Abstract
CD34+ hematopoietic stem cells are used clinically to support cytotoxic therapy, and recent studies raised hope that they could even serve as a cellular source for nonhematopoietic tissue engineering. Here, we examined in 18 volunteers the gene expressions of 1185 genes in highly enriched bone marrow CD34+(BM-CD34+) or granulocyte–colony-stimulating factor–mobilized peripheral blood CD34+(PB-CD34+) cells by means of cDNA array technology to identify molecular causes underlying the functional differences between circulating and sedentary hematopoietic stem and progenitor cells. In total, 65 genes were significantly differentially expressed. Greater cell cycle and DNA synthesis activity of BM-CD34+ than PB-CD34+ cells were reflected by the 2- to 5-fold higher expression of 9 genes involved in cell cycle progression, 11 genes regulating DNA synthesis, and cell cycle–initiating transcription factor E2F-1. Conversely, 9 other transcription factors, including the differentiation blocking GATA2 and N-myc, were expressed 2 to 3 times higher in PB-CD34+ cells than in BM-CD34+cells. Expression of 5 apoptosis driving genes was also 2 to 3 times greater in PB-CD34+ cells, reflecting a higher apoptotic activity. In summary, our study provides a gene expression profile of primary human CD34+ hematopoietic cells of the blood and marrow. Our data molecularly confirm and explain the finding that CD34+ cells residing in the bone marrow cycle more rapidly, whereas circulating CD34+ cells consist of a higher number of quiescent stem and progenitor cells. Moreover, our data provide novel molecular insight into stem cell physiology.
Introduction
Hematopoiesis is characterized by a well-regulated balance of self-renewal, differentiation, and migration of pluripotent CD34+ hematopoietic stem cells. CD34+ cells are capable of maintaining a life-long supply of the entire spectrum of hematopoietic cells dependent on systemic needs and are clinically used to support high-dose chemotherapy and radiotherapy in patients with malignant diseases. During steady-state hematopoiesis, small amounts of CD34+ cells are present in the peripheral blood, suggesting a continuous migration of hematopoietic stem cells between the bone marrow and other organs.1 Reconstitution after chemotherapy or the application of hematopoietic growth factors such as granulocyte–colony-stimulating factor (G-CSF) greatly facilitates the mobilization of CD34+ cells into the peripheral blood.2 Still, the exact mechanisms of stem cell mobilization and homing are not yet clear. Apparently, cytokines and adhesion molecules play an important role by mediating adhesive interactions of CD34+ cells with cellular and matrix components of the bone marrow stroma.1,3 Recent studies suggest that hematopoietic stem cells are, in addition to their hematopoietic potential, able to differentiate into nonhematopoietic cell types such as hepatocytes, cardiac muscle cells, vascular endothelial cells, and even neuronal cells.4-10 In the future, hematopoietic stem cells might serve as a therapeutic tool for the treatment of various degenerative disorders, going far beyond the actual employment as a means to reconstitute hematopoiesis and to develop immunotherapeutic effects after high-dose chemotherapy. It is still unclear which cellular subset of hematopoietic cells has the potential to trans-differentiate or which contains the claimed multipotent or even omnipotent progenitors. Possibly, CD34+cells or a subset of them represent these stem cells and are the cellular source of regeneration of various tissues. The molecular phenotype of CD34+ stem cells, the regulatory mechanisms directing their mode of living, and the molecular signals mediating mobilization, migration, and differentiation are only partially understood. A powerful tool to broaden the knowledge of molecular physiology of hematopoietic stem and progenitor cells is gene expression profiling by means of cDNA array technology. Numerous studies examined gene expression of human CD34+ cells, in most cases focusing on a limited number of genes that are hematologically interesting. Recently, Phillips et al11measured for the first time a gene expression profile of hematopoietic stem cells in mice. However, hematopoiesis of humans considerably differs from hematopoiesis in mice.12 In this study, we measured gene expression profiles of primary human CD34+stem cells using cDNA arrays covering 1185 genes to describe a broad molecular phenotype. CD34+ cells were derived from steady-state bone marrow or from peripheral blood after mobilization with G-CSF to compare these 2 sources of CD34+ cells and to figure out transcriptional changes that play a role in the regulation of self-renewal, differentiation, mobilization, and migration of human hematopoietic CD34+ stem and progenitor cells.
Materials and methods
Cells
After informed consent we obtained human peripheral blood mononuclear cells (PB-MNCs) from 10 healthy volunteers who donated hematopoietic stem cells for allogeneic transplantation. The donors received 12 μg human recombinant G-CSF per kilogram body weight for 4 to 5 days. Afterward cells were harvested using the CobeSpectra Apheresis System (Gambro BCT, Planegg-Martinsried, Germany). Donors mobilized between 5 × 106 and 18 × 106CD34+ cells per kilogram body weight, as determined by flow cytometry. Bone marrow mononuclear cells (BM-MNCs) were obtained from 6 healthy volunteers by bone marrow puncture. Two other bone marrow samples were derived from patients with nonhematologic malignancies in complete remission without bone marrow involvement. We purified PB-MNCs and BM-MNCs by density centrifugation using the lymphocyte separation medium Lymphoprep (Nycomed Pharma, Oslo, Norway) as previously described.13
Double immunomagnetic selection of CD34+ cells
CD34+ cells were positively selected using the midi-MACS immunomagnetic separation system (Miltenyi Biotec, Bergisch Gladbach, Germany) as previously described.14 In brief, 108 mononuclear cells were suspended in 300 μL sorting buffer (1× phosphate-buffered saline (PBS) supplemented with 2 mmol EDTA and 0.5% bovine serum albumin), 100 μL human immunoglobulin (Ig) FcR blocking antibody, and 100 μL monoclonal hapten-conjugated CD34 antibody (clone QBEND/10; Miltenyi Biotec) and were incubated for 15 minutes at 4°C. Cells were washed with 15 mL sorting buffer and centrifuged at 700g for 10 minutes. The pellet was resuspended in 400 μL sorting buffer, and cells were incubated with 100 μL paramagnetic microbeads conjugated to antihapten antibody (Miltenyi Biotec) per 108 cells for 15 minutes at 4°C. Cells were washed, resuspended in sorting buffer, passed through a 30-μm nylon mesh, and separated in a column exposed to the magnetic field of the MACS device. The column was washed 3 times with 3 mL sorting buffer and removed from the separator, and the retained cells were eluted with 5 mL sorting buffer using a plunger. Eluted cells were subjected to a second round of separation.
Immunofluorescence staining and flow cytometry
Immunomagnetically selected cells were stained with fluorescein isothiocyanate (FITC)– or phycoerythrin (PE)–conjugated monoclonal antibodies (mAbs) in 50 μL PBS at 4°C for 30 minutes. The following mAbs were used: CD34-FITC (clone 8G12; Becton Dickinson, Heidelberg, Germany), thrombin receptor-PE (clone SPAN12; Immunotech, Marseille, France), CD44-FITC (clone G44-26; PharMingen, Heidelberg, Germany), CD114-PE (clone LMM741; PharMingen), CD126-PE (clone M91; Immunotech). Isotype-identical monoclonal antibodies served as controls (IgG1 and IgG2b FITC/PE-conjugated). After antibody staining, cells were washed and suspended in PBS buffer. They were analyzed using a Becton Dickinson FACScan with a 2-W argon ion laser. Fluorescence was measured using 530/15 nm (FITC) and 575/36 nm (PE) bandpass filters. Data were analyzed using CellQuest software (Becton Dickinson) after gating on viable cells. Purity of separated CD34+ cells ranged between 90% and 99%. Expression levels of thrombin receptor, CD44, CD114, and CD126 were quantified in 3 BM-CD34+ and 3 PB-CD34+ samples using mean fluorescence intensity.
RNA isolation, reverse transcription, and hybridization to nylon cDNA arrays
Isolation of total RNA from 2 × 105 to 1 × 107 CD34+ cells was performed with the RNeasy Mini Kit (Qiagen AG, Hilden, Germany) according to the manufacturer's instructions. RNA yield and purity were measured photometrically. RNA samples were dissolved in 2 μL RNase free water. Atlas Human 1.2 I arrays (BD Biosciences Clontech, Heidelberg, Germany) were used for hybridization experiments. RNA amounts ranging between 600 ng and 5 μg were reversely transcribed with a gene-specific primer mix (CDS primer mix) and radioactively labeled with32P according to a modified protocol of the manufacturer's instructions. In brief, we used 35 μCi (1.295 MBq) α–32P-dATP per reaction and performed reverse transcription with 1.5 μL CDS primer mix instead of 1 μL per sample to increase sensitivity. The CDS primer mix was provided by the manufacturer, and it contained primers specific for the Atlas Human 1.2 I arrays. In contrast to the use of random hexamers, the gene-specific primer mix ensured that only cDNAs of those genes were synthesized that were present on our particular Atlas Human 1.2 I arrays. Sensitivity was increased because of decreased complexity of the resultant cDNA pool. We purified the labeled cDNA by column chromatography to remove unbound α–32P-dATP. Samples were denatured by incubation with 0.1 M NaOH for 20 minutes at 68°C and neutralized with 0.5 M NaH2PO4. Then human C0t-1-cDNA (Qiagen) was added to reduce unspecific cDNA binding. Furthermore, the cDNA arrays were prehybridized for 30 minutes with 0.5 mg salmon sperm DNA (Clontech) to reduce background binding to the nylon membrane. Radioactively labeled cDNA samples were then hybridized with the nylon cDNA arrays at 68°C and constant rotation in a hybridization oven (ThermoHybaid, Heidelberg, Germany) for 20 hours. After hybridization, high-stringency washing was performed according to the manufacturer's instructions. Washed arrays were enclosed in plastic wraps and exposed to PhosphorImager screens (FujiFilm, Düsseldorf, Germany) for 24 to 96 hours, depending on the activity of the sample. In this experimental setting, successful hybridizations could be performed starting with only 600 ng total RNA.
Quantification, normalization, and statistical analysis
The screens were scanned with a PhosphorImager (Fuji FLA-3000; FujiFilm) controlled by the BAS-Reader 3.01 software (Raytest, Straubenhardt, Germany). We measured dot intensities with the TINA 2.09g software (Raytest) and confirmed the results with ArrayVision software (Interfocus, Suffolk, United Kingdom). For comparison of signals from different experiments, we used a standard total intensity–based normalization strategy as previously described.15-18 In brief, after background subtraction, raw data were normalized by dividing each single-dot intensity by the median intensity of all dots of one experiment. Results were expressed as the ratio of background-corrected single-dot intensity to background-corrected median-dot intensity, which allowed comparison of independent samples differing in probe or hybridization strength. We compared the group of 8 BM-CD34+ cell samples with the group of 10 PB-CD34+ cell samples in search of statistically significant differential gene expression. For that purpose, we calculated for each gene the mean values of the BM and the PB groups, divided the BM mean by the PB mean, and obtained the ratio BM/PB. Values greater than 1 indicated a higher expression in bone marrow CD34+ cells, and values smaller than 1 indicated a higher expression in peripheral blood CD34+ cells. To check whether those differences were statistically significant, we used the Mann-Whitney U test, which is suitable for nonnormally distributed data. Only P values smaller than .01 were accepted as statistically significant.
