Key Points
Six novel candidate genes were identified for the occurrence of proteinuria and estimated glomerular filtration rate in sickle cell disease.
Functional annotation of these loci reveals biological mechanisms important not only to kidney function but also to sickle cell disease.
Abstract
Sickle cell disease nephropathy (SCDN), a common SCD complication, is strongly associated with mortality. Polygenic risk scores calculated from recent transethnic meta-analyses of urinary albumin-to-creatinine ratio and estimated glomerular filtration rate (eGFR) trended toward association with proteinuria and eGFR in SCD but the model fit was poor (R2 < 0.01), suggesting that there are likely unique genetic risk factors for SCDN. Therefore, we performed genome-wide association studies (GWAS) for 2 critical manifestations of SCDN, proteinuria and decreased eGFR, in 2 well-characterized adult SCD cohorts, representing, to the best of our knowledge, the largest SCDN sample to date. Meta-analysis identified 6 genome-wide significant associations (false discovery rate, q ≤ 0.05): 3 for proteinuria (CRYL1, VWF, and ADAMTS7) and 3 for eGFR (LRP1B, linc02288, and FPGT-TNNI3K/TNNI3K). These associations are independent of APOL1 risk and represent novel SCDN loci, many with evidence for regulatory function. Moreover, GWAS SNPs in CRYL1, VWF, ADAMTS7, and linc02288 are associated with gene expression in kidney and pathways important to both renal function and SCD biology, supporting the hypothesis that SCDN pathophysiology is distinct from other forms of kidney disease. Together, these findings provide new targets for functional follow-up that could be tested prospectively and potentially used to identify patients with SCD who are at risk, before onset of kidney dysfunction.
Introduction
Sickle cell disease (SCD) affects approximately 100 000 individuals in the United States,1 mostly with African ancestry. Caused by a mutation in the beta subunit of hemoglobin, SCD results in hemolytic anemia and damage to multiple organ systems via a complex set of pathophysiological mechanisms.2 SCD nephropathy (SCDN) affects 5% to 30% of patients with SCD and is associated with early mortality.3-7 SCDN is characterized by renal hypertrophy, hyposthenuria, proximal tubular dysfunction, glomerular abnormalities, and ultimately renal failure.8 Micro- or macroalbuminuria occurs in up to 68% of patients with SCD9 and 4% to 12% of adults with SCD progress to end-stage renal disease.4,10 SCDN detection relies on markers of relatively late-stage disease processes, namely proteinuria and decreased glomerular filtration rate (GFR). Consequently, it is difficult to identify those at risk before significant organ damage. Markers for early detection of renal dysfunction, such as genetic modifiers, are essential to guide development of effective treatments.
Although SCDN shares characteristics with other nephropathies, the underlying mechanisms of SCDN are unique.11,12 The hypoxic environment of the renal medulla promotes hemoglobin S polymerization, and repeated red cell sickling leads to ischemic injury and occluded medullary vasculature, manifesting as hematuria, papillary necrosis, and tubular acidosis. In addition, free heme can cause local inflammatory reactions and lead to renal failure.13 Damage-triggered release of vasodilating prostaglandins and nitric oxide then exacerbate hyperfiltration, an early marker of kidney stress that occurs at higher rates in SCD compared with the general population due to chronic hemolysis and anemia.14,15 Moreover, murine models of renal ischemia have shown that transgenic sickle mice display significantly greater renal injury than wild-type mice.16 Therefore, we hypothesize that the genetic underpinnings of SCDN may be distinct from other forms of kidney disease.
We previously reported the association of variants in APOL1 and MYH9 with proteinuria in patients with SCD,17 an association replicated in independent SCD cohorts.18-20 Reports from other nephropathies attributed this association to APOL121-24 owing to the absence of protein-altering MYH9 variants. However, using an in vivo zebrafish model, we demonstrated a functional interaction between myh9 and apol1 specifically under anemic stress,25 highlighting the importance of studying SCDN as a unique nephropathy.
Despite substantial effects, variants at the MYH9/APOL1 locus explain only a portion of SCDN risk,17 suggesting additional contributing genetic factors exist. Although other genetic variants have been associated with SCDN, these remain largely unreplicated.18-20,26,27 Large genetic studies of chronic kidney disease (CKD) and other renal phenotypes in non-SCD cohorts have uncovered many disease-associated loci,28-33 but their relevance to SCDN is unclear, particularly as most are from studies of primarily European ancestry.
To investigate the role of kidney-associated genetic loci in SCDN, we constructed polygenic risk scores (PRS) using 2 large meta-analyses of relevant renal phenotypes28,33 and tested them for association with proteinuria and estimated GFR (eGFR) in 2 cohorts of adult patients with SCD (Outcome Modifying Genes in SCD [OMG-SCD] and Walk-Treatment of Pulmonary Hypertension and SCD with Sildenafil Therapy [Walk-PHaSST]). Subsequently, we performed genome-wide association studies (GWAS) of proteinuria and eGFR to conduct, to the best of our knowledge, the largest meta-analyses of SCDN to date. Finally, functional annotation was performed to identify additional candidate single nucleotide polymorphisms (SNPs), perform gene-based analysis, and investigate potential regulatory roles for these novel loci.
Materials and methods
Participants and end points
Adult participants (aged ≥18 years) with renal outcomes and genetic data from 2 previously described SCD cohorts were utilized in this analysis, 576 from OMG-SCD6 and 502 from Walk-PHaSST.34 OMG-SCD participants were recruited for a cross-sectional observational study from 4 sickle cell centers in the southeastern United States. Walk-PHaSST participants were enrolled from 10 centers in the United States and United Kingdom during the screening phase of a trial of sildenafil. Participants provided informed consent and study protocols were approved by local institutional review boards or ethics committees.
At study enrollment, participants provided blood samples for genetic analyses and urine samples, collected at various times of day, for routine urinalysis at steady state. Baseline proteinuria was evaluated via dipstick analysis in OMG-SCD, and from either dipstick analysis or measured protein levels in Walk-PHaSST. Urine dipstick values of ≥1 (corresponding to ≥30 mg/dL) and measured urinary protein levels of ≥30 mg/dL were categorized as positive, whereas no evidence or trace amounts of urinary protein was considered negative.17 Baseline eGFR was calculated using the “Modification of Diet in Renal Disease” study definition.35 Patients were grouped according to hemoglobin beta genotype as a surrogate for SCD severity as follows: SS and Sβ0 thalassemia compared with SC and Sβ+ thalassemia.
Genotyping
DNA was extracted from blood using standard procedures, and genotyping was performed using Illumina Human610-Quad BeadChips (Illumina, Inc, San Diego, CA), at Duke University for OMG-SCD and at Boston University for Walk-PHaSST.36 Quality control procedures were performed using PLINK version 1.0737 and samples were excluded for call rates of <98%, sex discrepancies, and duplicates or first-degree relatives as determined by pairwise identity-by-descent estimates. SNPs meeting the following criteria were removed: call rates of <97%, minor allele frequency (MAF) of <5%, and Hardy-Weinberg equilibrium P values of <10–6. MYH9 rs16996672, APOL1 G1 (rs73885319/rs60910145), and APOL1 G2 (rs71785313) were genotyped in OMG-SCD, as previously described.17
Genetic imputation
A global reference panel from the 1000 Genomes Project38 was used to infer missing genotypes in both cohorts. Samples were prephased using SHAPEIT version 139 and genotypes imputed using IMPUTE2 version 2.1.2.40 Imputed SNPs with certainty of <90% were removed. The average imputation accuracy (comparing imputed calls to true genotypes) was 98.6% for OMG-SCD and 97.6% for Walk-PHaSST. Using only SNPs present in both cohorts with MAF of >5%, 2 887 736 SNPs were analyzed.
Statistical analysis
Differences of percentages and means between OMG-SCD and Walk-PHaSST were analyzed using χ2 tests and generalized linear models in SAS version 9.4 (SAS Institute, Cary, NC). Principal component analysis of genome-wide SNPs was performed using EIGENSOFT version 4.241 and population outliers were removed. Visual inspection of scree plots indicated that 2 principal components (PCs) were sufficient to control for population substructure in OMG-SCD; 6 were necessary in Walk-PHaSST (supplemental Figure 1). All statistical models for proteinuria included age, genotype PCs, hemoglobin genotype, and Duffy genotype42 as covariates; all models for eGFR included genotype PCs and hemoglobin genotype.
