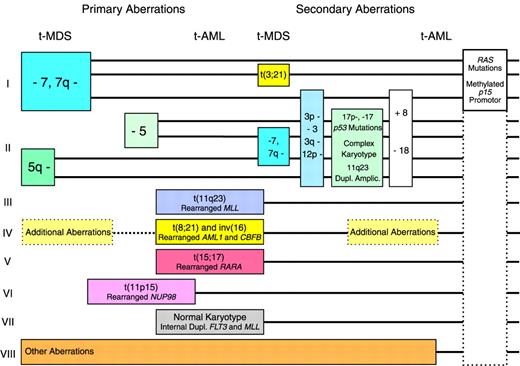
Revised model for cytogenetic and genetic pathways in t-MDS and t-AML
Pathway I in the revised model (Figure1) includes cases with deletions or loss of 7q or monosomy 7 but with normal chromosome 5. The abnormalities of chromosome 7 are the most commonly observed in t-MDS and t-AML following therapy with alkylating agents,5-7 but they are sometimes observed in only a subclone of cells or as evolutionary events during progression of the disease.14For this reason, other genetic abnormalities may be the important events in this pathway. Cases of t-MDS and t-AML with deletions or loss of 7q or monosomy 7 in many cases present additional chromosome aberrations, sometimes a balanced t(3;21)7 characteristic of t-MDS. Although the prognosis of patients belonging to pathway I is generally poor, some cases of t-MDS with monosomy 7 as the sole abnormality, only a modest cytopenia, and without an excess of blasts in the bone marrow may survive even for years on supportive therapy only.15 Mutations of the RASgenes16,17 and methylation of the of the p15gene promotor (D.H.C., J.P.-B., manuscript submitted) have been observed frequently in this pathway, but they probably do not represent primary genetic abnormalities in leukemic transformation. Mutations ofp53 are not very common.18 19
Pathway II includes cases of t-MDS and t-AML with deletions or loss of 5q or monosomy 5. These abnormalities have recently in most cases been unveiled by multicolor fluorescence in situ hybridization as unbalanced translocations to 5q.20,21 Defects of the long arm of chromosome 5 is the second most common cytogenetic abnormality of t-MDS and t-AML after therapy with alkylating agents. Abnormalities of chromosome 5 were in our series of patients always observed in all abnormal mitoses at diagnosis and were never seen as evolutionary events or in only a subclone.14 For this reason they were considered to represent more important changes in leukemogenesis than the defects of chromosome 7. Patients with abnormalities of chromosome 5 characteristically present many additional chromosome aberrations, including unidentified marker chromosomes, and sometimes deletions or loss of 7q or monosomy 7 also are observed. Mutations of the p53 gene are very common in this pathway. In 2 studies of 98 cases of t-MDS and t-AML, 20 of 26 patients with deletion or loss of 5q presented p53mutations, versus 7 of 72 patients with normal chromosome 518,19 (P < .001, χ2 test). The marker chromosomes in some cases have been identified as extensively deleted chromosome 5 or 720,21 and in other cases as derivative chromosomes with duplication or amplification of chromosome band 11q23, including the unrearranged MLLgene.22 Patients in pathway II have an extremely poor prognosis, particularly if p53 is mutated with loss of heterozygosity of the gene.19
Pathway III comprises patients with balanced translocations to chromosome band 11q23 characteristically presenting as overt t-AML. These translocations are often observed without additional chromosome abnormalities and are the most frequently observed aberrations in t-AML following therapy with the epipodophyllotoxins.8-10 Many patients observed with these abnormalities are children.9Patients with balanced translocations to 11q23 often obtain a complete remission following intensive antileukemic therapy, but the prognosis, even in children, is poor due to relapse of t-AML.23 In this pathway the MLL gene at 11q23 is chimerically rearranged with one of its many partner genes. An interesting example is the MLL-CBP fusion of the t(11;16) predominantly observed in t-AML.24 Mutations of p53 are infrequent in this and in the following pathways.19 25
Pathway IV in the former model was dominated by adult patients with overt t-AML and t(8;21).11,12 Subsequently, cases of t-AML with inv(16) also were observed related to therapy with topoisomerase II inhibitors,26-28 often an anthracycline. These patients present chimeric rearrangements of the core binding factor genesAML1(CBFA) at 21q22 and CBFB at 16q22, and they often have additional chromosome aberrations. Patients in this pathway show the best results following intensive antileukemic chemotherapy, and they often become long-term survivors.
Pathway V comprises cases of therapy-related acute promyelocytic leukemia with t(15;17) and chimeric rearrangement between thePML and the RARA genes. Such cases were first reported recurrently in Chinese patients treated with bimolane, a razoxane-related topoisomerase II inhibitor, for psoriasis.29 Subsequently, similar cases were observed after treatment with other topoisomerase II inhibitors, including anthracyclines, mitoxantrone,28,30-32 and radiotherapy only.33 Patients in this pathway often respond favorably to intensive antileukemic chemotherapy plus retinoic acid.
Pathway VI comprises more recently observed and rather rare cases of t-MDS and t-AML with different balanced translocations to chromosome band 11p15 and chimeric rearrangement between the NUP98 gene and its various partner genes.34-38 Almost half of the patients with these abnormalities developed leukemia after administration of topoisomerase II inhibitors, most often etoposide or anthracyclines or a combination of these drugs.