Cluster analysis
Cluster analysis was performed using Cluster and TreeView software.19 Data of all 18 experiments were imported in the Cluster software, median centered, and normalized. Afterward we performed hierarchical array clustering using an average linkage-clustering algorithm. The results, including array clusters and dendrograms, were displayed and analyzed using TreeView software.
Quantitative real-time reverse transcription–polymerase chain reaction
Total RNA was reversely transcribed using random hexamer primers, as previously described.20 For detection and quantification of mRNA levels of proteinase 3, ubiquitin–protein ligase (UBCH10), death-associated protein kinase (DAPK1), and caspase 4, the LightCycler technology (Roche Molecular Biochemicals, Mannheim, Germany) was used.21 For relative quantification, GAPDH mRNA served as external control. PCR reactions were performed using the LightCycler–FastStart DNA Master SYBR Green kit (Roche Molecular Biochemicals). PCR was carried out in a final volume of 20 μL using 0.5 μM each primer, 4 mM MgCl2, 2 μL supplied enzyme mix containing the reaction buffer, FastStart Taq DNA polymerase, and DNA double-strand specific SYBR Green I dye for detection of PCR products. PCR was performed in a LightCycler with a 480-second preincubation at 95°C followed by 45 cycles of 15 seconds at 95°C, 5 seconds at 63°C, and 20 seconds at 72°C. PCR products were then subjected to melting curve analysis with the LightCycler system and to conventional agarose gel electrophoresis to rule out synthesis of unspecific products. The following primers were used: GAPDH-specific—forward primer 5′-TCCATGACAACTTTGGTATCG-3′, reverse primer 5′-CTAATTCTAGTGGGTCAAGATGTAGC-3′; proteinase 3-specific—forward primer 5′-ACCGTGGTCACCTTCTTCTG-3′, reverse primer 5′-CCAGTCCACGTAGAGGGCTA-3′; ubiquitin-protein ligase-specific—forward primer 5′-GATGTCTGGCGATAAAGGGATT-3′, reverse primer 5′-AGGGCAGACCACTTTTCCTT-3′; death-associated protein kinase 1-specific—forward primer 5′-CACCAAGGAAACGCTACCTCT-3′, reverse primer 5′-TGGAAACCATCTGCTATGCAC-3′; caspase 4-specific—forward primer 5′-GCATTTGCTACCAGACCAGAG-3′, reverse primer 5′-GCCTGGACAATGACCTT-3′. Primers were designed and synthesized by TIB MOLBIOL (Berlin, Germany). Crossing points (CPs) of real-time PCR curves were determined by the LightCycler 3.5 software using the second derivative maximum method. For each target the BM- and PB-derived samples were measured in the same LightCycler run to directly compare the results. The difference of the CPs (ΔCP) of target and GAPDH control reflected the amount of target mRNA in each sample. Assuming an amplification efficacy of 1.9, reduction of the ΔCP by 1 cycle means that the amount of target mRNA is 1.9-fold increased. Two independent samples of BM-CD34+cells and 2 independent samples of PB-CD34+ cells were examined in duplicate.
Apoptosis assay
The apoptotic activity of CD34+ cells was measured using an Annexin V binding assay. After immunomagnetic selection, CD34+ cells were stained with the Annexin V–Fluos Staining Kit (Roche Molecular Biochemicals) according to the manufacturer's instructions. In brief, cells were incubated with the staining solution containing Annexin V–fluorescein and propidium iodide for 15 minutes at room temperature. Afterward stained cells were subjected to flow cytometry. After gating on propidium iodide–negative cells, the proportion of Annexin V–positive cells was assessed using CellQuest software (Becton Dickinson). Three independent samples of BM-CD34+ cells and 3 independent samples of PB-CD34+ cells were examined in duplicate.
Results
In this study, we measured gene expression profiles of CD34+ stem and progenitor cells using nylon cDNA arrays covering 1185 single genes. Highly enriched CD34+ cells (purity between 90% and 99%, Figure 1) of 18 volunteers were derived either from steady- state bone marrow (n = 8) or from peripheral blood after mobilization with G-CSF (n = 10). After RNA extraction, we performed reverse transcription with gene-specific primers instead of random primers, which greatly improved sensitivity. This experimental setting rendered the measurement of gene expression profiles of CD34+ cells derived from single samples. Our proceeding avoided the pooling of samples from different donors or amplification of cDNA, resulting in increased reproducibility and reliability of the gene expression data. Beyond that, our data allowed comparison between gene expression of steady-state bone marrow CD34+ cells and circulating CD34+ cells. We now describe the obtained gene expression data by dividing genes into functional groups. A representative selection of genes showing very high or differential gene expression is displayed in Figure 2. The complete gene expression data are available in a searchable database at the home page of the German Resource Center for Genome Research athttp://www.rzpd.de.22
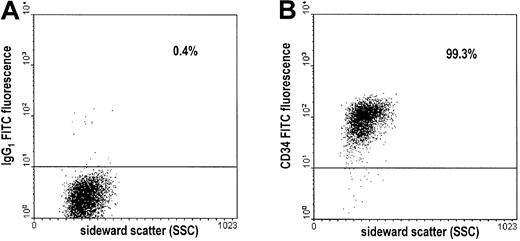
Assessment of the purity of CD34+ cells by flow cytometry.
After double immunomagnetic selection of CD34+ cells, cells were stained with FITC-conjugated antibodies and subjected to flow cytometry. Results of a representative experiment are shown. (A) Dot plot of cells stained with the FITC-conjugated, isotype-identical control antibody (IgG1 FITC). (B) Dot plot of cells stained with the FITC-conjugated CD34 antibody (CD34 FITC). Percentages of cells above the threshold intensity are indicated in both plots.
Assessment of the purity of CD34+ cells by flow cytometry.