Two large transethnic meta-GWAS, 1 for urinary albumin-to-creatinine ratio (UACR; PRSUACR) from the CKDGen Consortium,33 and 1 for eGFR (PRSeGFR) from the COGENT-Kidney Consortium,28 were used to construct PRSs, weighted by effect size, in both SCD cohorts using PRSice version 2.1.6.43 Default parameters for linkage disequilibrium (LD) clumping were employed. To determine the maximizing P value threshold from each meta-GWAS, we generated 1001 PRSs in OMG-SCD and Walk-PHaSST with thresholds ranging from P = .0001 to P = 1, in increments of .001, as described previously.44 Each PRS was then tested for association with proteinuria and eGFR using logistic and linear regression, respectively, in each cohort (R glm), followed by a fixed-effects meta-analysis in R metafor.45 A Bonferroni correction was applied to account for the 1001 thresholds examined (P = 5 × 10–5). The most significant PRS from meta-analysis was retained and forest plots were generated using metafor.
GWAS was performed in OMG-SCD and Walk-PHaSST using additive genetic models in PLINK to test for association between each SNP and proteinuria with logistic regression, and each SNP and eGFR using linear regression. Sex was also covaried in analysis of X chromosome SNPs using the --sex flag in PLINK. LD clumping was used to reduce redundancy from highly correlated SNPs in OMG-SCD, and the resulting set of independent SNPs was then interrogated in Walk-PHaSST, for each phenotype. Importantly, this process was conducted in reverse (independent SNPs identified in Walk-PHaSST and applied to OMG-SCD) and the results were largely the same, indicating that the 2 cohorts have similar ancestral make-up. Following LD clumping, the number of SNPs analyzed was 257 059 for proteinuria and 256 859 for eGFR. Meta-analysis was performed using METAL46 and false discovery rate (FDR) q-values were generated using R qvalue.47 FDR q-values ≤0.05 were deemed significant.
Post-GWAS annotation was performed using tools implemented in FUMA,48 including identification of candidate SNP sets in high LD with lead SNPs for each GWAS region regardless of MAF, culling known and predicted regulatory elements (RegulomeDB),49 and gene-based analysis using MAGMA.50 SNPs with FDR q-value ≤0.05 were input as lead SNPs and candidate SNP sets were identified using the 1000 Genomes African ancestry reference panel.38 To fully interrogate the genome-wide significant (GWS) regions, candidate SNP sets along with lead SNPs were queried for association with human kidney gene expression in the NephQTL database,51 a public resource consisting of expression quantitative trait loci (eQTL) in glomerular and tubulointerstitial human kidney tissue from individuals with nephrotic syndrome.
To evaluate the impact of the MYH9/APOL1 locus on the novel SCDN loci identified, we performed a sensitivity analysis, which included either MYH9 rs16996672, APOL1 G1, APOL1 G2, or the APOL1 G1/G2 recessive model (G1/G2) as covariates in the aforementioned regression models for proteinuria and eGFR.
Finally, we utilized a subset of OMG-SCD participants (n = 193) with available longitudinal eGFR to assess rapid renal decline, as previously described.52 Briefly, trajectories of eGFR decline were obtained from beta estimates of steady-state eGFR regressed on time (years) for each individual, and rapid eGFR decline was defined by a slope of ≥3 mL/min per 1.73 m2. Here, we tested the proteinuria and eGFR GWS hits for association with rapid renal decline using logistic regression and controlling for genotype PCs and hemoglobin genotype in SAS version 9.4. A Bonferroni correction was applied to adjust for multiple testing (n = 6 SNPs, pbon = 8 × 10–3).
Results
Study participants
Participant characteristics are shown in Table 1. OMG-SCD participants were younger (33.9 years vs 37.6 years, P < 10–4) and had a higher percentage of hemoglobin SS or Sß0 disease (84.55% vs 72.46%, P < 10–4) compared with those in Walk-PHaSST. The cohorts did not differ by sex (P = .94). OMG-SCD participants had more proteinuria (26.0% vs 18.1%, P = .01) but did not differ from Walk-PHaSST participants by mean eGFR (P = .41) or percentage of eGFR <60 (P = .37). Age, hemoglobin genotype, and mean eGFR only differed by cohort in patients without proteinuria.
Relevance of kidney-associated loci to SCDN
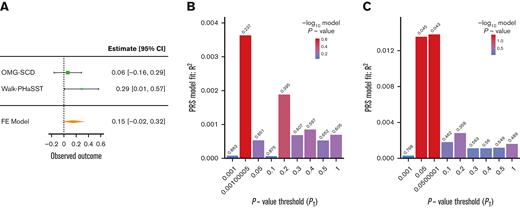
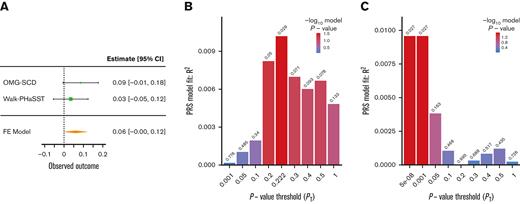
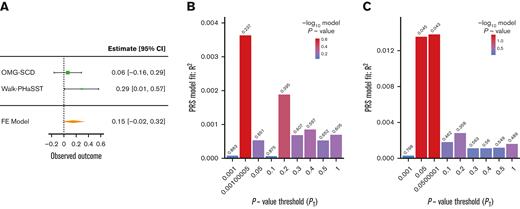
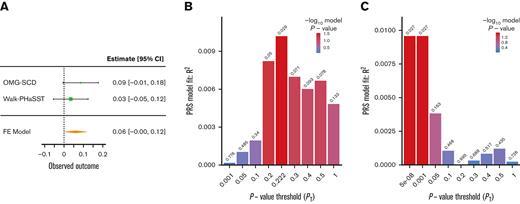
Two PRSs generated using summary statistics from large transethnic GWAS for UACR (PRSUACR)33 and eGFR (PRSeGFR)28 were evaluated for association with proteinuria and eGFR in SCD cohorts. Both PRSs trended toward association with the respective outcome, PRSUACR with proteinuria (P = .09 at PT = 0.05) and PRSeGFR with eGFR (P = .07 at PT = 0.579) in meta-analysis (Figures 1A and 2A). These results indicate that the UACR GWAS SNPs from CKDGen most predictive of proteinuria in our cohorts were those with P < .05 (PT = 0.05) but that prediction in SCD was not significant (P = .09). Likewise, the eGFR GWAS SNPs from COGENT-Kidney most predictive of eGFR in our cohorts were those with P < .579 (PT = 0.579) but the prediction in SCD was not significant (P = .07). Although each model displayed suggestive evidence of association, the model fit (R2) for each phenotype was poor in both OMG-SCD (PRSUACR and proteinuria, R2 = 4 × 10–3; PRSeGFR and eGFR, R2 = 0.01; Figures 1B and 2B) and Walk-PHaSST (PRSUACR and proteinuria, R2 = 0.01; PRSeGFR and eGFR, R2 = 0.01; Figures 1C and 2C).
PRS analysis for proteinuria using summary statistics from the CKDGen Consortium UACR GWAS. (A) Forest plot depicting the FE proteinuria meta-analysis of OMG-SCD and Walk-PHaSST generated using metafor at the maximizing P value threshold (PT) = .05. (B) Bar plot depicting model fit (R2) of the CKDGen Consortium UACR PRS predicting proteinuria in OMG-SCD at a wide range of P value thresholds. (C) Bar plot depicting model fit (R2) of the CKDGen Consortium UACR PRS predicting proteinuria in Walk-PHaSST at a wide range of P value thresholds. Model fit for proteinuria (depicted in B-C) was estimated using Nagelkerke pseudo R2. FE, fixed effects.
PRS analysis for proteinuria using summary statistics from the CKDGen Consortium UACR GWAS. (A) Forest plot depicting the FE proteinuria meta-analysis of OMG-SCD and Walk-PHaSST generated using metafor at the maximizing P value threshold (PT) = .05. (B) Bar plot depicting model fit (R2) of the CKDGen Consortium UACR PRS predicting proteinuria in OMG-SCD at a wide range of P value thresholds. (C) Bar plot depicting model fit (R2) of the CKDGen Consortium UACR PRS predicting proteinuria in Walk-PHaSST at a wide range of P value thresholds. Model fit for proteinuria (depicted in B-C) was estimated using Nagelkerke pseudo R2. FE, fixed effects.
PRS analysis for eGFR using summary statistics from the COGENT-Kidney Consortium eGFR GWAS. (A) Forest plot depicting the FE eGFR meta-analysis of OMG-SCD and Walk-PHaSST generated using metafor at the maximizing P value threshold (PT) = .579. (B) Bar plot depicting model fit (R2) of the COGENT-Kidney Consortium eGFR PRS predicting eGFR in OMG-SCD at a wide range of P value thresholds. (C) Bar plot depicting model fit (R2) of the COGENT-Kidney Consortium eGFR PRS predicting eGFR in Walk-PHaSST at a wide range of P value thresholds. FE, fixed effects.