An important, still unsolved question is the extent to which each individual balanced chromosome aberration in pathways III to VI is associated with a specific topoisomerase II inhibitor. In a review of the literature of 442 cases of t-MDS and t-AML with balanced chromosome aberrations,33 it was shown that balanced translocations to 11q23, previous therapy with epipodophyllotoxins, and younger age were all significantly interrelated. When taking age into account in a multivariate analysis, there was no longer a significant association between previous therapy with epipodophyllotoxins and t-AML with translocations to 11q23. Thus, age and perhaps other factors such as race could explain the associations observed between specific drugs and specific cytogenetic subgroups of t-MDS and t-AML.
Pathway VII includes the subgroup of 10% to 15% of patients with t-MDS or t-AML and a normal karyotype, a subgroup well known for many years.5-7 As in AML de novo,39 it has been shown by multicolor fluorescence in situ hybridization that patients with t-MDS or t-AML and a normal karyotype only rarely present new cryptic cytogenetic rearrangements undetected by conventional G-banding.21 Unlike most patients with therapy-related leukemia and unbalanced chromosome aberrations, cases with a normal karyotype for unknown reasons predominantly present as overt t-AML.7 They have not been shown to be associated with any specific type of previous therapy, and they often respond to intensive antileukemic chemotherapy.7 Recently, we observed internal tandem duplications of the FLT3 or of the MLLgene in 4 of 6 patients with overt t-AML and a normal karyotype, whereas only 1 of another 76 patients with t-MDS or t-AML presented anMLL duplication.40 Both types of duplication were apparently not related to any specific type of previous therapy. Internal tandem duplications of the FLT3 and theMLL genes have previously been demonstrated mainly in cases of AML de novo with a normal karyotype,41-45 and theMLL duplications have been suggested to develop by endogenous recombinations at Alu repeats. In conclusion, patients with internal tandem duplications of FLT3 and MLL and perhaps also other patients in this pathway with a normal karyotype could represent sporadic and incidental cases of AML de novo unrelated to previous mutagenic exposures.
Pathway VIII includes patients with various chromosome aberrations uncharacteristic of t-MDS and t-AML. Some of these may likewise represent incidental cases of MDS and AML de novo; others may later turn out as recurrent but more rare cytogenetic abnormalities of t-MDS and t-AML.
Mouse models of human MDS and AML
Classification of t-MDS and t-AML in different genetic pathways may seem premature because major parts of leukemogenesis are still poorly understood. For instance, the specific genetic abnormalities directly cooperating with or predisposing to deletions or loss of the long arms of chromosomes 5 and 7 are still completely unknown. As far as the recurrent balanced chromosome aberrations in pathways III to V are concerned, recently developed mouse models support that their chimeric fusion genes are directly involved in leukemogenesis.46-56 It has been possible by various techniques to introduce these genes with appropriate promoters in mouse bone marrow precursors, and in many instances this resulted in the development of leukemia in a high percentage of the animals. Although the technique used has varied from study to study, and although caution must be taken in comparing these highly experimental models with human leukemia,57 at least 3 important observations have been made.
First, leukemias in mice develop after a latent period that sometimes is rather long, indicating that additional genetic events are required for leukemic transformation. Second, the phenotype of the leukemias observed in mice is sometimes very similar to its human counterpart. Thus, introduction of the PML-RARA fusion gene in mouse bone marrow progenitors resulted in the development of retinoic acid–sensitive promyelocytic leukemia,46-49 and introduction of 2 different MLL fusion genes resulted in the development of leukemias with monocytic differentiation or myelomonocytic leukemias.50,51 Finally, the potential to induce leukemias in mice seems to vary between the different chimeric genes. Thus, the AML1-MDS1-EVI1 fusion gene, primarily observed in the t(3;21) at blastic transformation of chronic myeloid leukemia and subsequently in t-MDS, often induced leukemia,52 whereas another 2 rearrangements of core binding factor genes, the AML1-ETO from the t(8;21)53,54 and the CBFB-MYH11 from the inv(16),55,56 so far have been shown only to induce defective and dysplastic hematopoiesis or impaired neutrophil maturation. The transforming potential of these transcripts has been suggested as more restricted,54 and additional exposure to a single sublethal dose of N-ethyl-N-nitrosourea was in one of these studies required for subsequent development of overt leukemia.56 The persistence of the AML1-ETOfusion transcript in bone marrow cells from most patients in long-term remission after intensive chemotherapy or even bone marrow transplantation of AML with t(8;21)58-60 further supports that the transcript, even if detected in vivo in human bone marrow cells, is not firmly associated with the development of overt leukemia.
Conclusion
The proposed model of genetic pathways in t-MDS and t-AML relates 3 types of etiology—alkylating agents, topoisomerase II inhibitors, and spontaneous endogenous recombinations unrelated to exogenous mutagenic exposures—to a complicated framework of cytogenetic and genetic abnormalities and clinical characteristics of these diseases. The diversity of cytogenetic and genetic abnormalities of t-MDS and t-AML is remarkable. Whereas most of the recurrent chromosome aberrations now must be considered as identified, many invisible genetic changes still remain to be discovered. Such new abnormalities could result in a further revision of the model. So far, methylation of the p15 promotor is the only abnormality observed in a high percentage of patients with t-MDS and t-AML and in their de novo counterparts. New techniques such as the microarray equipment for studies of gene expression may facilitate detection of additional and more general genetic abnormalities of MDS and AML.
Supported by grants from the Danish Cancer Society, HS Forskningspulje 1997, and Anders Hasselbalch's Foundation.
© 2002 by the American Society of Hematology
References
Author notes
Jens Pedersen-Bjergaard, Cytogenetic Laboratory, Section 4052, Rigshospitalet, Blegdamsvej 9, 2100 Copenhagen Ø, Denmark.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal