Key Points
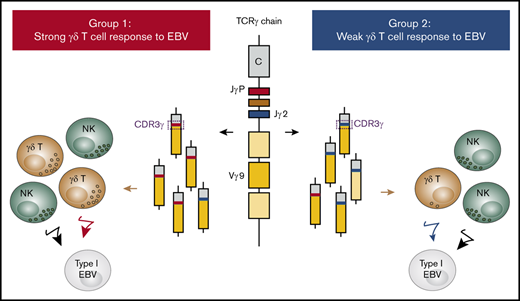
One-half of the studied human population makes a strong γδ T-cell response to EBV and has γ-chain repertoires enriched for JγP CDR3s.
The other half makes a weak γδ T-cell response to EBV and has γ-chain repertoires enriched for Jγ2 CDR3s.
Abstract
Humans form 2 groups based on their innate immunity to Epstein-Barr virus (EBV). Group 1 makes a strong natural killer (NK)–cell and γδ T-cell response, whereas group 2 makes a strong NK-cell response, but a weak γδ T-cell response. To investigate the underlying basis for this difference in γδ T-cell immunity to EBV, we used next-generation sequencing to compare the γδ T-cell receptor (TCR) repertoires of groups 1 and 2. In the absence of EBV, group 1 TCRγ chains are enriched for complementarity determining region 3 (CDR3s) containing JγP, whereas group 2 TCRγ chains are enriched for CDR3s containing Jγ2. In group 1 donors, EBV activates many γδ T cells expressing Vγ9JγP, inducing proliferation that produces a large population of activated effector cells. The TCRs using Vγ9JγP are closely related to the TCRs of γδ T cells that respond to phosphoantigens. In group 2 donors, EBV activates a small subpopulation of γδ T cells, most expressing Vγ9JγP. In conclusion, we find that differences in the TCRγ-chain repertoire underlie the differential response of group 1 and group 2 to EBV.
Introduction
Epstein-Barr virus (EBV) is a herpesvirus that infects over 90% of the human population worldwide. It is commonly contracted during early childhood and then establishes an asymptomatic lifelong infection. In a minority of individuals, EBV infection leads to one of a number of malignant or nonmalignant diseases.1,2
EBV infects epithelial cells and B cells. In vivo, EBV infection is usually controlled by the human host’s immune system. Consequently, only some latently EBV-infected B cells survive to become memory B cells. This form of EBV latency, termed “latency I,” resembles that present in Burkitt lymphoma, an EBV-associated malignancy.3 In this investigation, we used an EBV+ Burkitt lymphoma cell line to study the innate immune response to EBV infection. With this in vitro model, we previously showed that innate immunity to EBV divides healthy humans into 2 groups.4 For group 1 donors, their natural killer (NK) cells and γδ T cells both make a strong response to EBV, whereas group 2 donors make a strong NK-cell response but a weak γδ T-cell response. Indeed, after EBV stimulation in vitro, the proliferation of γδ T cells from group 1 donors is ∼10-fold greater than that of the γδ T cells from group 2 donors. This qualitative difference in response does not correlate with sex, HLA type, or previous exposure to either EBV or cytomegalovirus (CMV).4
The γδ T cells represent 1% to 5% of peripheral blood lymphocytes. Among them, up to 75% carry a Vδ2+ T-cell receptor (TCR). Vδ2+ T cells are activated by phosphorylated nonpeptide antigens.5 These antigens, commonly called phosphoantigens (pAgs), accumulate in EBV-infected cells that maintain a type I EBV infection,4 which explains the capacity of these cells to activate Vδ2+ T cells. Consistent with this finding, Xiang et al have shown that Vδ2+ T cells expanded in vitro with pAgs are able to prevent EBV-induced malignancy in humanized mice.6
The human γ-chain locus is located on chromosome 7 and comprises 2 constant region genes (Cγ1 and Cγ2), 5 joining segments (Jγ1, Jγ2, JγP, JγP1, and JγP2) and 14 variable gene segments, of which only 6 (Vγ2, Vγ3, Vγ4, Vγ5, Vγ8, Vγ9) are functional.7,8 The human δ-chain locus lies within the α-chain locus on chromosome 14. It comprises 8 V segments (Vδ1-8) of which only Vδ1, Vδ2, and Vδ3 are bona fide Vδ segments. The other 5 segments are Vα segments that are rarely used in δ-chain rearrangements. The δ-chain locus also contains 3 D segments (Dδ1-3), 4 J segments (Jδ1-4), and 1 C region gene.9,10 The complementarity determining region 3 (CDR3) loops of the δ chain have the highest potential for diversity and are longer than the γ-chain CDR3 loops, which are short and constrained.11 A majority of γδ T cells in peripheral blood have TCRs with γ chains encoded by the Vγ9, JγP, and Cγ1 gene segments and Vδ chains encoded by Vδ2.12 Despite this bias, there is no apparent restriction to the pairing of Vγ9JγPCγ1 with other δ chains. At birth, Vδ2+ T cells comprise a small proportion of T cells in the thymus as well as of peripheral blood T cells. Thus, the expansion and dominance of Vδ2+ T cells in the blood of adult humans is likely due to selective pressures mediated by pathogen-derived pAgs.13,14
The γδ T-cell response to pAgs is defined by the γ-chain CDR3.15,16 Previous studies by other investigators showed that γδ T-cell clones, making a strong response to pAgs, express a JγP gene segment, whereas γδ T-cell clones using other Jγ segments are less responsive.15,17 One possible cause of the distinctive γδ T-cell responses made by group 1 and group 2 donors to EBV is that their γδ T cells have different receptor repertoires. To test this hypothesis, we used a next-generation-sequencing approach to determine and compare the γ and δ TCR repertoires of group 1 and group 2 donors. Comparison of the γδ T-cell repertoires was also made before and after stimulation with EBV.