PRS analysis for eGFR using summary statistics from the COGENT-Kidney Consortium eGFR GWAS. (A) Forest plot depicting the FE eGFR meta-analysis of OMG-SCD and Walk-PHaSST generated using metafor at the maximizing P value threshold (PT) = .579. (B) Bar plot depicting model fit (R2) of the COGENT-Kidney Consortium eGFR PRS predicting eGFR in OMG-SCD at a wide range of P value thresholds. (C) Bar plot depicting model fit (R2) of the COGENT-Kidney Consortium eGFR PRS predicting eGFR in Walk-PHaSST at a wide range of P value thresholds. FE, fixed effects.
GWAS for novel SCDN loci
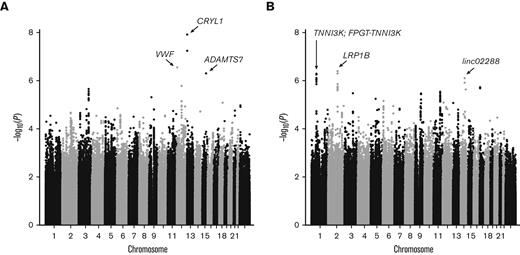
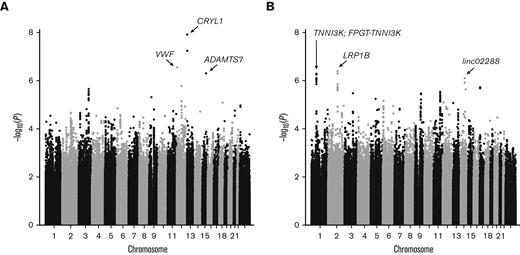
Owing to little overlap between known kidney disease loci and SCDN, we performed discovery GWAS of proteinuria and eGFR in OMG-SCD and Walk-PHaSST. No systematic bias was observed as the genomic inflation factors were well controlled (λGC = 1.02 and 1.01 for proteinuria and eGFR, respectively; supplemental Figure 2A-B). Meta-analysis results are depicted in Figure 3. Three GWS associations were identified for proteinuria: rs9315599 in CRYL1 (P = 7.13 × 10–9, q = 1.2 × 10–3), rs2238104 in VWF (P = 2.86 × 10–7, q = 0.02), and rs3743057 in ADAMTS7 (P = 5.09 × 10–7, q = 0.03). Functional annotation identified 4 candidate SNPs for CRYL1 and 10 candidate SNPs for ADAMTS7 (2 of which reside in nearby MORF4L1, supplemental Table 1). No additional SNPs tagged the VWF locus. GWAS for eGFR identified 3 GWS regions upon meta-analysis: rs1968911 in LRP1B (P = 4.14 × 10–7, q = 0.05), rs4903539 in linc02288 (P = 8.11 × 10–7, q = 0.05), and rs7526762 in FPGT-TNNI3K/TNNI3K (P = 9.12 × 10–7, q = 0.05). Functional annotation revealed 38 candidate SNPs for LRP1B, 4 SNPs for linc02288, and 15 SNPs for FPGT-TNNI3K/TNNI3K (supplemental Table 1). Cohort-specific results for the GWS loci are provided in supplemental Table 2.
Manhattan plots of GWAS meta-analysis. Results for (A) proteinuria and (B) eGFR. GWS loci are labeled with arrows pointing to the lead SNP for each locus. Genes harboring GWS loci (q ≤ 0.05) are noted.
Manhattan plots of GWAS meta-analysis. Results for (A) proteinuria and (B) eGFR. GWS loci are labeled with arrows pointing to the lead SNP for each locus. Genes harboring GWS loci (q ≤ 0.05) are noted.
Novel SCDN loci are independent of APOL1-associated risk
The GWS associations persisted in OMG-SCD even after controlling for MYH9 or APOL1 risk variants (Table 2). MYH9 rs16996672, APOL1 G1, and APOL1 G1/G2 were all associated with proteinuria but not as strongly as the CRYL1 or ADAMTS7 SNPs when included together in a linear model. VWF SNP rs2238104 remained associated with proteinuria when controlling for MYH9 or APOL1 risk variants but its effect was attenuated. APOL1 G2 was not associated with proteinuria when any of the GWS SNPs were included. Conversely, none of the MYH9, APOL1 G2, or APOL1 G1/G2 variants were associated with eGFR when included in models with FPGT-TNNI3K/TNNI3K, LRP1B, or linc02288 SNPs. APOL1 G1 was associated with eGFR, and its effect was stronger than that of the FPGT-TNNI3K/TNNI3K SNP but not as strong as the LRP1B and linc02288 SNPs when included in the same linear model. These results clearly support the presence of additional genetic risk factors in SCDN, beyond those explained by MYH9 and APOL1.
Putative functionality of SCDN loci
To evaluate putative functionality of the GWS loci, we queried publicly available databases (RegulomeDB and NephQTL) for known regulatory function and kidney-specific eQTLs (Table 3). CRYL1 SNP rs4770035, which is in high LD with lead SNP rs9315599, disrupts several transcription factor (TF) binding sites, including FOXl1. It is also an eQTL for nearby GJB2 in kidney tubulointerstitium (P = 8.6 × 10–3). In VWF, rs2238104 is an eQTL for CHD4 and CD9 in glomerulus (P = 7.1 × 10–3 and P = .02, respectively) and GAPDH in tubulointerstitium (P = .01). The lead SNP for ADAMTS7, rs3743057, is an eQTL for ANKRD34C and MORF4L1 in tubulointerstitium (P = .01 and P = .05, respectively). Finally, rs7182809 in MORF4L1, an LD SNP for the ADAMTS7 locus, overlaps several TF binding sites in HEK293 cells, TF binding motifs for MAFK and RREB1, and exhibits a chromatin state of strong transcription in kidney.
GWS loci identified for eGFR also display evidence for regulatory function. rs12373750, an LD SNP for LRP1B, overlaps TF binding motifs for MAFB and MAFK. In linc02288, lead SNP rs4903539 resides in an open chromatin peak and exhibits a chromatin state of strong transcription. Finally, rs4903539 disrupts several TF binding motifs including NFATC1, RARA, RARB, and RARG and is an eQTL for NGB in tubulointerstitium (P = .04).
Novel SCDN loci and rapid renal decline
We assessed the novel GWS loci for association with rapid renal decline, another kidney disease manifestation that we have described previously52 (supplemental Table 3). In addition to increased risk for proteinuria, OMG-SCD participants with CRYL1 rs9315599 risk alleles were 3.8 times as likely as those without risk alleles to have rapidly declining eGFR (P = 4 × 10–3), an association surpassing correction for multiple testing. Also, those with the LRP1B rs1968911 G allele, which is associated with lower eGFR, were less likely to display rapid renal decline (odds ratio, 0.41, P = .01). Concordant with this finding, patients displaying hyperfiltration at baseline (eGFR >130 mL/min per 1.73 m2) had a steeper rate of eGFR decline compared with those with lower baseline eGFR (−2.92 mL/min per 1.73 m2 vs −1.44 mL/min per 1.73 m2, respectively, P = .02). No other GWS SNPs were associated with rapid renal decline (P > .05).
Discussion
We provide evidence for several new SCDN genes based on, to the best of our knowledge, the largest meta-analysis of adults with SCD to date. The strongest association was rs9315599 in crystallin lambda-1 (CRYL1) with risk for proteinuria. CRYL1 encodes an enzyme that catalyzes the dehydrogenation of L-gulonate into dehydro-L-gulonate and is highly expressed in human kidney.53 L-gulonate and its precursor, D-glucuronic acid, have been previously associated with polycystic kidney disease, CKD, and end-stage renal disease.54-56 Functional annotation supports a regulatory role for CRYL1 in kidney function. rs4770035, which is in high LD with rs9315599, disrupts binding of FOXl1, a TF implicated in renal cell carcinoma (RCC) in humans57,58 and differentiation of nephron progenitors to podocytes in mice.59 Renal medullary carcinoma, a subtype of RCC, is strongly associated with sickle cell trait and SCD.60 In addition, the rs4770035 C allele, which increased risk for proteinuria, also increases expression of GJB2 in kidney tubulointerstitium, a protein associated with sepsis-induced acute injury of renal tubular epithelial cells.61 These data suggest that the CRYL1 GWS SNP may affect kidney function through regulation of nearby GJB2.
We also report association between proteinuria and rs2238104 in VWF, the gene encoding von Willebrand factor that, along with ADAMTS13, is involved in platelet adhesion and thrombosis. Not only implicated in vaso-occlusive crises,62 VWF and ADAMTS13 are critical to acute ischemia-reperfusion kidney injury in mice.63,64 Furthermore, thrombospondin-1, which interacts with VWF and ADAMTS13 to promote vascular adhesion, is also increased during vaso-occlusive crises.65,66 We previously showed that genetic variants in THBS1, the gene encoding thrombospondin-1, are associated with pulmonary hypertension in SCD, a condition comorbid with SCDN.67 In addition, VWF variant rs2238104 is an eQTL for CHD4 and CD9 in glomerulus. CHD4 has been identified as a γ-globin silencer68,69 controlling the switch from fetal to adult hemoglobin, an event significantly associated with disease severity and survival in SCD.3,70,71 Regulation of hemoglobin switching may be important to kidney function, as low hemoglobin F has been associated with microalbuminuria in children with SCD.72 rs2231804 is also an eQTL for CD9, a tetraspanin implicated in focal segmental glomerulosclerosis and crescentic glomerulonephritis such that CD9 expression in parietal epithelial cells is associated with disease in mouse and human, and Cd9 gene editing in mouse can thwart glomerular damage in these nephropathies.73 Therefore, the association between rs2231804 in VWF and proteinuria may reflect its regulatory effect on nearby genes important not only to kidney function but also to SCD hallmarks, such as vaso-occlusion and hemoglobin switching.
Finally, we identified an association between proteinuria and rs3743057 in ADAMTS7. LD at this locus includes SNPs in nearby MORF4L1, whose expression levels in kidney tubulointerstitium are associated with rs3743057. Moreover, rs7182809, which is in high LD with rs3743057, disrupts binding of MAFK and RREB1, TFs previously implicated in renal disease.74,75 Several studies have reported robust association between genetic variation at chromosome 15q25 (ADAMTS7-MORF4L1) and coronary artery disease,76-79 a well-known comorbidity of CKD.80,81 Indeed, ADAMTS7 expression is increased in elderly mice with angiotensin II–mediated kidney injury.82
In the eGFR GWAS, SNPs in lipoprotein receptor–related protein 1b (LRP1B), a putative tumor suppressor gene previously associated with RCC,83,84 and linc02288, a long noncoding RNA, also display evidence for a regulatory role in SCDN. rs12373750, an LD SNP for LRP1B rs1968911, falls in TF binding motifs for MAFB and MAFK, factors previously associated with focal segmental glomerulosclerosis and diabetic nephropathy.74,85 rs4903539 in linc02288 might alter the binding motifs for TFs integral to kidney function, such as NFATC1,86 and retinoic acid receptors RARA, RARB, and RARG.87,88 More research is needed to understand the potential role that these TFs might play in SCDN.
Many genes previously reported from SCDN candidate gene studies18-20,26 were nominally significant in our study (supplemental Tables 4 and 5). SNPs in BMPR1B, CUBN, PKD1L2, and MYH9 were associated with both proteinuria and eGFR, whereas SNPs in CD163 and APOL1 were associated with only proteinuria, and SNPs in HMOX1 were associated with only eGFR. No association was detected for SNPs in AGGF1, CYP4B1, or TOR2A. Failure to consistently replicate previous associations could be because of power limitations in the SCD cohorts but it also underscores the complex genetic architecture of SCDN. None of the proteinuria GWS loci identified in this study were associated with eGFR; none of the eGFR GWS loci were associated with proteinuria. Of the top hits, only rs9315599 in CRYL1 and rs1968911 in LRP1B were associated with rapid renal decline in a smaller OMG-SCD subset with longitudinal eGFR. As each renal phenotype represents a different time point in the progression through kidney impairment to renal failure, we theorize there are likely different genetic modifiers of SCDN acting at different stages. Larger sample sizes are needed to disentangle the complex genetic mechanisms of SCDN.
PRSs calculated using effect sizes from the non-SCD CKDGen33 and COGENT-Kidney28 Consortiums trended toward association with proteinuria and eGFR in our SCD cohorts, supporting the polygenic nature of renal dysfunction. However, goodness-of-fit statistics indicated these PRSs are poor predictors of SCDN, possibly owing to several factors. First, PRS analysis is highly dependent on genetic architecture, LD, and allele frequencies of the base and target data sets.89-91 Although the COGENT-Kidney data set had the largest number of African ancestry subjects of any available eGFR GWAS, it still only amounted to 2.63% of the sample. Likewise, the CKDGen data set comprised only 1.2% African ancestry participants. This likely influenced our nonreplication of individual lead SNPs from the CKDGen UACR and COGENT-Kidney eGFR GWAS (supplemental Tables 6 and 7). Notably, the GWS SNPs identified have very different allele frequencies across ancestral populations. For example, the CRYL1 rs9315599 G allele frequency is 42.71% in non-Finnish Europeans but only 8.78% in African/African Americans (gnomAD version 2.1.1). Second, participants in the CKDGen and COGENT-Kidney Consortium GWAS did not have SCD. As previously noted, SCDN may arise via different mechanisms than other nephropathies, and patients with SCDN have a steeper rate of renal decline compared with other African Americans.52 Recently, Khan et al92 sought to optimize a CKD PRS for participants of African ancestry that includes APOL1 risk genotypes. When tested in our SCD cohort, we still did not observe a significant association with eGFR (P = .19). Similarly, controlling for APOL1 genotypes in models associating PRSUACR with proteinuria and PRSeGFR with eGFR made no appreciable difference in OMG-SCD. Moreover, we have shown that the novel loci identified display stronger association with SCDN than the well-established APOL1 risk variants, suggesting SCDN is governed by distinct genetic drivers. In fact, many loci identified here have been implicated not only in kidney dysfunction but also in biological processes integral to SCD.
Research focused on individuals of African ancestry is lacking.93 Because individuals of African ancestry are ∼4 times as likely as individuals of European ancestry to progress to kidney failure,94 it is imperative that future studies prioritize collection of African ancestry participants. Recently, the first African GWAS of kidney function was performed in Uganda,32 but the sample size is dramatically smaller than the CKD consortia studies performed in mostly European ancestry individuals (3288 vs 1 201 909).31 Interestingly, one of the most significant eGFR associations identified in East Africans was rs141845179 in hemoglobin beta (HBB), which tags the sickle cell mutation (rs334). Moreover, rs334 was the most significant SNP in the African ancestry subset of the UACR GWAS utilized in PRS analysis here. Consistent with these findings, hemoglobin beta genotype is significantly associated with proteinuria and eGFR in our cohorts, and thus was included as a covariate in all models. Not restricted to homozygous SCD, African Americans with sickle cell trait also display increased risk for CKD, albuminuria, and eGFR decline.95 Larger genetic studies of African ancestry renal disease are necessary not only to further our understanding of the racial disparities observed in kidney failure but also to address the potentially distinct biomechanisms of SCDN.
This study is not without limitations. Because these cohorts utilized previously collected data, we were restricted to using proteinuria largely measured via dipstick at 1 time point. Future studies should include more rigorous outcomes, such as UACR, and repeated tests. Second, replication in additional adult SCD cohorts of similar ancestral background is essential to corroborate the associations reported in this study. Moreover, analysis of rare genetic variants at these loci could uncover signals in protein-coding regions. Finally, molecular follow-up should be performed to determine the functional consequences, if any, of the novel SCDN loci reported here.
In summary, patients with SCD are living longer but increasingly experience significant complications, including SCDN. Patients would benefit greatly from earlier detection making interventions more efficacious. Here, we report 6 GWS loci for markers of renal dysfunction in the largest analysis of SCDN to date. Using publicly available human kidney data, we have showed that GWS SNPs could be regulating expression of genes in pathways that are important not only to renal function but also SCD. These results provide new targets for functional follow-up that could be tested prospectively and potentially used for identification of patients with SCD that are at risk, before onset of kidney dysfunction. These findings also further the prospect of prophylactic therapeutics designed to forestall progression to clinically significant kidney disease.
Acknowledgments
The authors thank all the individuals for participating in this study.
This work was supported, in part, by grants 2015131 and 2012126 from the Doris Duke Charitable Foundation, R01HL68959 and R01HL079915 from the National Heart, Lung and Blood Institute, and DK110104 and DK124836 from the National Institute of Diabetes and Digestive and Kidney Diseases. The Walk-PHaSST cohort was collected as a clinical trial (#NCT00492531) funded with federal funds from the National Heart, Lung, and Blood Institute, National Institutes of Health, and Department of Health and Human Services under contract HHSN268200617182C.
Authorship
Contribution: A.E.A.-K. and M.J.T. designed the research study; V.R.G., Y.Z., and M.T.G. generated the Walk-PHaSST data; K.L.S. generated the OMG-SCD genotype data; M.E.G., K.N.E., K.L.S., A.E.A.-K., and M.J.T. analyzed the data; M.E.G. drafted the manuscript; and all authors edited and approved the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Allison E. Ashley-Koch, Duke Molecular Physiology Institute, 300 N Duke St, Durham, NC 27710; e-mail: allison.ashleykoch@duke.edu.
References
Author notes
The cohorts used in this analysis contain legacy samples that have IRB restrictions on sharing. For data access, please contact allison.ashleykoch@duke.edu for OMG-SCD and gladwinmt@upmc.edu or zhanyx@upmc.edu for Walk-PHaSST.
The full-text version of this article contains a data supplement.