After double immunomagnetic selection of CD34+ cells, cells were stained with FITC-conjugated antibodies and subjected to flow cytometry. Results of a representative experiment are shown. (A) Dot plot of cells stained with the FITC-conjugated, isotype-identical control antibody (IgG1 FITC). (B) Dot plot of cells stained with the FITC-conjugated CD34 antibody (CD34 FITC). Percentages of cells above the threshold intensity are indicated in both plots.
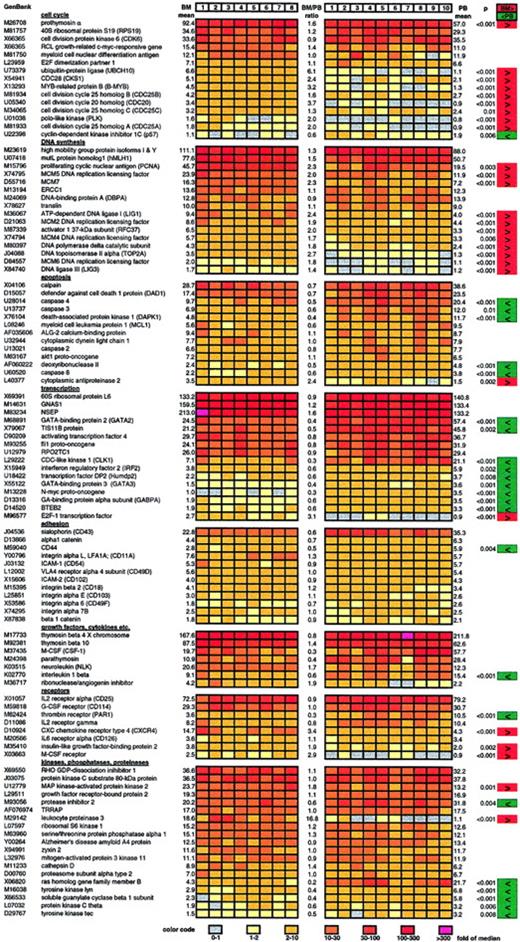
Representative selection of highly or differentially expressed genes in bone marrow and circulating CD34+ cells.
Results of the gene expression analysis of 8 samples of BM-CD34+ cells (left) and 10 samples of PB-CD34+ cells (right) are displayed. Relative gene expression (normalized to median) is color coded. The color code can be found at the bottom of the figure. The mean of the 8 bone marrow–derived samples (BM mean), the mean of the 10 peripheral blood–derived samples (PB mean), and the ratio of those 2 mean values (BM/PB ratio) are indicated for each gene. Genes that are expressed at a significantly higher level in BM-CD34+ cells than in PB-CD34+ cells are marked in red. Genes with lower expression in BM-CD34+ cells are marked in green.P values resulting from the Mann-Whitney U test are also indicated for all significantly differentially expressed genes.
Representative selection of highly or differentially expressed genes in bone marrow and circulating CD34+ cells.
Results of the gene expression analysis of 8 samples of BM-CD34+ cells (left) and 10 samples of PB-CD34+ cells (right) are displayed. Relative gene expression (normalized to median) is color coded. The color code can be found at the bottom of the figure. The mean of the 8 bone marrow–derived samples (BM mean), the mean of the 10 peripheral blood–derived samples (PB mean), and the ratio of those 2 mean values (BM/PB ratio) are indicated for each gene. Genes that are expressed at a significantly higher level in BM-CD34+ cells than in PB-CD34+ cells are marked in red. Genes with lower expression in BM-CD34+ cells are marked in green.P values resulting from the Mann-Whitney U test are also indicated for all significantly differentially expressed genes.
Gene expression profiles: bone marrow versus peripheral blood CD34+ cells
A considerable number of cell cycle–promoting genes was significantly higher expressed in bone marrow CD34+ cells than in circulating CD34+ cells, as assessed by examination of 92 genes involved in cell cycle regulation, among them cyclins, kinases, and kinase inhibitors. We found that the expression of 9 genes known to drive cell cycle progression was significantly elevated 2- to 6-fold in BM-CD34+ cells than in PB-CD34+ cells (Figure 2). On the other hand, expression of the cell cycle inhibitor p57 was 1.7-fold greater in PB-CD34+ cells than in BM-CD34+ cells.
Moreover, the expression of genes executing DNA synthesis was also distinctly greater in BM-CD34+ cells than in PB-CD34+ cells. In detail, the expression of 11 of 66 examined genes involved in DNA synthesis and replication was significantly 1.4- to 2.7-fold higher in BM-CD34+ cells than in PB-CD34+ cells (Figure 2).
Focusing on 79 apoptosis-related genes—such as death receptors and associated proteins, death kinases, caspases, and bcl family proteins—we found the expression of caspases 3, 4, and 8, death-associated protein kinase 1, and deoxyribonuclease II to be significantly 2- to 3-fold higher in G-CSF–mobilized PB-CD34+ cells, suggesting an increased apoptotic rate compared with steady- state BM-CD34+ cells (Figure 2). Consistent with that, expression of the antiapoptotic cytoplasmic antiproteinase 2 was 2.4-fold higher in BM-CD34+ cells.
We examined 134 transcription-related genes and measured the differential expression of 10 of them when comparing BM- and PB-CD34+ cells. Most were expressed 2- to 3-fold more often in PB-CD34+ cells than in BM-CD34+ cells (Figure 2). Only mRNA level of transcription factor E2F-1, which is known to initiate cell cycle entry, was significantly higher in BM-CD34+ cells.
Expression of 12 of 68 examined genes encoding for cell surface molecules involved in adhesion processes was detected in CD34+ cells (Figure 2). Only CD44 was significantly differentially expressed—2-fold higher in PB-CD34+cells than in BM-CD34+ cells.