Materials and methods
Cells and cell culture
Blood samples were collected from CMV-seronegative healthy adult volunteer donors and purchased as anonymized leukoreduction system chambers from the Stanford Blood Center. The CMV status of the donors was determined serologically at the Stanford Blood Center. Peripheral blood mononuclear cells (PBMCs) were isolated by density gradient centrifugation (Ficoll-Paque PLUS; GE Healthcare) as recommended by the manufacturer and then cryopreserved in fetal bovine serum (FBS; Gemini) containing 10% dimethyl sulfoxide (EMD Millipore).
Cryopreserved PBMCs were thawed, suspended in RPMI 1640 medium (Corning Cellgro), and centrifuged for 5 minutes at 1500 rpm. Cell pellets were resuspended in RPMI 1640 containing 10% heat-inactivated FBS and then kept overnight at 37°C. This period of culture allowed the PBMCs to recover from the freeze-thaw before they were subjected to in vitro experimentation.
The EBV+ Akata cell line was kindly provided by J. Sample (Pennsylvania State University, State College, PA). Akata cells were cultured in RPMI 1640 containing heat-inactivated FBS (10%), l-glutamine (5 mM), and streptomycin/penicillin (100 IU/100 µg/mL; Thermo Fisher Scientific).
Assay of γδ T-cell proliferation in response to EBV and IPP
Five million PBMCs were cultured with the EBV+ Akata cells, at an effector-to-target cell ratio of 10:1 in a 24-well plate. Cells were cultured in RPMI 1640 containing heat-inactivated FBS (5%), heat-inactivated human AB serum (5%) (Sigma Aldrich), l-glutamine (2 mM), streptomycin/penicillin (100 IU/100 µg/mL), and recombinant human interleukin 2 (200 IU/mL). Interleukin 2 was obtained from Maurice Gately (Hoffmann-La Roche Inc) through the AIDS Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health (NIAID, NIH). Cells were passaged on reaching confluence and supplemented with fresh medium every other day for a period of 10 days. In some experiments, 200 000 PBMCs were stimulated with 100 µM exogenous isopentenyl pyrophosphate (IPP; Santa Cruz Biotechnology) and then cultured in 96-well plates. Cells were passaged on reaching confluence and then supplemented with fresh medium every 2 days for a period of 10 days.
Antibodies and analysis by flow cytometry
To assess the proportions of Vδ1+ T cells and Vδ2+ T cells, PBMCs were stained with the monoclonal antibody conjugates: anti-TCRVδ1 (REA173; Miltenyi Biotec), anti-TCRVδ2 (123R3; Miltenyi Biotec), anti-TCRVγ9 (B3; BioLegend), and anti-CD3 (UCHT1; BioLegend). Data were collected with an Accuri C6 instrument (Becton Dickinson) and analyzed using FlowJo software version 10.1.
γδ T-cell sorting and RNA extraction
Two hundred thousand γδ T cells were purified from unstimulated PBMCs, EBV-stimulated PBMCs, and IPP-stimulated PBMCs, using the TCRγ/δ+ T Cell Isolation kit (Miltenyi Biotec) as recommended by the manufacturer. From these preparations of γδ T cells, total RNA was extracted using the RNeasy Mini kit (Qiagen) according to the manufacturer’s instructions.
TCR-repertoire analysis
Extracted RNA was used as the template to amplify clone-specific rearrangements of the TCRγ and the TCRδ genes, using the iR-Profile Reagent kit as recommended by the manufacturer (iRepertoire Inc). Briefly, RNA was subjected to reverse transcription polymerase chain reaction using human γ- and δ-multiplex primers targeting each of the V and C genes by capturing segments of ∼380 bp in length. This achieved full coverage of the γ- and δ-chain CDR1, CDR2, and CDR3 regions. The products from the first polymerase chain reaction (PCR) were subjected to a second PCR, performed with universal primers and giving exponential amplification. PCR products were then purified using the PCR2 Clean-up Mix according to the manufacturer’s recommendation (iRepertoire). DNA concentrations were assessed by Qubit fluorescent-based methodology, and library generation was judged successful if the size of the amplicons ranged between 380 bp and 550 bp. As each library had a distinctive barcode, 10 libraries were pooled in equal amounts to give 1 sample. To provide accurate DNA concentrations and good quality controls prior to sequencing, the pooled libraries were assessed by bioanalysis with an Agilent Bioanalyser. Further preparation for loading the libraries into the MiSeq system was performed according to the “Denature and Dilute Libraries Guide” provided by Illumina. A ready-to-use PhiX library was added to the pooled libraries as a control to improve run efficiency. Libraries were sequenced using a MiSeq next-generation sequencer with a reagent kit v2 500 cycle, as recommended by the manufacturer (Illumina).
The vast amount of raw TCR data obtained from each library was analyzed using iRweb software (iRepertoire). After removing errors made during amplification and sequencing, this software gives filtered DNA sequences. It also provides a DNA-sequence alignment for all gene combinations and the sequences of the protein they encode. This identifies the CDR regions and the length of their CDR3s. The software also provides the total number of all CDR3s obtained from the filtered sequences as well as the number of distinct CDR3s.
B-cell purification and detection of EBV genomic DNA
B cells were purified from the PBMCs of healthy volunteer donors using the Dynabeads Isolation kit as recommended by the manufacturer (Life Technologies). Genomic DNA was isolated from peripheral blood B cells using the QIAamp DNA Blood Mini kit (Qiagen). A part of the EBNA1 gene was amplified using PCR (forward primer, 5′-GGT AGA AGG CCA TTT TTC CAC-3′; reverse primer, 5′-CTC CAT CGT CAA AGC TGC AC-3′) as described.18
Statistical analyses
Data were analyzed using Prism software. P values were calculated using the unpaired Student t test. Only P < .05 was considered significant.
Results
Group 1 and group 2 individuals have similar TCRδ-chain repertoires
Humans divide into 2 distinctive groups based on their γδ T-cell response to EBV. Group 1 makes a strong response, group 2 a weak response.4 To investigate the basis for this difference, we first used flow cytometry to compare the abundance of Vδ2+ T cells as well as Vδ1+ T cells, the predominant γδ T-cell subpopulations,19 in unstimulated PBMCs. Cells from 10 group 1 and 10 group 2 individuals were subjected to analysis. All individuals have comparable proportions of circulating Vδ2+ T cells. Vδ1+ T cells, which also have similar frequencies in groups 1 and 2 (Figure 1A), represent most of the Vδ2− γδ T cells. A small minority of Vδ1+ T cells express Vγ9, whereas most Vδ2+ T cells express Vγ9 (Figure 1B).
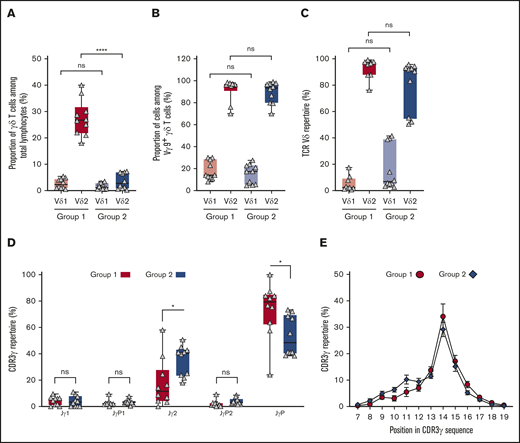
Group 1 and group 2 donors have different TCRγ chains and similar TCRδ chains. (A) Vδ1+ (light red, light blue) and Vδ2+ (dark red, dark blue) T-cell proportions in total lymphocytes of group 1 (N = 10; red) and group 2 (N = 10; blue). (B) Vγ9Vδ1+ and Vγ9Vδ2+ T-cell proportions within γδ T cells of group 1 (N = 10; red) and group 2 (N = 10; blue). (C) Frequency of Vδ1 (light red, light blue) and Vδ2 (dark red, dark blue) usage in γδ T cells of group 1 (N = 10; red) and group 2 (N = 10; blue). (D) Frequency of Jδ1 in γδ T cells of group 1 (G1; N = 10; red) and group 2 (G2; N = 10; blue). (E) Kullback-Leibler sequence logos showing residue preferences for the top 20 CDR3δ2 sequences. Due to length variations, the analyses considered the first 10 residues for each CDR3δ2. Data are for representative group 1 and group 2 individuals. Black arrows denote position 5 in the CDR3δ2 sequences. Each logo consists of amino acid stacks, 1 for each position in the sequence. The stack height indicates the extent of sequence conservation; the residue height indicates its relative frequency. Residue colors correspond to biochemical properties: acidic (red), basic (blue), hydrophobic (black), polar (green). Logos were generated using WebLogo.28 (F) Frequency of Vγ9 usage in group 1 (G1; N = 10; red) and group 2 (G2; N = 10; blue). (G) Frequency of Jγ usage in group 1 (N = 13; red) and group 2 (N = 12; blue). (H) Proportions of CDR3γ of different length in group 1 (N = 13; red circles) and group 2 (N = 12; blue diamonds) donors’ TCRγ repertoires. Mean values and standard errors of the mean are given. (I) Proportions of CDR3 Jγ2-Vγ9 among CDR3 Jγ2 in group 1 (G1; N = 13; red) and group 2 (G2; N = 12; blue) donors’ TCRγ repertoires. Statistical significance between groups was assessed using the Student t test (*P = .02; **P = .003; ***P ≤ .0006). ns, not significant.
Group 1 and group 2 donors have different TCRγ chains and similar TCRδ chains. (A) Vδ1+ (light red, light blue) and Vδ2+ (dark red, dark blue) T-cell proportions in total lymphocytes of group 1 (N = 10; red) and group 2 (N = 10; blue). (B) Vγ9Vδ1+ and Vγ9Vδ2+ T-cell proportions within γδ T cells of group 1 (N = 10; red) and group 2 (N = 10; blue). (C) Frequency of Vδ1 (light red, light blue) and Vδ2 (dark red, dark blue) usage in γδ T cells of group 1 (N = 10; red) and group 2 (N = 10; blue). (D) Frequency of Jδ1 in γδ T cells of group 1 (G1; N = 10; red) and group 2 (G2; N = 10; blue). (E) Kullback-Leibler sequence logos showing residue preferences for the top 20 CDR3δ2 sequences. Due to length variations, the analyses considered the first 10 residues for each CDR3δ2. Data are for representative group 1 and group 2 individuals. Black arrows denote position 5 in the CDR3δ2 sequences. Each logo consists of amino acid stacks, 1 for each position in the sequence. The stack height indicates the extent of sequence conservation; the residue height indicates its relative frequency. Residue colors correspond to biochemical properties: acidic (red), basic (blue), hydrophobic (black), polar (green). Logos were generated using WebLogo.28 (F) Frequency of Vγ9 usage in group 1 (G1; N = 10; red) and group 2 (G2; N = 10; blue). (G) Frequency of Jγ usage in group 1 (N = 13; red) and group 2 (N = 12; blue). (H) Proportions of CDR3γ of different length in group 1 (N = 13; red circles) and group 2 (N = 12; blue diamonds) donors’ TCRγ repertoires. Mean values and standard errors of the mean are given. (I) Proportions of CDR3 Jγ2-Vγ9 among CDR3 Jγ2 in group 1 (G1; N = 13; red) and group 2 (G2; N = 12; blue) donors’ TCRγ repertoires. Statistical significance between groups was assessed using the Student t test (*P = .02; **P = .003; ***P ≤ .0006). ns, not significant.
For each donor, analysis of TCR repertoire was performed on a population of unstimulated γδ T cells. For TCRδ, a mean sequencing read depth of 847 930 ± 33 819 was obtained, giving an average of 4643 ± 1333 unique CDR3 sequences per donor. For TCRγ, the mean sequencing read depth was 692 668 ± 34 455, giving an average of 1625 ± 370 unique CDR3 sequences per donor. Analysis of the D50 diversity (the proportion of dominant unique TCRs that represent 50% of the repertoire), showed no significant difference between group 1 and group 2 for either the TCRδ or TCRγ chain.
For all individuals, the TCRδ chains consist predominantly of a Vδ2 variable region associated with a Jδ1 joining region (Figure 1C-D). This combination of gene segments is consistent with the results of previous studies by others19-22 and with our flow cytometry analysis (Figure 1A). Thus, we find that there is little difference between group 1 and group 2 in their use of Vδ gene segments.
A key feature of the γδ TCRs that recognize pAgs is a hydrophobic residue at position 97 of the δ chain.14,16 Corresponding to position 5 of the CDR3δ segment, this residue is hydrophobic in the 20 most frequent CDR3δ2 sequences in both donor groups (Figure 1E). These sequences represent >50% of the CDR3δ2 repertoires for almost all donors. Thus, group 1 and group 2 have an abundance of CDR3δ2 segments with the potential to recognize pAgs.
Overall, similarity of the TCRδ repertoires in groups 1 and 2 argues against differences in TCRδ repertoire being a factor determining the differential γδ T-cell response to EBV.
Group 1 and group 2 differ in their TCRγ-chain repertoires
Recognition of pAgs is critically dependent on CDR3γ, which exhibits less sequence and length diversity than CDR3δ.15,16 Because γδ T cells expressing the Vγ9JγP chain respond to pAgs and EBV-infected Daudi cells,15 the TCRγ-chain repertoires were compared. No significant difference in CDR3 Vγ9 usage was seen for group 1 and group 2 (Figure 1F). In contrast, γδ T cells using JγP are abundant in group 1, whereas Jγ2 is dominant in group 2. Performing analysis on a further 5 individuals gave similar results. The combined analysis of 13 group 1 donors and 12 group 2 donors is given in Figure 1G. Comparison of CDR3γ length revealed a group 1 bias for CDR3s of 14-aa residues and a group 2 bias for 10 residue CDR3s (Figure 1H). This difference in CDR3 length is consistent with the previous observation that CDR3s of the Vγ9Jγ2 chain can be of shorter length (6-14 residues) than CDR3s of the Vγ9JγP chain (11-14 residues).16
It is likely that a subset of the γδ T cells using Jγ2 does not express Vδ2, but another Vδ, in particular Vδ1. A low frequency of Vδ1 pairing with Vγ9 was observed (Figure 1B).23 Nonetheless, in a majority of group 1 and group 2 individuals, >50% of the Jγ2 CDR3s contain Vγ9 (Figure 1I). As Vγ9-containing γ chains usually pair with a δ chain containing Vδ2,19,22 most γδ TCRs that have Jγ2 could therefore derive from Vδ2+ T cells.
This analysis has uncovered a striking difference in the TCRγ repertoires of group 1 and group 2 individuals. TCRγ-chain repertoires of group 1 are enriched for CDR3s containing JγP, whereas those of group 2 are enriched for CDR3s containing Jγ2. This finding provides an explanation for the bimodal γδ T-cell response to EBV that we previously reported.4
In responding to EBV, γδ T cells preferentially use the JγP segment
TCRγ- and TCRδ-chain repertoire analyses were performed on γδ T-cell populations isolated from PBMCs that had been stimulated during a 10-day culture with Akata, an EBV-infected B-cell line of type 1 latency. As previously seen,4 in response to EBV, the population of γδ T cells from group 1 donors increased >500-fold. This effect is ∼10-fold greater than seen for group 2 donors. There was no such increase for Vδ1+ T cells (Figure 2A). Most Vδ2+ T cells express Vγ9, whereas only a small fraction of Vδ1+ T cells express Vγ9 (Figure 2B). Analysis of the δ-chain repertoires confirmed the results obtained by flow cytometry. Thus, Vδ2 is expressed by the majority of proliferating γδ T cells (Figure 2C). As observed for cells that were not stimulated with EBV, CDR3δ exhibited high diversity, and no CDR3s were common to all of the donors.
In responding to EBV, γδ T cells preferentially use the JγP segment. Two hundred thousand γδ T cells were isolated from PBMCs after stimulation with Akata, an EBV-transformed B cell line. (A) Vδ1+ (light red, light blue) and Vδ2+ T-cell proportions (dark red, dark blue) within EBV-stimulated lymphocytes of group 1 (N = 10; red) and group 2 (N = 10; blue). (B) Vγ9Vδ1+ and Vγ9Vδ2+ T-cell proportions within EBV-stimulated γδ T cells of group 1 (N = 10; red) and group 2 (N = 10; blue). (C) Frequency of Vδ1 (light red, light blue) and Vδ2 (dark red, dark blue) usage in EBV-stimulated γδ T cells of group 1 (N = 10; red) and group 2 (N = 10; blue). (D) Frequency of Jγ usage by EBV-stimulated γδ T cells of group 1 (N = 10; red) and group 2 (N = 10; blue). (E) Proportions of CDR3γ of different length in group 1 (N = 10; red circles) and group 2 (N = 10; blue diamonds) donors’ TCRγ repertoires. Means and standard errors of the mean are given. Statistical significance was assessed using the Student t test (*P = .02; ****P ≤ .0001).
In responding to EBV, γδ T cells preferentially use the JγP segment. Two hundred thousand γδ T cells were isolated from PBMCs after stimulation with Akata, an EBV-transformed B cell line. (A) Vδ1+ (light red, light blue) and Vδ2+ T-cell proportions (dark red, dark blue) within EBV-stimulated lymphocytes of group 1 (N = 10; red) and group 2 (N = 10; blue). (B) Vγ9Vδ1+ and Vγ9Vδ2+ T-cell proportions within EBV-stimulated γδ T cells of group 1 (N = 10; red) and group 2 (N = 10; blue). (C) Frequency of Vδ1 (light red, light blue) and Vδ2 (dark red, dark blue) usage in EBV-stimulated γδ T cells of group 1 (N = 10; red) and group 2 (N = 10; blue). (D) Frequency of Jγ usage by EBV-stimulated γδ T cells of group 1 (N = 10; red) and group 2 (N = 10; blue). (E) Proportions of CDR3γ of different length in group 1 (N = 10; red circles) and group 2 (N = 10; blue diamonds) donors’ TCRγ repertoires. Means and standard errors of the mean are given. Statistical significance was assessed using the Student t test (*P = .02; ****P ≤ .0001).
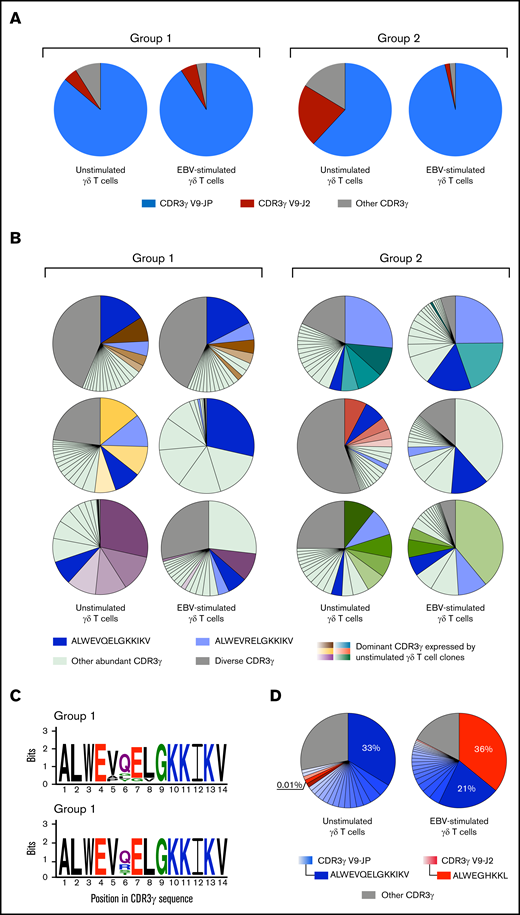
Analysis of the γ-chain repertoires for group 1 and group 2 individuals showed that EBV stimulates γδ T-cell clones characterized by a preferential expression of CDR3s using JγP segments of 14 aa (Figure 2D-E). The use of JγP was almost always combined with Vγ9 (Figure 3A), as observed in a previous study showing how Vγ9JγP is used by γδ T cells in responding to type I EBV-transformed Daudi cells or pAgs.15 All individuals express the Vγ9JγP public CDR3s.15,24,25 Of these, ALWEVQELGKKIKV and ALWEVRELGKKIKV are among the top 20 most abundant CDR3s for most individuals, both in the absence and presence of EBV (Figure 3B). Other abundant CDR3γ expressed by unstimulated γδ T cells are not always among those most frequently expressed by EBV-stimulated γδ T cells (Figure 3B). Thus, EBV triggers a γδ T-cell response that is mediated by a particular subset of CDR3s.
Most γδ T-cell clones that respond to EBV express a Vγ9JγP CDR3. Two hundred thousand γδ T cells were isolated from PBMCs before and after stimulation with Akata, an EBV-transformed B cell line. (A) Pie charts show the γ-chain CDR3 repertoires of unstimulated γδ T cells and EBV-stimulated γδ T cells from 1 representative group 1 donor and 1 representative group 2 donor. CDR3γ V9-JP sequences are represented in blue and CDR3γ V9-J2 sequences are represented in red. The gray zone represents the combination of all other unique CDR3s within the CDR3γ repertoires. (B) Pie charts show the distribution of the 20 most abundant γ-chain CDR3s of unstimulated γδ T cells and EBV-stimulated γδ T cells, from 3 representative group 1 donors and 3 representative group 2 donors. The public CDR3s ALWEVQELGKKIKV (dark blue) and ALWEVRELGKKIKV (light blue) are shown. For each donor, the 5 most dominant CDR3s expressed by unstimulated γδ T cells are shown in different shades of color and, when required, in dark blue or light blue for ALWEVQELGKKIKV and ALWEVRELGKKIKV, respectively. To track these same CDR3s in the EBV-stimulated γδ T-cell repertoires, we depicted them in similar colors. The other 15 CDR3s are colored in light green and the remaining unique CDR3s are shaded in gray. (C) Kullback-Leibler sequence logos show the residue preferences for the dominant 14-mer CDR3γ sequences from EBV-stimulated γδ T cells of group 1 (N = 10 CDR3γ) and group 2 (N = 10 CDR3γ) donors. Populations of γδ T cells were isolated from EBV-stimulated PBMCs. Each logo consists of amino acid stacks, 1 for each position. The stack height indicates the sequence conservation; the residue height indicates its relative frequency. Residue colors correspond to biochemical properties: acidic (red), basic (blue), hydrophobic (black), polar (green). Logos were generated using WebLogo.28 (D) The charts illustrate the distribution of the 20 most abundant γ-chain CDR3s among unstimulated γδ T cells and EBV-stimulated γδ T cells from a group 1 donor: donor 1. Populations of γδ T cells were isolated from unstimulated PBMCs and EBV-stimulated PBMCs. CDR3γ V9-JP sequences are represented by different shades of blue and CDR3γ V9-J2 are represented by different shades of red. The gray zone indicates a combination of all the other unique CDR3s within the CDR3γ repertoires.