Examining gene expression of 150 hormones, growth factors, cytokines, chemokines, and their receptors, we found 38 to be expressed. Macrophage-specific colony-stimulating factor receptor (M-CSF-R), CXC chemokine receptor type 4 (CXCR4), and insulinlike growth factor–binding protein 2 were expressed 2- to 3.4-fold higher in BM-CD34+ cells than in PB-CD34+ cells. On the other hand, the expression of thrombin receptor (PAR1) and interleukin (IL)-1β was 3.3-fold and 1.8-fold higher in PB-CD34+ cells.
We also examined differential gene expression of intracellular signal transducers, kinase network members, intracellular protein phosphatases, G-proteins, adenylate–guanylate cyclases, diesterases, and proteinases. We found that gene expression of ras homolog gene family member B, tyrosine kinase lyn, guanylate cyclase β 1, protein kinase C θ, and tyrosine kinase tec was elevated in PB-CD34+ cells in comparison with BM-CD34+cells. Conversely, the expression of MAP kinase–activated protein kinase 2 was greater in BM-CD34+ cells.
Expression of leukocyte proteinase 3 was strong in BM-CD34+cells, but it was not detectable at all in PB-CD34+ cells. On the contrary, the expression of a corresponding antagonist, protease inhibitor 2, was distinctly decreased in BM-CD34+ cells.
Cluster analysis
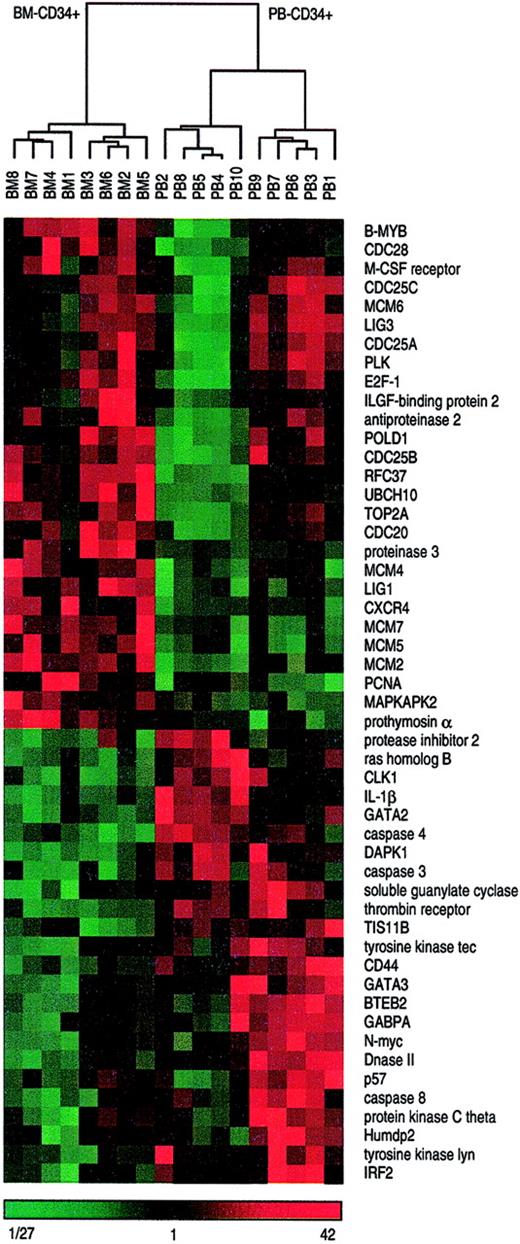
We performed hierarchical array clustering of the 18 CD34+ cell samples using an average linkage clustering algorithm that resulted in 2 clearly distinguishable array clusters, one containing the 8 BM-CD34+ samples and the other containing the 10 PB-CD34+ samples. Another array clustering was made focusing on 51 differentially expressed genes. This resulted in similar, but even more distinct, array clusters. In the BM cluster the samples were similar, whereas within the PB cluster 2 subgroups could be distinguished (Figure3). These 2 clusters did not correlate with age, sex, G-CSF dose, or mobilization kinetics of the donors.
Hierarchical cluster analysis distinguishes bone marrow– and peripheral blood–derived CD34+ cells.
All 18 CD34+ cells samples derived either from bone marrow or from peripheral blood were subjected to hierarchical cluster analysis on the basis of 51 differentially expressed genes displayed in Figure 2. Median-centered and normalized data are displayed using a color code, shown at the bottom of the figure. Red fields indicate higher values than median, green fields indicate lower values than median, and black fields indicate values that are equal to the median. The dendrogram visualizes the correlation of the different subgroups. Branch nodes connect closely related samples. The branch length indicates the degree of relationship. The shorter a branch is the more similar are the connected samples. Two main clusters are clearly distinguishable, the left one containing all 8 BM-CD34+cell samples and the right one containing all 10 PB-CD34+cell samples. Moreover, the right main branch distinctly separates 2 subclusters each with 5 PB-CD34+ cell samples.
Hierarchical cluster analysis distinguishes bone marrow– and peripheral blood–derived CD34+ cells.
All 18 CD34+ cells samples derived either from bone marrow or from peripheral blood were subjected to hierarchical cluster analysis on the basis of 51 differentially expressed genes displayed in Figure 2. Median-centered and normalized data are displayed using a color code, shown at the bottom of the figure. Red fields indicate higher values than median, green fields indicate lower values than median, and black fields indicate values that are equal to the median. The dendrogram visualizes the correlation of the different subgroups. Branch nodes connect closely related samples. The branch length indicates the degree of relationship. The shorter a branch is the more similar are the connected samples. Two main clusters are clearly distinguishable, the left one containing all 8 BM-CD34+cell samples and the right one containing all 10 PB-CD34+cell samples. Moreover, the right main branch distinctly separates 2 subclusters each with 5 PB-CD34+ cell samples.
Corroboration of cDNA array data
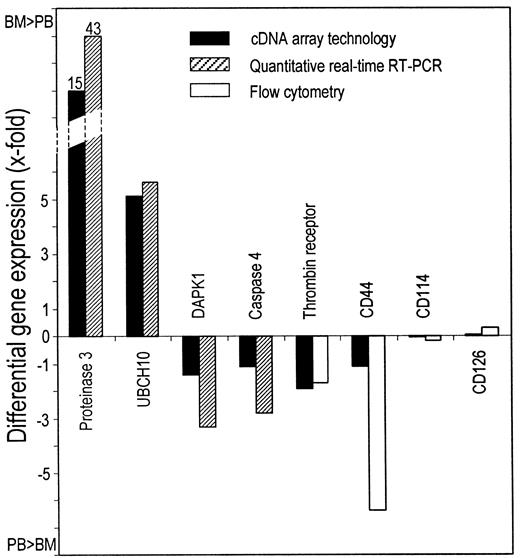
To corroborate the data obtained by cDNA array technology, we surveyed the expression of 8 genes by quantitative real-time reverse transcription–polymerase chain reaction (RT-PCR) or fluorescence-activated cell sorter analysis and performed an apoptosis assay to check for functional impact. Quantitative real-time RT-PCR confirmed that leukocyte proteinase 3 (LP3) and ubiquitin-protein ligase (UBCH10) were more markedly expressed in BM-CD34+cells than in PB-CD34+ cells, whereas DAPK1 and caspase 4 were expressed more in PB-CD34+ cells (Figure4). Assuming an amplification efficacy of 1.9, the expression of LP3 and UBCH10 was 44- and 6.6-fold higher in BM-CD34+ cells, and the expression of DAPK1 and caspase 4 was 4.3- and 3.8-fold higher in PB-CD34+ cells (Figure 5). We also examined differential gene expression by flow cytometry with fluorochrome-conjugated monoclonal antibodies directed against the 4 different surface molecules—thrombin receptor, CD44, CD114 (G-CSF receptor), and CD126 (IL-6 receptor α). Thrombin receptor and CD44 expression were 2.7-fold and 7.4-fold higher on PB-CD34+ cells (P = 0.03 and P = .02), whereas no significant difference could be detected for CD114 and CD126. All these findings were in line with the cDNA array data (Figure 5).
Corroboration of differential mRNA expression by quantitative real-time RT-PCR.
Real-time RT-PCR curves for 4 different genes (leukocyte proteinase 3, ubiquitin protein-ligase [UBCH10], DAPK1, and caspase 4) with the respective GAPDH controls are displayed. A representative BM-CD34+ sample is shown on the left, and a representative PB-CD34+ sample is shown on the right side. Analysis of each sample was performed in duplicate. The differences of the CPs (ΔCP) of target and GAPDH are indicated. The smaller the ΔCP, the higher the relative abundance of the target's mRNA.
Corroboration of differential mRNA expression by quantitative real-time RT-PCR.
Real-time RT-PCR curves for 4 different genes (leukocyte proteinase 3, ubiquitin protein-ligase [UBCH10], DAPK1, and caspase 4) with the respective GAPDH controls are displayed. A representative BM-CD34+ sample is shown on the left, and a representative PB-CD34+ sample is shown on the right side. Analysis of each sample was performed in duplicate. The differences of the CPs (ΔCP) of target and GAPDH are indicated. The smaller the ΔCP, the higher the relative abundance of the target's mRNA.
Comparison of differential gene expression measured by cDNA array technology and either quantitative real-time RT or flow cytometry.
Upward-pointing bars indicate a higher expression in BM-CD34+ than in PB-CD34+ cells. Downward-pointing bars indicate a higher expression in PB-CD34+ than in BM-CD34+ cells. Mean values of 2 (real-time RT-PCR) and 3 (flow cytometry) independent experiments in duplicate are indicated.
Comparison of differential gene expression measured by cDNA array technology and either quantitative real-time RT or flow cytometry.
Upward-pointing bars indicate a higher expression in BM-CD34+ than in PB-CD34+ cells. Downward-pointing bars indicate a higher expression in PB-CD34+ than in BM-CD34+ cells. Mean values of 2 (real-time RT-PCR) and 3 (flow cytometry) independent experiments in duplicate are indicated.
To check whether the higher expression of proapoptotic genes in PB-CD34+ cells had functional impact, we measured apoptosis using an Annexin V binding assay. Among the BM-CD34+ cells, a small fraction of 3.7% (± 0.7 SD) showed apoptotic activity, whereas 7.2% (± 1.7 SD) of the PB-CD34+ cells were apoptotic (data not shown). This difference, though small, was statistically significant (P = .03).
Discussion
Human CD34+ hematopoietic stem cells represent a heterogeneous cell population composed of early pluripotent hematopoietic stem cells and numerous lineage-committed hematopoietic progenitors in different developmental stages.23 CD34+ cells are mainly found in the bone marrow, but a small number of CD34+ cells appear in the peripheral blood.24 Under certain circumstances, such as reconstitution after cytotoxic chemotherapy or cytokine stimulation, the number of circulating CD34+ cells increases markedly. It is not surprising that CD34+ cells from bone marrow and peripheral blood, though they are developmentally and functionally closely related, must have different abilities to fulfill their different duties. However, the transcriptional changes in CD34+ cells dependent on their environment and different physiological tasks are unknown. Thus, comparative analysis of gene expression of CD34+ cells from the 2 compartments, bone marrow and peripheral blood, is an approach to determine molecular causes underlying their functional differences.
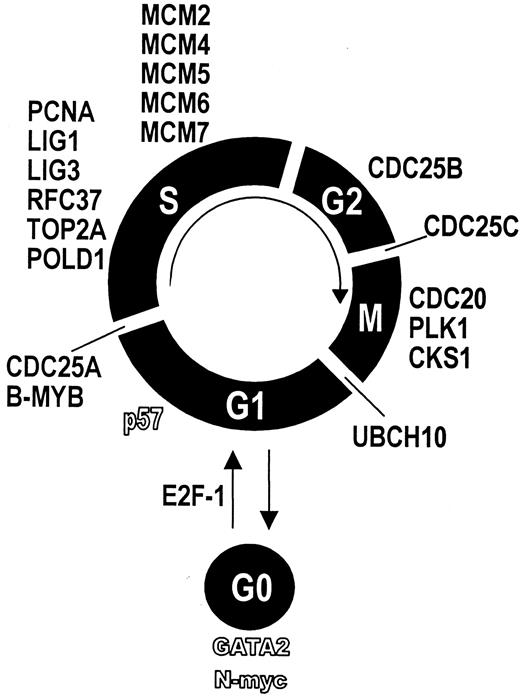
Applying cDNA array technology, we found that bone marrow CD34+ cells expressed 9 cell cycle driving genes and 11 genes required for DNA synthesis at distinctly higher levels than circulating CD34+ cells. This indicates a higher cycling activity of BM-CD34+ cells than PB-CD34+ cells. For each phase of the cell cycle, we found genes that were expressed significantly greater in BM-CD34+ cells (Figure6). For instance, CDC25A is necessary for G1-S transition by activating cyclin E-CDK2.25B-MYB is a transcription factor required for S-phase entry.26,27 The serine–threonine kinase PLK plays an important role in late G2 phase and during M phase. Furthermore, CDC20 is crucially involved in M-phase progression.28 Finally, ubiquitin-protein ligase (UBCH10) is essential for M-phase exit.29 The expression of genes that are involved in DNA synthesis and replication and, thus, necessary for the S phase of cell cycle was also higher in BM-CD34+ cells. Ligase 1, for example, is known to mediate DNA replication by interacting with proliferating cell nuclear antigen,30 both of which were expressed at a higher level in BM-CD34+ cells. Mini-chromosome maintenance proteins (MCM) also crucially take part in DNA replication.31 Interestingly, the cell cycle–initiating transcription factor E2F-1 showed greater expression in BM-CD34+ cells. E2F-1 is known to promote cell cycle progression through the activation of MCM genes.32 Thus, up-regulation of the E2F-1 transcription factor might be a molecular switch that initiates the proliferation of hematopoietic stem and progenitor cells. On the other hand, the cell cycle inhibitor p57 showed a lower expression in BM-CD34+ cells than PB-CD34 cells. In summary, these results can serve as a molecular explanation for the finding that BM-CD34+ cells are more rapidly cycling, whereas mobilized hematopoietic stem cells are more deeply arrested in G0.33 34
Differentially expressed genes and their relation to cell cycle.
Genes whose expression was higher in BM-CD34+ cells are displayed in bold font. Genes whose expression was lower in BM-CD34+ cells are displayed by hollow characters. Each gene is assigned to the respective cell cycle phase it is known to regulate.
Differentially expressed genes and their relation to cell cycle.
Genes whose expression was higher in BM-CD34+ cells are displayed in bold font. Genes whose expression was lower in BM-CD34+ cells are displayed by hollow characters. Each gene is assigned to the respective cell cycle phase it is known to regulate.
Five proapoptotic genes, including caspases 3, 4, and 8, were expressed to a higher degree, whereas gene expression of the antiapoptotic cytoplasmic antiproteinase 2 was decreased in circulating CD34+ cells, suggesting greater apoptotic activity for these cells than for BM-CD34+ cells. This could be functionally confirmed by an apoptosis assay showing a significantly increased proportion of apoptotic CD34+ cells in PB. Circulating CD34+ cells might be susceptible to anoikis, a special kind of detachment-induced, caspase-mediated apoptosis that has been proven to play a role in several nonhematopoietic cell types but also in the myeloid leukemia cell line HL-60.35-37 Anoikis might serve as a physiological mechanism to prevent uncontrolled proliferation once a hematopoietic stem cell has left the bone marrow.
Several transcription-related genes were differentially expressed between BM- and PB-CD34+ cells. For instance, GATA2 and N-myc were expressed more in PB-CD34+ cells. GATA2 is known to play a crucial role in the transcriptional regulation of early hematopoiesis by blocking differentiation and allowing self-renewal.38 N-myc expression also keeps cells in a determined state, and its down-regulation leads to progress through differentiation.39 Expression of GATA2, N-myc, and other transcription factors, which was higher in PB-CD34+ cells, might cause an arrest of differentiation in circulating CD34+ cells, irrespective of their developmental state.
The expression of adhesion molecules in CD34+ cells, as found in our study, is in line with previous studies,1,40which show the validity of our data. Only CD44 was significantly differentially expressed, namely higher, in PB-CD34+ cells. This finding confirms a recent study41 and is feasible because CD44 is known to play a role in the homing of CD34+cells.42 For all other genes encoding for adhesion molecules, we could not detect significant differential expression. This is in contrast to previous studies that measured higher protein levels of adhesion molecules such as very late antigen-4 (VLA-4) or leukocyte function antigen-1 (LFA-1) on BM-CD34+ cells compared to circulating CD34+ cells.1 40 This may be due to posttranscriptional regulation of those surface molecules.