Most γδ T-cell clones that respond to EBV express a Vγ9JγP CDR3. Two hundred thousand γδ T cells were isolated from PBMCs before and after stimulation with Akata, an EBV-transformed B cell line. (A) Pie charts show the γ-chain CDR3 repertoires of unstimulated γδ T cells and EBV-stimulated γδ T cells from 1 representative group 1 donor and 1 representative group 2 donor. CDR3γ V9-JP sequences are represented in blue and CDR3γ V9-J2 sequences are represented in red. The gray zone represents the combination of all other unique CDR3s within the CDR3γ repertoires. (B) Pie charts show the distribution of the 20 most abundant γ-chain CDR3s of unstimulated γδ T cells and EBV-stimulated γδ T cells, from 3 representative group 1 donors and 3 representative group 2 donors. The public CDR3s ALWEVQELGKKIKV (dark blue) and ALWEVRELGKKIKV (light blue) are shown. For each donor, the 5 most dominant CDR3s expressed by unstimulated γδ T cells are shown in different shades of color and, when required, in dark blue or light blue for ALWEVQELGKKIKV and ALWEVRELGKKIKV, respectively. To track these same CDR3s in the EBV-stimulated γδ T-cell repertoires, we depicted them in similar colors. The other 15 CDR3s are colored in light green and the remaining unique CDR3s are shaded in gray. (C) Kullback-Leibler sequence logos show the residue preferences for the dominant 14-mer CDR3γ sequences from EBV-stimulated γδ T cells of group 1 (N = 10 CDR3γ) and group 2 (N = 10 CDR3γ) donors. Populations of γδ T cells were isolated from EBV-stimulated PBMCs. Each logo consists of amino acid stacks, 1 for each position. The stack height indicates the sequence conservation; the residue height indicates its relative frequency. Residue colors correspond to biochemical properties: acidic (red), basic (blue), hydrophobic (black), polar (green). Logos were generated using WebLogo.28 (D) The charts illustrate the distribution of the 20 most abundant γ-chain CDR3s among unstimulated γδ T cells and EBV-stimulated γδ T cells from a group 1 donor: donor 1. Populations of γδ T cells were isolated from unstimulated PBMCs and EBV-stimulated PBMCs. CDR3γ V9-JP sequences are represented by different shades of blue and CDR3γ V9-J2 are represented by different shades of red. The gray zone indicates a combination of all the other unique CDR3s within the CDR3γ repertoires.
To characterize the CDR3γ most frequently used in responding to EBV, we compared the dominant 14-mer CDR3γ (10 CDR3s for each group) from the EBV-stimulated cells. This analysis showed considerable similarities between the sequences (Figure 3C). Both group 1 and group 2 CDR3s have lysine residues in their C-terminal region. These residues are necessary for pAg recognition by Vδ2+ T cells.16,26 In position 5, valine was present in most CDR3s of group 1 and all of the group 2 CDR3s. Alanine and proline were also present at position 5 of the group 1 CDR3s. Position 6 is more diverse, with glutamine present in >50% of the sequences, followed in frequency by glycine, leucine, and tyrosine in group 1, and by arginine, serine, and threonine in group 2. In group 2 CDR3s, positions 7 and 8 have invariant glutamic acid and leucine, respectively, whereas in group 1, glycine and valine also make a minor contribution at these positions (Figure 3C). The greater diversity of CDR3γ in group 1 than in group 2 likely arises from the predominance of Vγ9JγP CDR3s in group 1 prior to stimulation with EBV (Figure 1G).
In comparison with the Vγ9JγP CDR3s, Vγ9Jγ2 CDR3s are less common and their expression by EBV-stimulated γδ T cells is limited (Figure 3A). An exceptional case is donor 1, a group 1 individual whose response to EBV raised the frequency of ALWEGHKKL, a Vγ9Jγ2 CDR3, by 3600-fold (from 0.01% to 36%). A decrease in the frequency of the public ALWEVQELGKKIKV CDR3 from 33% to 21% was also observed. For donor 1, ALWEGHKKL replaced ALWEVQELGKKIKV as the most abundant CDR3γ (Figure 3D). CDR3δ-repertoire analysis of the EBV-stimulated γδ T cells of donor 1 showed that the 60 most frequent CDR3δs all have a Vδ2 chain. We can therefore conclude with confidence that the dominant γδ T-cell clone combines a ALWEGHKKL CDR3γ chain with a Vδ2 chain and comprises >10 million cells.
In conclusion, our data show that, for most donors, the γδ T cells responding to EBV preferentially express CDR3s with Vγ9JγP chains and are, therefore, similar to the γδ TCRs that recognize and respond to pAgs.
Discussion
Previously, we showed that a panel of healthy human blood donors evenly divided into 2 groups based upon their innate immune response to EBV. For group 1 donors, NK cells and γδ T cells both make a robust response to EBV, whereas the group 2 donors combine a robust NK-cell response with a weak γδ T-cell response. For group 2 donors, the number of responding γδ T cells is only 10% of that observed for group 1 donors.4 Here, we used flow cytometry and next-generation sequencing to study the γδ TCR repertoires of our donor cohort. We identified a striking qualitative difference between the 2 donor groups.
The key difference between group 1 and 2 individuals is in their TCRγ repertoire. Group 1 has TCRγ chains enriched for CDR3s containing JγP, whereas group 2 has TCRγ chains enriched for CDR3s containing Jγ2.
In contrast to the striking differences in their TCRγ-chain repertoire, the group 1 and 2 donors have similar δ-chain repertoires. Although some group 1 donors have more Vδ2+ T cells than group 2 donors, this difference is not significant and, in most group 1 and group 2 donors, Vδ2 T cells represent a minority (∼0.2% to 1.5%) of the circulating lymphocytes. Moreover, all individuals have an abundance of CDR3δ2 segments containing a hydrophobic residue at position 97 of the δ chain that permits the recognition of pAgs.
Donor 1 made a strong response to EBV with a clone of γδ T cells that expresses ALWEGHKKL, a Vγ9Jγ2 CDR3. This CDR3γ was only expressed by donor 1 and it contributed to a stronger anti-EBV immune response than most CDR3s with Vγ9JγP. For all other donors, EBV stimulation induced a proliferation of γδ T-cell clones that preferentially express Vγ9JγP CDR3s. Type I EBV-infected B cells maintain a high concentration of endogenous IPP, explaining their capacity to activate Vγ9Vδ2 T cells.4 We therefore expected to detect an immune response to EBV that is dominated by γδ T-cell clones expressing Vγ9JγP CDR3s.15 In contrast, 4 γδ T-cell clones expressing a Vγ9Jγ2 chain did not respond either to Daudi cells or mycobacteria.15 Therefore, the difference we observe in the γ-chain repertoire between group 1 and group 2 donors can explain why group 2 donors make a weaker γδ T-cell response to EBV than group 1. Because all of the donors are CMV-seronegative and most of them are EBV+, the group 1 and 2 difference in the γ-chain repertoire is not a consequence of CMV or EBV infections. Instead, increasing evidence points to a pAg-driven process that selects particular TCRγ and δ junctional motifs.14,27 The preferential use of JγP by group 1 could reflect their history of exposure and response to pAgs. Polymorphisms in γ, δ, and other genes affecting γδ T-cell development and function are other possible contributors to the differential response of group 1 and group 2.
In conclusion, our study describes a key dimorphism in the TCRγ-chain repertoires of healthy individuals that correlates with their immune responses to EBV. Those who make a strong response to EBV have an abundance of TCRγ chains containing JγP CDR3s, whereas those making a weak response have far fewer JγP CDR3s and an abundance of Jγ2-containing CDR3s. That γδ T cells responding to EBV preferentially express CDR3s with Vγ9JγP is consistent with this finding. Moreover, the dimorphism we see in the γδ TCR repertoire can explain the clinical outcome of primary EBV infections, as well as the susceptibility of immunocompromised individuals to EBV-induced malignancy. We identified one Vγ9Jγ2 CDR3 and several Vγ9JγP CDR3s that are highly responsive to EBV. These findings have implication for any γδ T-cell–based immunotherapy for EBV-mediated diseases, in particular in the generation of chimeric antigen receptor T cells.
Data-sharing requests may be e-mailed to the corresponding author, Zakia Djaoud, at zdjaoud@stanford.edu.
Acknowledgments
The authors thank J. Sample for the EBV+ Akata cells, and L. Guethlein and P. Mérieau for advice.
This work was supported by National Institutes of Health, National Institute of Allergy and Infectious Diseases grant R01 AI136952 (P.P.).
Authorship
Contribution: Z.D. conceived the project and designed and performed the experiments; and Z.D. and P.P. interpreted the data and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Zakia Djaoud, Stanford University School of Medicine, Fairchild D159, 299 Campus Dr West, Stanford, CA 94305; e-mail: zdjaoud@stanford.edu.