We found greater expression of the CXC chemokine receptor 4 (CXCR4) on BM-CD34+ cells than on PB-CD34+ cells. The ligand of CXCR4 is stromal cell–derived factor 1 (SDF1), which is expressed by stromal cells of the bone marrow. Our data support the model that SDF1-CXCR4 interaction is crucial for adhesion and differentiation of hematopoietic progenitors in bone marrow.43 44
Thrombin receptor (PAR1) seems to be important for circulating CD34+ cells; we measured a 3.3-fold higher expression in PB-CD34+ cells than in BM-CD34+ cells. Thrombin receptor is a 7-transmembrane, G-protein–coupled cell surface receptor that plays an important role in monocyte chemotaxis and phagocytosis.45 46 The higher expression of thrombin receptor in mobilized CD34+ cells suggests a chemotaxis-mediating function not only for monocytes but also for CD34+ cells. Released thrombin may guide CD34+hematopoietic stem cells to sites of cellular damage or increased consumption to facilitate regeneration processes.
Interestingly, proteinase 3 was strongly expressed by BM-CD34+ cells but not by PB-CD34+ cells, whereas protease inhibitor 2 was expressed at a 2-fold higher level by PB-CD34+ cells. Proteinase 3 is expressed by neutrophils and monocytes, and it is capable of cleaving connective tissue protein, such as elastin.47 Protease inhibitor 2 is able to inhibit proteinase 3.48 The combination of greater expression of proteinase 3 and lower expression of the corresponding protease inhibitor 2 in BM-CD34+ cells compared to PB-CD34+ cells suggests that high proteinase 3 activity is essential for CD34+ cells localized in the bone marrow. The ability to cleave connective tissue protein might be necessary for CD34+ cells to produce and to maintain a suitable microenvironment for hematopoiesis within the bone marrow stroma. Little is known about the other differentially expressed genes displayed in Figure 2. Further studies will be necessary to clarify the impact of those genes on the physiology of CD34+ cells.
Hierarchical cluster analysis showed that bone marrow and circulating CD34+ cells can be clearly distinguished mathematically by their gene expression profiles, and it supports the significance of the measured transcriptional differences. The close relation of the samples within the bone marrow cluster shows that the inter-individual differences of gene expression patterns of normal bone marrow CD34+ cells are relatively small. The presence of 2 subgroups within the peripheral blood cluster suggests that there might be 2 or more molecularly distinguishable mechanisms of stem cell mobilization. A correlation of the subgroups with age, sex, G-CSF dose, or mobilization kinetics could not be found. However, the 10 examined samples of PB-CD34+ cells comprise too small a population from which to draw reliable conclusions regarding these 2 subclusters.
In our array experiments, we performed RT with gene-specific primers instead of random primers. This greatly improved the sensitivity and rendered the measurement of gene expression profiles of CD34+ cells derived from single samples. Our experimental setting avoided pooling of samples from different donors or amplification of cDNA. We used cDNA arrays from different batches to check for false cDNA annotations or mistakes during the spotting procedure. Those measures resulted in increased reproducibility and reliability, reflected by small inter-individual differences of the gene expression data. Furthermore, the examination of 18 independent samples led to highly significant results (P < .01), even if the mean difference of the bone marrow and peripheral blood–derived samples was smaller than 2-fold. The validity of our data was further verified by corroboration experiments using quantitative real-time RT-PCR and flow cytometry for selected genes, which confirmed the gene expression data obtained by cDNA array technology on mRNA and on the protein level.
In summary, our study and our database provide a gene expression profile of primary human CD34+ hematopoietic stem cells and may serve as a basis for further research in the field of functional genomics of human hematopoietic stem cells. Comparison of gene expression of BM-CD34+ cells and PB-CD34+ cells molecularly confirms and explains the model that CD34+cells residing in the bone marrow are cycling more rapidly, whereas circulating CD34+ cells consist of a higher number of quiescent stem and progenitor cells. Moreover, our data give novel molecular insight into stem cell biology. An improved understanding of stem cell physiology might be helpful for the clinical use of CD34+ cells to support cytotoxic therapy and for the development of new therapeutic approaches using hematopoietic stem cells as a cellular source for tissue engineering.
Supported by the Leukämie-Liga eV, Düsseldorf, Germany.
U.S. and R.K. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Ulrich Steidl, Klinik für Hämatologie, Onkologie und Klinische Immunologie, Universität Düsseldorf, Moorenstrasse 5, D-40225 Düsseldorf, Germany; e-mail: usteidl@usteidl.de.

![Fig. 4. Corroboration of differential mRNA expression by quantitative real-time RT-PCR. / Real-time RT-PCR curves for 4 different genes (leukocyte proteinase 3, ubiquitin protein-ligase [UBCH10], DAPK1, and caspase 4) with the respective GAPDH controls are displayed. A representative BM-CD34+ sample is shown on the left, and a representative PB-CD34+ sample is shown on the right side. Analysis of each sample was performed in duplicate. The differences of the CPs (ΔCP) of target and GAPDH are indicated. The smaller the ΔCP, the higher the relative abundance of the target's mRNA.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/99/6/10.1182_blood.v99.6.2037/6/m_h80622274004.jpeg?Expires=1767764389&Signature=Lqa6x0e4X7oVKbUf5IAKqHIYAbxjkcXZCMQ4F1FhmO8CuqC~xsKL0579L~d9fffs8lNG7OTRtfw7KudhbhIMfQ~cEsVa5yUjY1Nr7JzB-0CCtuz1nUnlDF7p7qlXy0pFaB0nUyae7-UmSLI3U61BW5jroHWUL6x~9dAPhEacZ5SUNPAjG7uTccW~WLj0jSmYTpIRZPgSeZ8TiHDDPcMApPyfRyQ8eTlHvNxLi7ZsCQXk8AP5MuQTzsGc-EUZzzcebf6PWBTHv9vfAcs-NYJqXEOBNo1vP5-HlMK4M0lGEJlk6NpR2I8QxRY62DzMH6pWFB5xFPe16fgYveD6D3FLpQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal