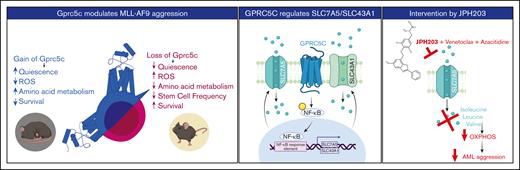
Gprc5c drives AML progression by regulating the BCAAs transporters SLC7A5 and SLC43A1.
Inhibition of SLC7A5 using JPH203 elicits strong antileukemic effects in AML, which are strengthened by venetoclax and azacitidine treatment.
Visual Abstract
Leukemia stem cells (LSCs) share numerous features with healthy hematopoietic stem cells (HSCs). G-protein coupled receptor family C group 5 member C (GPRC5C) is a regulator of HSC dormancy. However, GPRC5C functionality in acute myeloid leukemia (AML) is yet to be determined. Within patient AML cohorts, high GPRC5C levels correlated with poorer survival. Ectopic Gprc5c expression increased AML aggression through the activation of NF-κB, which resulted in an altered metabolic state with increased levels of intracellular branched-chain amino acids (BCAAs). This onco-metabolic profile was reversed upon loss of Gprc5c, which also abrogated the leukemia-initiating potential. Targeting the BCAA transporter SLC7A5 with JPH203 inhibited oxidative phosphorylation and elicited strong antileukemia effects, specifically in mouse and patient AML samples while sparing healthy bone marrow cells. This antileukemia effect was strengthened in the presence of venetoclax and azacitidine. Our results indicate that the GPRC5C–NF-κB-SLC7A5–BCAAs axis is a therapeutic target that can compromise leukemia stem cell function in AML.
Introduction
Leukemia progression is fueled by self-renewing and relapse-causing leukemia stem cells (LSCs) that exhibit a stem cell-like gene expression signature.1,2 Similar to dormant hematopoietic stem cells (HSCs), LSCs rarely divide and reside in a long-term quiescent, low-metabolic state, which confers resistance to cytotoxic agents that primarily target replicating cells.3 The reversibility of the dormant to the active form of LSCs drives acute myeloid leukemia (AML) recurrence months to years after initial remission.4
The unique metabolic profile of LSCs can be exploited for pharmacological therapies. For instance, LSCs have low reactive oxygen species (ROS) levels and rely on amino acid metabolism to sustain oxidative phosphorylation (OXPHOS). In contrast, HSCs mainly rely on glycolysis and increase OXPHOS during differentiation.5-7 This onco-metabolic status makes LSCs an ideal cellular target for venetoclax and azacitidine, drugs that decrease amino acid uptake and reduce OXPHOS.3,8 Specific amino acids have been linked to increased aggressiveness in multiple hematologic malignancies.9-11 The neutral amino acid transporter Slc1a5 has been implicated as an antileukemia target. The loss of Slc1a5 in AML slightly reduced glutamine abundance, but the levels of branched-chain amino acids (BCAAs), cysteine, and threonine were more severely reduced.12 Branched-chain amino acid transaminase 1 (BCAT1) is a cytosolic aminotransferase for BCAAs. In AML and chronic myeloid leukemia, BCAT1 expression predicted disease outcomes. Diminished expression of BCAT1 reduced disease burden but could be rescued with BCAA supplementation.9,10 Recently, valine dietary restriction was identified as a mechanism to reduce T-cell acute lymphoblast leukemia (T-ALL) burden by blocking the translation of subunits of the electron transport chain (ETC) complex I, thereby inhibiting OXPHOS.13 The genetic or pharmacological inhibition of Slc7a5, which is responsible for the transport of BCAAs, is sufficient to reduce the aggressiveness of T-ALL.14 However, the role of the BCAA amino acid transporters in AML remains elusive.
Recently, we identified the transmembrane receptor GPRC5C as a key regulator of mouse and human HSCs15,16. However, the functionality of GPRC5C in AML is yet to be studied. Here, we show that GPRC5C regulates leukemia by increasing intracellular BCAAs through NF-κB-mediated transcription of SLC7A5 and SLC43A1. Treatment with the SLC7A5 inhibitor JPH203 in combination with venetoclax and azacitidine decreased the in vitro leukemia burden. Collectively, these results provide a preclinical rationale for the development of JPH203 as a novel therapeutic option for AML.
Methods
Human ethics
Samples were obtained during routine diagnostic procedures after obtaining written consent from patients or parents/guardians in the case of minors. All analysis of human data were carried out in accordance with the Declaration of Helsinki. This study was approved by the Institutional Ethical Review Board of Hospital Clinic of Barcelona (HCB/2018/0020).
Mouse models
All the mice were bred in-house in individually ventilated cages in an animal facility at the Max Planck Institute of Immunobiology and Epigenetics. The mice were euthanized by cervical dislocation according to the German guidelines. Animal procedures were performed according to protocols approved by the German authorities, Regierungspräsidium Freiburg (the euthanizing of animals for scientific purposes according to section 4 (3) of the German Animal Protection Act; animal protocol numbers G-18/142, G-19/74, and G20/124).
Generation of MLL-AF9 mice
To generate MLL-AF9-Gprc5c-OE mice, lineage–Sca1+ckit+ (LSK) cells were transduced with MLL-AF9–puro fusion oncogene. After 72 hours of puromycin selection, lentivirus for Gprc5c-OE-GFP was introduced. Similarly, to generate MLL-AF9-Gprc5c–knockout (KO), LSK cells from Gprc5c-KO mice were isolated and transduced with MLL-AF9–green fluorescent protein (MLL-AF9–GFP). After successful transduction, 2.5 × 105 GFP+ cells were sorted and injected into recipient mice that were irradiated with 6 Gy.
Isolation of mouse LSKs
Murine bone marrow (BM) cells were isolated from the femur, tibia, hip bone, and vertebra by gentle crushing in phosphate-buffered saline (PBS) using a mortar and pestle. Red blood cell lysis was performed using Ammonium-Chloride-Potassium (ACK) Lysing Buffer (Thermo Fisher Scientific) for 5 minutes at room temperature (RT). Dynabeads Untouched Mouse CD4 Cells Kit (Thermo Fisher Scientific) was used for lineage-negative enrichment, according to the manufacturer’s protocol. Briefly, the BM was stained with a 1:4 dilution of the lineage cocktail for 30 to 60 minutes at 4°C on a rotating wheel. Labeled cells were then incubated for 20 minutes with 400 μL per mouse of washed polyclonal sheep anti-rat IgG-coated Dynabeads. Depletion of lineage cells was performed using a magnet. Lineage-depleted BM cells were stained with lineage markers (Gr1, CD11b, B220, Ter119, CD4, and CD8a), ckit, Sca1, CD150, CD48, and CD34 and sorted on a FACSAria Fusion Flow Cytometer (BD Biosciences) for downstream analysis.
Cell cycle analysis (Ki-67)
The stained cells were fixed with BD Cytofix/Cytoperm Buffer (BD Biosciences) for 10 minutes at 4°C. Next, the cells were stained with intracellular Ki-67 at 1:100 (BD Biosciences) for 45 to 90 minutes at 4°C. Before cell cycle analysis on a BD LSRFortessa (BD Biosciences), the cells were stained in the dark with Hoechst 33342 or 4′,6-diamidino-2-phenylindole (Thermo Fisher Scientific) for 30 minutes at RT.
CellROX
Cells were incubated at 37°C with CellROX DeepRed at 1/500 (Thermo Fisher Scientific) in their corresponding media for 30 minutes. Subsequently, the cells were washed 3 times in PBS and stained for fluorescence-activated cell sorting (FACS) analysis using a BD LSRFortessa.
Annexin V staining
The cells (between 5 × 104 and 1 × 105) were treated as specified in the experimental scheme. After treatment, the cells were washed with annexin V staining buffer, stained with annexin V antibody in allophycocyanin (1/100) for 15 minutes at RT, and then washed. Before FACS analysis, the cells were stained with 4′,6-diamidino-2-phenylindole (1/1000), and CountBright Absolute Counting Beads (Thermo Fisher Scientific) were added.
Staining with pNF-κB
To fix and permeabilize the cells before immunofluorescent staining, the cells were treated with the Transcription Factor Buffer Set (BD Pharmingen) according to the manufacturer’s protocol. The cells were stained with pNF-κB (abcam number ab86299) for 1 hour, followed by staining with a secondary antibody with Alexa Fluor 594(Thermo Fisher no. A-11012).
Caspase-3 staining
Cells were fixed with BD Cytofix/Cytoperm Buffer for 10 minutes at 4°C. Next, cells were stained with intracellular capase-3 phycoerythrin (PE) at 1:100 (BD Biosciences; no. 570186) in PermWash solution (BD Biosciences) for 45 to 90 minutes at 4°C, and washed.
Single-cell division
Single cells were subjected to FACS into individual wells on 72-well Terasaki plates (Greiner Bio-One) containing 10 μL of the desired cell media. At 24 hours after sorting, the cells were evaluated for division under a light microscope.
Quantitative reverse transcription PCRanalysis
For real-time polymerase chain reaction (PCR), total RNA from 1000 to 2000 cells was isolated using the Arcturus PicoPuro RNA Isolation Kit (Thermo Fisher Scientific) according to the manufacturer’s guidelines. First-strand synthesis was reverse-transcribed using the SuperScript VILO complementary DNA (cDNA) Synthesis Kit (Thermo Fisher Scientific) according to the manufacturer’s guidelines. For quantitative reverse transcription PCR (qRT-PCR) analysis, Fast SYBR Green Master Mix was used on a StepONE Real-Time PCR System (Thermo Fisher Scientific) or QuantStudio 6 Flex (Thermo Fisher Scientific). RNA expression was normalized to Oaz1 and/or B2m housekeeping gene expression and presented as relative quantification (ratio = 2−ΔΔCT).
Virus production and primary cell infection
Lentiviral particles were produced in HEK293T cells. A total of 15 μg of the lentiviral vector was transiently packaged with 9 μg psPAX2 and 6 μg pMD2.G with jetPRIME (Polyplus) per 150 mm tissue culture dish. Transduced HEK293T cells were 80% confluent. Viral supernatant was collected at 48 hours after transfection. Virus concentration was achieved by ultracentrifugation through a 20% sucrose cushion for 120 minutes at 4°C and 30 000 rpm. For the induction of cells, concentrated viral particles were spinoculated at 1000 g for 90 minutes. Before spinoculation, the viral supernatant was removed, and the desired media was added with protamine sulfate.
For the production of retrovirus (MLL-MLL-AF9–Puro or GFP), 75% to 85% confluent Plat-E cells were transduced with 30 μg of retroviral vector in a 150 mm tissue culture dish. The viral supernatant was collected 48 hours after transfection and was filtered. The filtered viral supernatants were directly added to the cells and then spinoculated as described earlier.
Cell culture media
THP1, U937, and NB4 cells were cultured in RMPI 1640 (Thermo Fisher) with 10% fetal bovine serum (FBS). KG1 cells were cultured in Iscove modified Dulbecco medium (IMDM) with 20% FBS, and K562 cells were cultured in IMDM with 10% FBS. Samples of patient with primary AML were cultured in IMDM, 15% bovine serum albumin, insulin, and transferrin (StemCell Technologies), 100 ng/mL stem cell factor (SCF), 50 ng/mL FMS-like tyrosine kinase 3 ligand, 20 ng/mL interleukin-3, 20 ng/mL granulocyte colony-stimulating factor (PeproTech) 10−4 M β-mercaptoethanol, 500 nM SR1 (Stemcell Technologies), and 500 nM UM729 (Stemcell Technologies). Mobilized peripheral blood cells were cultured in StemSpan-ACF (Stemcell Technologies) containing SCF at 100 ng/mL, thrombopoietin at 50 ng/mL, FLT3-L at 100 ng/mL, Glutamax 1×, low density lipoprotein at 10 μg/mL, and 35 nM UM171.
Construction of lentiviral shRNAs targeting Slc7a5 and Slc43a1
Short hairpin RNA (shRNA) vectors targeting GPRC5C were designed according to the report by Pelossof et al.17
shRenilla: AAGGTATATTGCTGTTGACAGTGAGCGCAGGAATTATAATGCTTATCTATAGTGAAGCCACAGATGTATAGATAAGCATTATAATTCCTATGCCTACTGCCTCGGACTTCAAGGGGCTA
shSlc7a5.1565: TGCTGTTGACAGTGAGCGCCTGTAGGTTTTTATTTTCAAATAGTGAAGCCACAGATGTATTTGAAAATAAAAACCTACAGATGCCTACTGCCTCGGA
shSlc7a5.1569: TGCTGTTGACAGTGAGCGCAGGTTTTTATTTTCAAATCAATAGTGAAGCCACAGATGTATTGATTTGAAAATAAAAACCTATGCCTACTGCCTCGGA
shSlc43a1.1809: TGCTGTTGACAGTGAGCGCTCTGTGGATTTATAAATACTATAGTGAAGCCACAGATGTATAGTATTTATAAATCCACAGAATGCCTACTGCCTCGGA
shSlc43a1.1810: TGCTGTTGACAGTGAGCGCCTGTGGATTTATAAATACTAATAGTGAAGCCACAGATGTATTAGTATTTATAAATCCACAGATGCCTACTGCCTCGGA
Preparation of liver/spleen sections
Sections of the liver were isolated and fixed in 4% paraformaldehyde overnight at RT and subsequently embedded in paraffin. After fixation, the organs were embedded using PolyFreeze (Millipore Sigma) and frozen at 80°C. The sections were stained with hematoxylin and eosin for histological analysis.
Seahorse XF cell mito stress test
Mitochondrial stress tests were performed to measure oxygen consumption using the Seahorse Xfe96 Analyzer (Agilent) according to the manufacturer’s protocol. Cultivated cells were transferred to a poly-D-lysine–coated (Sigma), 96-well seahorse plate and incubated for at least 1 hour in a CO2-free incubator in XF Seahorse medium (XF Dulbecco's Modified Eagle Medium, 10mM glucose, 1mM pyruvate, and 2mM glutamine) (all Agilent) at 37°C. A Seahorse Xfe96 Analyzer was used to inject 10 μM oligomycin (Sigma), 15 μM Carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (Sigma), and rotenone 1 μM (Sigma) + antimycin A 10 μM (Sigma). The Seahorse Xfe96 Analyzer was used to measure the oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) after each injection.
Metabolomics
A total of 250 000 cells were washed in 3% glycerol solution and extracted in 10 μl of 80% acetonitrile or 100 μL of 80% cold methanol containing 13C yeast extract as an internal standard. The samples in methanol were vacuum concentrated. Metabolite quantification by liquid chromatography–tandem mass spectrometry was carried out using an Agilent 1290 Infinity II ultra-high-performance liquid chromatography in line with an Aglient 6495 QQQ-MS operating in MRM mode.
Calculation of CDI
A CDI value < 1 is synergistic, CDI = 1 is additive, and CDI > 1 indicates that the drugs are antagonistic.
Bulk RNA-seq
The RNA of 1000 freshly sorted cells was extracted using the Arcturus PicoPure kit (Applied Biosystems), and DNase treatment was performed (Qiagen). Subsequently, cDNA libraries were prepared using the SMART-Seq version 4 Ultra Low Input RNA kit (Takara Bio) with 12 cycles of amplification. Further, NEBNext Ultra II FS DNA kit and NEBNext Multiplex Oligos (New England BioLabs) were used to generate unique dual-barcoded sequencing libraries from cDNA libraries. To this end, 5 ng of cDNA library was fragmented for 22.5 minutes, adapters were ligated, and libraries were amplified for 8 cycles. RNA sequencing (RNA-seq) libraries were sequenced at a depth of 60 million reads, with a 100 bp paired end on a NovaSeq platform (Illumina).
Bulk RNA-seq analysis method: low-level processing
Raw fastq files were mapped against the mm10 reference genome using the mRNA-seq tool from the bioinformatics pipeline snakePipes.19 “Alignment” mode was used for mapping of sequenced reads using STAR (version STAR_2.7.4a),20 followed by the quantification of expression counts by featureCounts.21 DeepTools QC (version 3.3.2)22 was used for quality checking. Genes with an average expression higher than 100 counts in at least 1 condition were selected for further analysis. Differential expression was generated with DESeq223; results with a false discovery rate < 0.1 and a log2–fold change threshold ≥ 0.5 were considered significant.
Bulk RNA-seq analysis method: downstream analysis
The results of the differential expression calculated using DESeq2 package were visualized using the EnhancedVolcano. For evaluation of the expression of the previously published signatures of upregulated and downregulated genes in LSCs (Gal et al46, Gentles et al51) in the pairwise comparison between MLL-AF9-Gprc5c-Overexpression (AF9-Gpr-OE) and MLL-AF9-Control, Gene Set Enrichment Analysis (GSEA) was performed with the GSEA function of the clusterProfiler R package, and the enrichment profiles were plotted using the gseaplot2 function from the enrichplot R package.
Statistics and reproducibility
Statistical analysis was performed using the unpaired Student t test or 2-way analysis of variance with correction for multiple comparisons (using Tukey method or Šidák correction), unless otherwise stated. All data are presented as mean ± standard deviation, unless otherwise stated. GraphPad Prism version 9 was used for statistical analysis.
Kaplan-Meier curve based on GPRC5C expression
GPRC5C expression data, assessed by RNA sequencing, were available for 342 diagnostic samples included in the Leucegene cohort of patients with AML, as previously described.24 These cases included adult patients with de novo AML (excluding acute promyelocytic leukemia) who were treated with curative intent. The 75th expression percentile was used as the threshold value to dichotomize GPRC5C expression (high: ≥75th percentile; low: <75th percentile). GPRC5C expression data, assessed by RNA sequencing, were available for 172 patients from The Cancer Genome Atlas. The Kaplan-Meier survival curve was generated using Bloodspot.
Results
Elevated GPRC5C expression correlates with poor survival
High expression of GPRC5C construed shorter overall survival in both the Leucegene and BEAT AML cohorts (Figure 1A supplemental Figure 1A).24,25 Along these lines, GPRC5C was enriched in patients with the highest frequency of LSCs, and paired sample analysis showed a significant increase in GPRC5C levels upon disease relapse (supplemental Figure 1B,C)50. In-depth analysis of the European LeukemiaNet (ELN) 2017 risk or cytogenetic risk categorization revealed an overrepresentation of GPRC5C high-expressing patients within the most adverse groups (supplemental Figure 1D). Lastly, single-cell RNA sequencing data revealed that GPRC5C expression was enriched in AML cells with 17 LSC gene signature2,26 (supplemental Figure 1E). In summary, the transcriptomic data justified further investigation of GPRC5C in AML.
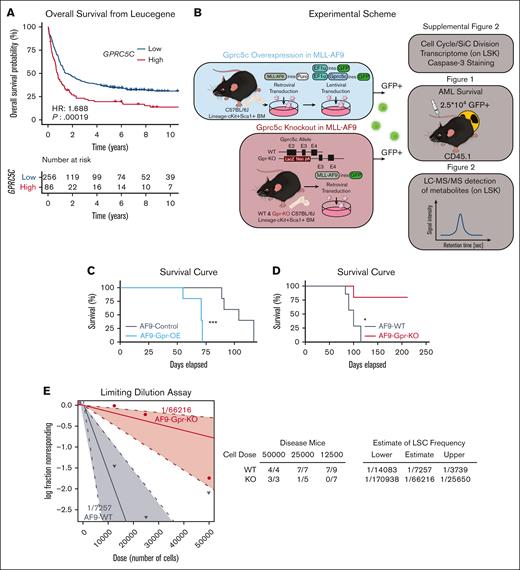
Gprc5c expression levels regulate the development of leukemia. (A) Overall survival according to GPRC5C expression in the Leucegene cohort. High expression levels of GPRC5C (GPRC5C high: ≥75th percentile, red curve) compared with low expression levels (GPRC5C low: <75th percentile, blue curve) are displayed by Kaplan-Meier curves. The P-value was obtained using the log-rank test for the comparison of survival curves. (B) Blue box: schematic representation of the experimental design to overexpress and assess the impact of increased Gprc5c levels in MLL-AF9. Red box: schematic representation of the experimental design to evaluate the effect of the loss of Gprc5c in MLL-AF9. (C) Survival curve depicting AML penetrance upon the gain of Gprc5c in MLL-AF9; n = 9. (D) Survival curve depicting AML penetrance upon loss of Gprc5c in MLL-AF9. Combination of 2 independent experiments; n = 5 to 7. (E) Limiting dilution assays for MLL-AF9-WT and MLL-AF9-Gprc5c-KO. Combination of 2 independent experiments. All represented by mean ± standard deviation. (C-D) Mantel-Cox test, ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P <.0001. For all experiments, at least 2 independent experiments were performed. ns, not significant.
Gprc5c expression levels regulate the development of leukemia. (A) Overall survival according to GPRC5C expression in the Leucegene cohort. High expression levels of GPRC5C (GPRC5C high: ≥75th percentile, red curve) compared with low expression levels (GPRC5C low: <75th percentile, blue curve) are displayed by Kaplan-Meier curves. The P-value was obtained using the log-rank test for the comparison of survival curves. (B) Blue box: schematic representation of the experimental design to overexpress and assess the impact of increased Gprc5c levels in MLL-AF9. Red box: schematic representation of the experimental design to evaluate the effect of the loss of Gprc5c in MLL-AF9. (C) Survival curve depicting AML penetrance upon the gain of Gprc5c in MLL-AF9; n = 9. (D) Survival curve depicting AML penetrance upon loss of Gprc5c in MLL-AF9. Combination of 2 independent experiments; n = 5 to 7. (E) Limiting dilution assays for MLL-AF9-WT and MLL-AF9-Gprc5c-KO. Combination of 2 independent experiments. All represented by mean ± standard deviation. (C-D) Mantel-Cox test, ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P <.0001. For all experiments, at least 2 independent experiments were performed. ns, not significant.
Gprc5c overexpression drives progression of MLL-AF9 driven AML (alias KMT2A::MLLT3)
Next, we investigated enhanced Gprc5c expression in AML using an engineered MLL-AF9 model (Figure 1B; supplemental Figure 2A). LSCs are defined by their quiescent phenotype compared with their more proliferative blast counterpart.27 Cell cycle analysis showed a mild increase in cells in the G0 phase in MLL-AF9-Gpr-OE compared with MLL-AF9 control cells (supplemental Figure 2B), which was further supported by delayed cell division and reduced Cdk6 expression (supplemental Figure 2C,D). Together, this indicated that Gprc5c induced a quiescent phenotype. Additionally, LSCs are characterized by lower ROS levels than those in blasts.3 When ROS levels were measured using CellROX, we observed reduced ROS levels in MLL-AF9-Gpr-OE compared with those in MLL-AF9-control cells (supplemental Figure 2E). Lastly, we measured the fitness of MLL-AF9-Gpr-OE cells by caspase-3 staining, which revealed that MLL-AF9-Gpr-OE enhanced in vitro survival (supplemental Figure 2F).
We generated a transplantation model of MLL-AF9-Gpr-OE; these mice experienced an early onset of terminal AML compared with MLL-AF9-control leukemia mice (Figure 1C; supplemental Figure 2G). From MLL-AF9-Gpr-OE leukemia cells, we isolated LSK cells for transcriptome analysis. In line with our previous findings, we observed a downregulation of proapoptotic genes (Fas and Cybb), whereas prosurvival genes (Traf1, Bcl2,28 and Ilrar129) were upregulated. In addition, LSC-associated genes (Cd93, Sox4, and Havcr2 [Tim3]) were highly expressed in MLL-AF9-Gpr-OE (supplemental Figure 2H).30-32 GSEA revealed that the MLL-AF9-Gpr-OE reflected an enhanced LSC-like phenotype (supplemental Figure 2I). Taken together, these data demonstrated that MLL-AF9-Gpr-OE accelerated leukemia progression by mimicking an LSC-like phenotype.
Gprc5c KO delays disease development in MLL-AF9–driven AML
To further explore the role of Gprc5c in the pathogenesis of AML, we transduced the MLL-AF9 fusion oncogene into Gprc5c-KO (MLL-AF9-Gpr-KO) and wild-type (MLL-AF9-WT) BM cells (Figure 1B). Cell cycle analysis and single-cell division assays revealed that the loss of Gprc5c led to a mild but significant acceleration in proliferation (supplemental Figure 2J,K). Additionally, we observed higher ROS levels and poorer in vitro survival upon the loss of Gprc5c (supplemental Figure 2L,M). We probed the stemness and survival genes of the MLL-AF9-Gpr-KO cells. In contrast to MLL-AF9-WT, we observed that genes associated with prosurvival (Bcl2), and stemness in LSCs (Cd93 and Sox4) were downregulated in MLL-AF9-Gpr-KO (supplemental Figure 2N). These features suggested that MLL-AF9-Gpr-KO best resembled blast cells instead of LSCs.
The transplantation model of MLL-AF9-Gpr-KO revealed that loss of Gprc5c delayed disease development. Here, only 1 of 5 MLL-AF9-Gpr-KO mice reached end-stage disease, whereas all 7 MLL-AF9-WT mice succumbed to AML (Figure 1D). The limiting dilution assay estimated an LSC frequency of 1in 7257 in MLL-AF9-WT cells compared with 1 in 66 216 in MLL-AF9-Gpr-KO cells (Figure 1E). These results suggest that Gprc5c is essential for the development of MLL-AF9-driven leukemia and loss of Gprc5c expression impairs leukemia formation.
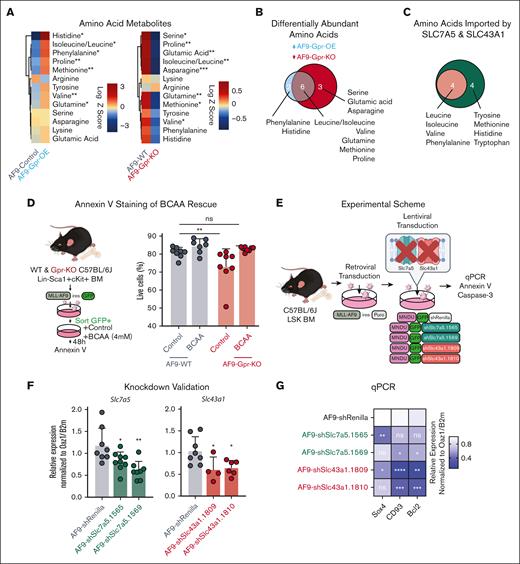
Gprc5c modulates BCAA metabolism in AML
Previously, ROS-low LSCs have been linked to higher amino acid metabolism.8 Because we had detected differential ROS levels upon the gain or loss of Gprc5c, we decided to perform liquid chromatography–tandem mass spectrometry to measure amino acids in the LSK population (Figure 1B). An increase in amino acids was detected in the MLL-AF9-Gpr-OE cells. In contrast, MLL-AF9-Gpr-KO had a reduction in several amino acids (Figure 2A). The amino acids significantly differential in MLL-AF9-Gpr-OE and MLL-AF9-Gpr-KO included BCAAs (leucine, isoleucine, and valine), glutamine, methionine, and proline (Figure 2B). To complement metabolomics, we examined the transcriptome of MLL-AF9-Gpr-OE, which revealed that Slc7a5 expression had a minor but nonsignificant increase, whereas Slc43a1 expression was significantly increased (supplemental Figure 2H). In contrast, MLL-AF9-Gpr-KO significantly downregulated the expressions of both Slc43a1 and Slc7a5 (supplemental Figure 2N). Slc7a5 and Slc43a1 are amino acid transporters that share an affinity for several amino acids and Slc7a5 is a high-affinity transporter for the BCAAs phenylalanine, tyrosine, methionine, histidine, and tryptophan,33 and Slc43a1 is more selective and only transports BCAAs and phenylalanine34 (Figure 2C). The differential expression of Slc7a5 and Slc43a1 might account for the differential abundance of the BCAAs upon loss of Gprc5c gain. Interestingly, supplementation with BCAAs restored survival, which was previously reduced in the absence of Gprc5c signaling, although we cannot exclude additional pathways that play a role in this context (Figure 2D; supplemental Figure 3A).
Taken together, these data suggested that Gprc5c could regulate the expression of Slc7a5 and Slc43a1 to control the importation of the BCAAs that are critical for survival.
BCAA transporters Slc43a1 and Slc7a5 are critical for leukemic viability
To further investigate the amino acid transporters Slc43a1 and Slc7a5, we compared LSK, lineage-negative Sca1+ckit− (LSK−), and more mature myeloid blast–like cells (Gr1+CD11b+) from diseased MLL-AF9 mice. Higher Slc7a5 and Slc43a1 expression was detected in LSKs than in more differentiated LSK− and Gr1+CD11b+ cells (supplemental Figure 3B,C). In line with this, we observed that several amino acid metabolites, including BCAAs, were enriched in the LSK compartment (supplemental Figure 3D).
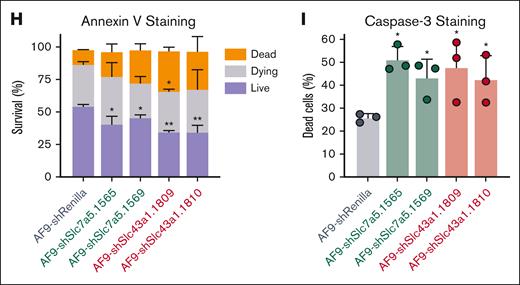
We hypothesized that targeting Slc43a1 and Slc7a5 using a shRNA vector would decrease the fitness of the MLL-AF9 cells (Figure 2E,F). Indeed, the loss of Slc7a5 or Slc43a1 in MLL-AF9 led to the downregulation of prosurvival (Bcl2) and leukemia stem-associated genes (Sox4 and CD93; Figure 2G). Annexin V and caspase-3 staining showed that loss of Slc7a5 and Slc43a1 reduced AML viability (Figure 2H,I).
In healthy mice LSK and LSK–, we observed that Slc7a5 and Slc43a1 were increased in differentiated blood populations, which contrasts with our observations in diseased MLL-AF9 mice (supplemental Figure 3F). Similarly, in the transcriptome of GPR-OE in healthy human mobilized peripheral blood, SLC7A5 expression was not altered in GPR-OE, but SLC43A1 expression was slightly decreased in GPR-OE (supplemental Figure 3F),15 in contrast to our observations in MLL-AF9-Gpr-OE.
These data indicate that the loss of the BCAA transporters SLC7A5 and SLC43A1 impedes the fitness of LSCs. In healthy HSCs, SLC7A5 and SLC43A1 expression was enriched within mature progenitor populations, suggesting that these transporters may play different roles in disease and healthy hematopoiesis.
GPRC5C enhances BCAA transporters expression through NF-κB signaling
Previously, NF-κB signaling was identified to regulate the expression of SLC7A5.35 In agreement, treatment of the leukemia cell line THP1 with an IkappaB kinase (IKK inhibitor), which prevents the release of NF-κB from IκB, decreased the expression of SLC7A5. Interestingly, we also observed downregulation of SLC43A1 upon IKK inhibition, suggesting that NF-κB may also regulate the transcription of SLC43A1 (supplemental Figure 4A).
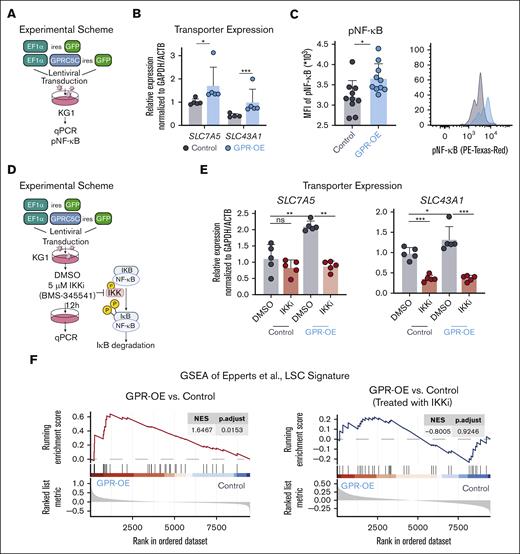
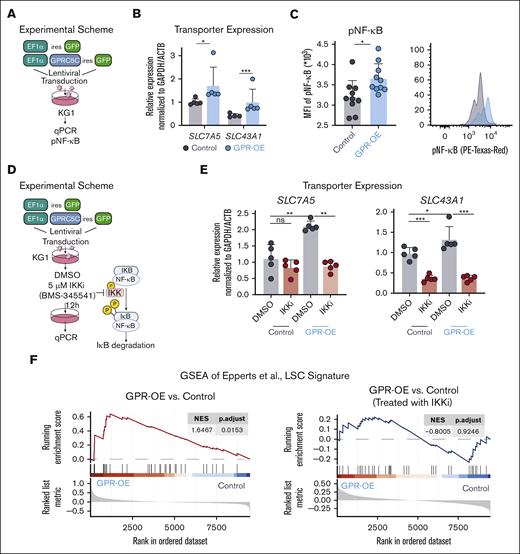
To further investigate NF-κB signaling through GPRC5C, we engineered the leukemia cell line KG1 to overexpress GPRC5C (Figure 3A). Upon GPRC5C overexpression, KG1 cells increased SLC7A5 and SLC43A1 expression (Figure 3B). GSEA revealed enrichment of NF-κB target genes upon GPRC5C expression. In agreement with this, we observed increased phosphorylation of NF-κB upon GPR-OE, which indicated increased activation of NF-κB signaling (Figure 3C; supplemental 4B). However, when NF-κB signaling was blocked by IKK inhibition, GPRC5C could not induce the expression of SLC7A5 or SLC43A1 (Figure 3D,E). Along these lines, the ability of GPRC5C to reprogram cells toward an LSC phenotype was lost when NF-κB activation was prevented by IKK inhibition (Figure 3F).
GPRC5C induces the expression of SLC7A5 and SLC43A1 via NF-κB signaling. (A) Experimental scheme to overexpress GPRC5C in KG1. (B) qPCR of SLC7A5 and SLC43A1 expression after GPRC5C-OE in KG1. Normalized to housekeeping gene GAPDH/ACTB and KG1-control; n = 5. (C) Mean fluorescence intensity measurement of pNF-κB in KG1-control and KG1- GPRC5C -OE cells with FACS plot representation; n = 10. (D) Experimental scheme of IKKi inhibition to prevent NF-κB activation in KG1-Control and KG1-GPRC5C-OE cells. (E) qPCR of SLC7A5 and SLC43A1 expression after GPRC5C -OE in KG1 with or without treatment with IKK inhibitor. Normalized to housekeeping gene GAPDH/ACTB and KG1-control; n = 5. (F) GSEA of the LSC signature from Eppert et al in KG1-control and KG1- GPRC5C-OE cells with or without treatment with IKK inhibitor (BMS-345541). GSEA was performed with BH-adjusted P values after the adaptive multilevel splitting Monte Carlo approach. All represented by mean ± standard deviation. Unpaired Student t test was performed unless otherwise indicated ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗ P < 0.0001. For all experiments, at least 2 independent experiments were performed. BH, Benjamini-Hochberg correction; DMSO, dimethyl sulfoxide; MFI, mean fluorescence intensity; IKKi, IκB kinase inhibitor.
GPRC5C induces the expression of SLC7A5 and SLC43A1 via NF-κB signaling. (A) Experimental scheme to overexpress GPRC5C in KG1. (B) qPCR of SLC7A5 and SLC43A1 expression after GPRC5C-OE in KG1. Normalized to housekeeping gene GAPDH/ACTB and KG1-control; n = 5. (C) Mean fluorescence intensity measurement of pNF-κB in KG1-control and KG1- GPRC5C -OE cells with FACS plot representation; n = 10. (D) Experimental scheme of IKKi inhibition to prevent NF-κB activation in KG1-Control and KG1-GPRC5C-OE cells. (E) qPCR of SLC7A5 and SLC43A1 expression after GPRC5C -OE in KG1 with or without treatment with IKK inhibitor. Normalized to housekeeping gene GAPDH/ACTB and KG1-control; n = 5. (F) GSEA of the LSC signature from Eppert et al in KG1-control and KG1- GPRC5C-OE cells with or without treatment with IKK inhibitor (BMS-345541). GSEA was performed with BH-adjusted P values after the adaptive multilevel splitting Monte Carlo approach. All represented by mean ± standard deviation. Unpaired Student t test was performed unless otherwise indicated ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗ P < 0.0001. For all experiments, at least 2 independent experiments were performed. BH, Benjamini-Hochberg correction; DMSO, dimethyl sulfoxide; MFI, mean fluorescence intensity; IKKi, IκB kinase inhibitor.
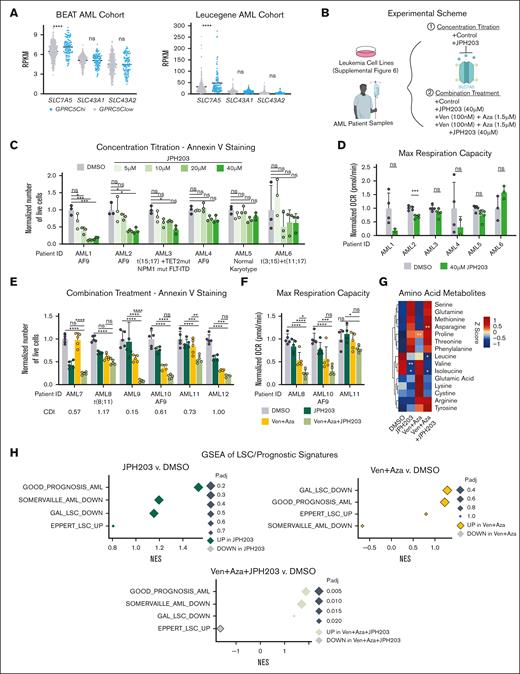
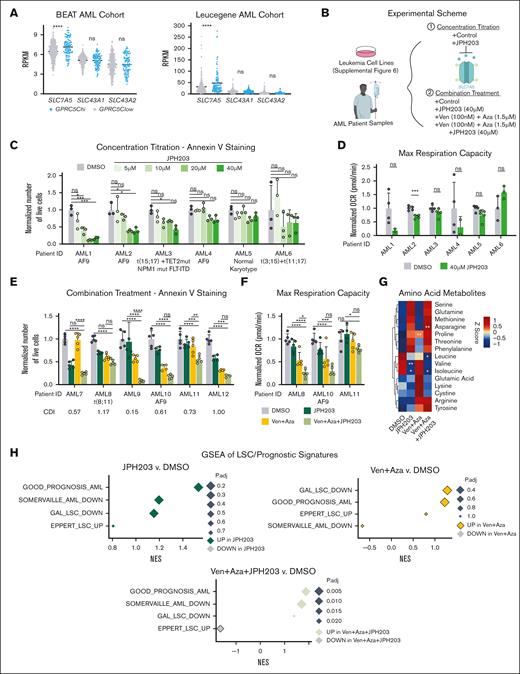
JPH203 combined with Ven + Aza increased antileukemia efficacy in AML. (A) Dot plot showing reads per kilobase of transcript per million mapped reads of SLC7A5, SLC43A1, and SLC43A2 stratified based on GPRC5Chi(GPRC5C high: ≥75th percentile) and GPRC5Clow (GPRC5C low: <75th percentile), expression from the BEAT AML and the Leucegene AML cohorts. (B) Experimental scheme to assess treatment with JPH203 + Ven + Aza in leukemia cell lines and BM samples from patients with de novo AML. (C) Absolute number of live cells, as determined by annexin V staining in specimens of patients with AML treated with JPH203 for 24 hours. Normalized to the DMSO control of each AML specimen; n = 3 technical replicates per AML specimen. (D) Maximum respiration measured by the seahorse assay of patient cells with AML treated with JPH203 for 12 hours. Normalized to the DMSO control of each AML specimen; n = 2 to 5 technical replicates per AML specimen. (E) Absolute cell count of live cells as determined by annexin V staining in patient specimens with AML treated with Ven + Aza + JPH203 and measured at 24 hours. Normalized to the DMSO control of each AML specimen. CDI was calculated for each AML specimen. CDI value < 1 (synergistic), CDI = 1 (additive), and CDI > 1 (antagonistic); n = 3 to 5 technical replicates per AML specimen. (F) Oxygen consumption levels measured by the seahorse assay of specimens of patients with AML treated with Ven + Aza + JPH203 for 12 hours. Normalized to the DMSO control of each AML specimen; n = 5 technical replicate per AML specimen. (G) Heat map of amino acids metabolites. Z-score centering of the mean value of metabolite abundance per specimen from patients with AML; n = 3 specimens from patients with AML. (H) GSEA of LSC/prognostic signatures under treatment conditions compared with the DMSO control. GSEA was performed with BH-adjusted P values after the adaptive multilevel splitting Monte Carlo approach. GAL_LSC_DOWN,46 GOOD_PROGNOSIS_AML,47 EPPERT_LSC_UP,48 and SOMERVAILLE_AML_DOWN.49 All represented by mean ± standard deviation. (A,G) Unpaired Student t test; (C-F) 2-way analysis of variance. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. For all experiments, at least 2 independent experiments were performed.
JPH203 combined with Ven + Aza increased antileukemia efficacy in AML. (A) Dot plot showing reads per kilobase of transcript per million mapped reads of SLC7A5, SLC43A1, and SLC43A2 stratified based on GPRC5Chi(GPRC5C high: ≥75th percentile) and GPRC5Clow (GPRC5C low: <75th percentile), expression from the BEAT AML and the Leucegene AML cohorts. (B) Experimental scheme to assess treatment with JPH203 + Ven + Aza in leukemia cell lines and BM samples from patients with de novo AML. (C) Absolute number of live cells, as determined by annexin V staining in specimens of patients with AML treated with JPH203 for 24 hours. Normalized to the DMSO control of each AML specimen; n = 3 technical replicates per AML specimen. (D) Maximum respiration measured by the seahorse assay of patient cells with AML treated with JPH203 for 12 hours. Normalized to the DMSO control of each AML specimen; n = 2 to 5 technical replicates per AML specimen. (E) Absolute cell count of live cells as determined by annexin V staining in patient specimens with AML treated with Ven + Aza + JPH203 and measured at 24 hours. Normalized to the DMSO control of each AML specimen. CDI was calculated for each AML specimen. CDI value < 1 (synergistic), CDI = 1 (additive), and CDI > 1 (antagonistic); n = 3 to 5 technical replicates per AML specimen. (F) Oxygen consumption levels measured by the seahorse assay of specimens of patients with AML treated with Ven + Aza + JPH203 for 12 hours. Normalized to the DMSO control of each AML specimen; n = 5 technical replicate per AML specimen. (G) Heat map of amino acids metabolites. Z-score centering of the mean value of metabolite abundance per specimen from patients with AML; n = 3 specimens from patients with AML. (H) GSEA of LSC/prognostic signatures under treatment conditions compared with the DMSO control. GSEA was performed with BH-adjusted P values after the adaptive multilevel splitting Monte Carlo approach. GAL_LSC_DOWN,46 GOOD_PROGNOSIS_AML,47 EPPERT_LSC_UP,48 and SOMERVAILLE_AML_DOWN.49 All represented by mean ± standard deviation. (A,G) Unpaired Student t test; (C-F) 2-way analysis of variance. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. For all experiments, at least 2 independent experiments were performed.
Together, these data suggested that GPRC5C enhances the activation NF-κB to mediate an increase in SLC7A5 and SLC43A1.
JPH203 treatment is an effective antileukemia treatment
From the BEAT AML and Leucegene cohorts, we observed that patients with GPRC5Chi had higher SLC7A5 expression than patients with GPRC5Clow25,36(Figure 4A). However, SLC43A1 was not differentially expressed, which might be attributed to the higher expression of SLC7A5, suggesting that it may act as the main importer of BCAAs in AML and emphasizing the importance of targeting SLC7A5 to trigger disease regression (Figure 4A). Of note, examination of the BEAT AML cohort revealed no difference in the survival of patients with AML with higher or lower expression of SLC7A5 or SLC43A1c (supplemental Figure 5A). Next, we treated mouse MLL-AF9 cells with 2-amino-2-norbornanecarboxylic acid (BCH), an inhibitor of SLC7A5 and SLC43A1. BCH treatment decreased the survival of MLL-AF9 cells, but healthy lineage-depleted BM cells were not affected (supplemental Figure 6A-E). We additionally tested JPH203, a potent and selective inhibitor of Slc7a5. Like BCH, JPH203 induced the apoptosis of MLL-AF9 cells while sparing healthy BM cells (supplemental Figure 6F-I). We decided to proceed with JPH203 because it was able to achieve a much more potent effect at a lower concentration. Because LSCs were found to be highly dependent on OXPHOS,3 we measured the oxygen consumption. JPH203 did not alter the baseline OXPHOS level, but the spare respiratory capacity was significantly reduced (supplemental Figure 6J). These data show that MLL-AF9 leukemia cells require a high level of Slc7a5-mediated amino acid influx for survival and to regulate their cellular ability to respond to energy demands.
Next, we investigated the effect of the SLC7A5-specific inhibitor, JPH203, on human-derived material. We assessed the sensitivity of JPH203 using a set of leukemia cell lines that represented a spectrum of mutations in AML (Figure 4B). After 96 hours of in vitro treatment, reduced cell survival was detected; this effect was mainly observed in THP1, preceded by NB4, U937, and K562 cell lines. No changes were observed in the survival of the KG1 cells (supplemental Figure 7A). Cell lines that responded to JPH203 treatment (THP1, U937, NB4, and K562) also showed reduced baseline OXPHOS level and maximal respiration capacity (supplemental Figure 7B). We expanded our investigation of JPH203-induced apoptosis to samples of patients with AML from diverse mutational backgrounds (Figure 4B). JPH203 decreased in vitro survival and the maximal respiration capacity to varying degrees, depending on the AML specimen (Figure 4C,D). These results suggest that JPH203 induces apoptosis by targeting the mitochondrial capacity for energy production.
Combination treatment (JPH203 with venetoclax and azacitidine) rewires metabolism to diminish AML viability
To enhance the antileukemia activity of JPH203, we proposed a combination treatment of JPH203 (40 μM) with venetoclax (100 nM) and azacitidine (1.5 μM) (Ven + Aza) (Figure 4B). Ven + Aza was chosen for its success in clinical trials to induce long-term remission in patients with AML.37-39 Combination treatment (Ven + Aza + JPH203) further decreased THP1, NB4, U937, and K562 cell survival and OXPHOS. These results suggested that Ven + Aza + JPH203 reduced the respiratory capacity to hinder the survival of leukemia cell lines (supplemental Figure 7C,D). KG1, which was resistant to JPH203, did not show JPH203 sensitivity in the presence of Ven + Aza (supplemental Figure 7C,D). However, ectopic GPRC5C expression in KG1 was sufficient to confer sensitivity to Ven + Aza + JPH203 (supplemental Figure 7E).
To further investigate the mechanism of action, we performed transcriptomic and metabolomic analyses of the THP1 treated with the combination of Ven + Aza + JPH203 (supplemental Figure 8A). The combination of Ven + Aza + JPH203 upregulated amino acid transporters and suppressed DNA damage repair and cell cycle (supplemental Figure 8B). More importantly, metabolomic analysis revealed that Ven + Aza + JPH203 led to the most substantial reduction in amino acid abundance compared with only Ven + Aza or JPH203 (supplemental Figure 8C). BCAAs can be metabolized to acetyl coenzyme A (acetyl-CoA) or succinyl-CoA, which fuels the tricarboxylic acid (TCA) cycle to produce reduced NAD and FADH2, and then oxidized in the ETC to generate adenosine triphosphate.40 Examination of metabolites associated with the TCA cycle revealed that Ven + Aza + JPH203 treatment reduced the levels of succinic acid, citric acid, malic acid, and furmaric acid (supplemental Figure 8C). Metabolites generated from the TCA cycle were used to fuel the ETC to produce adenosine triphosphate, which was also reduced upon Ven + Aza + JPH203 treatment. Additionally, GSEA revealed the downregulation of components of ETC complex I (supplemental Figure 8D).
Similarly, in samples of patients with AML, the Ven + Aza + JPH203 combinatory treatment was associated with a robust and significant reduction in AML survival by targeting the respiratory capacity. We calculated the CDI for each patient sample and found that the degree of response was highly varied (Figure 4E,F). In most specimens of patients with AML, the addition of JPH203 to Ven + Aza exerted synergism. In contrast, the addition of JPH203 to Ven + Aza was not associated with increased toxicity in healthy human BM cells (supplemental Figure 7F). When we examined the metabolome of AML cells preceding our treatment regimens, we observed a wide variation in the degree of intracellular amino acid abundance. However, only the reduction in BCAAs was consistent among all 3 AML specimens that were treated with JPH203 alone or Ven + Aza + JPH203 (Figure 4G). GSEA revealed that the combinatory treatment of Ven + Aza + JPH203 reduced the expression of genes associated with LSCs and increased the expression of genes associated with a good prognosis (Figure 4H).
To summarize, we identified JPH203 as a novel inhibitor of AML, when administered in combination with venetoclax and azacytidine, to prevent OXPHOS, leading to leukemic cell death.
Discussion
Recently, we isolated a quiescent human HSC population using surface GPRC5C levels.15 We showed that GPRC5C is essential for HSC function through genetic loss- and gain-of-function analyses. Data from multiple AML cohorts suggested that GPRC5C expression could modulate AML aggression. Indeed, ectopic Gprc5c in engineered MLL-AF9 cells mimicked the LSC phenotype and accelerated disease development. In contrast, these features were reversed when Gprc5c expression was absent (Figure 5, left panel). Transcriptome data support the notion that GPRC5C may function as an oncogene in AMLs beyond MLL-AF9. However, future studies should investigate whether GPRC5C is functionally crucial in AMLs of other cytogenetics.
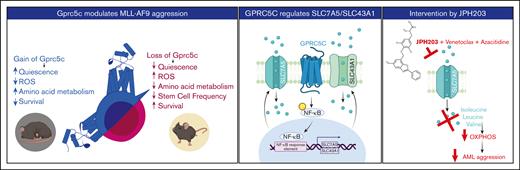
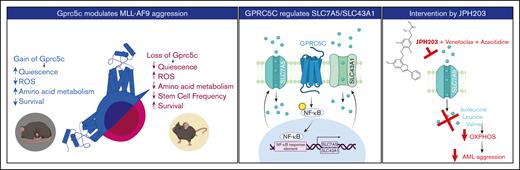
Graphic abstract. Left panel - GPRC5C modulates AML aggression. Middle panel - GPRC5C signals through NF-κB to enhance the expression of SLC7A5 and SLC43A1. Right panel - Inhibition of SLC7A5 with JPH203 targets the respiratory capacity of leukemic cells, hindering their viability. This effect is strengthen by the addition of venetoclax and azacitidine.
Graphic abstract. Left panel - GPRC5C modulates AML aggression. Middle panel - GPRC5C signals through NF-κB to enhance the expression of SLC7A5 and SLC43A1. Right panel - Inhibition of SLC7A5 with JPH203 targets the respiratory capacity of leukemic cells, hindering their viability. This effect is strengthen by the addition of venetoclax and azacitidine.
LSCs contain a high abundance of amino acids that are essential for survival.8 Here we found an association between GPRC5C expression and amino acid metabolism in AML. In particular, we observed that high GPRC5C levels activated NF-κB, leading to the increased transcription of SLC7A5 and SLC43A1. The loss of Slc7a5 has been shown to have a proapoptotic effect in SKM-1, a myelodysplastic syndrome cell line, and in T-ALL cell lines.13,41 However, relatively little is known about the role of Slc43a1, especially in AML. By targeting these 2 BCAA transporters separately, we observed decreased survival of leukemic cells, but whether targeting these BCAA transporters simultaneously has a synergistic effect on AML survival still needs to be investigated.
JPH203 is an inhibitor of SLC7A5 and was shown to be a safe and effective treatment in a phase 1 human study of advanced solid tumors.42 Although treatment with Ven + Aza has been successful in most patients with AML, there are still patients for whom the combination is ineffective.38,39,43 Although our study would benefit from assessing additional human AML samples, here, we showed that the combination of Ven + Aza + JPH203 was not only effective at eradicating leukemia activity by targeting oxidative respiration but also that samples from patients who did not respond to Ven + Aza responded well to JPH203 in vitro. It would be interesting in future studies to further address the mechanism of action within JPH203 + Ven + Aza. LSCs can escape venetoclax eradication through the upregulation of fatty acid oxidation (FAO), making amino acid metabolism redundant.44 BCAA and its intermediate metabolite, acetyl-CoA, are involved in driving FAO and the TCA cycle.45 Treatment with JPH203 can potentially inhibit both FAO and OXPHOS simultaneously, interfering with a backup metabolic machinery. Unfortunately, there are no specific inhibitors of SLC43A1; therefore, we cannot determine whether the phenotype observed is specifically from the loss of BCAAs because SLC7A5 has an affinity for tryptophan, phenylalanine, and histidine as well.
In conclusion, we show that GPRC5C alters the metabolic profiles of leukemia cells by generating a dependence on amino acid metabolism to orchestrate an enhanced aggressive disease. Combining JPH203 with Ven + Aza reverses this phenotype, selectively eradicating leukemia cells while sparing healthy hematopoietic cells. Future studies should address the effectiveness of the here suggested treatment in vivo. Collectively, these results provide a preclinical rationale to further develop JPH203 as a novel therapeutic option for AML.
Acknowledgments
The authors thank S. Hobitz, K. Schuldes, L. Bischer, and A. Würch from the Max Planck Institute of Immunology and Epigenetics (MPI-IE) Flow Cytometry Core Facility for their assistance and D. Ruf, K. Eichmann, C. Zedlick, H. Kwiatkowski and all members of the MPI-IE Laboratory Animal Core Facility for excellent animal welfare and husbandry. The authors thank the MPI-IE Deep Sequencing Core Facility for their assistance. H.T. thanks the Core-to-Core Program Advanced Research Networks “Integrative approach for normal and leukemic stem cells.”
This work was supported by the Max Planck Society, ERC-Stg-2017 (VitASTEM, 759206), the Behrens-Weise-Foundation, the German Research Foundation under the German Excellence Strategy (CIBSS-EXC-2189; project ID 390939984), SFB1425 (project no. 422681845), SFB992 (project no. 192904750; B07), SFB1479 (P05), the European Union’s Horizon 2020 Research and Innovation Programme under the Marie Skłodowska-Curie Actions Grant (agreement 813091), and José Carreras Leukämie-Stiftung all to N.C.-W.
Authorship
Contribution: Y.W.Z. and N.C.-W. contributed to conceptualization of the study; Y.W.Z., T.V.-H., J.M., M.-E.L., M.C.R.-M., N.O., N. Karantzelis, J.R., N. Karabacz, K.J., T.M., K. Schönberger., K. Shoumariyeh., J.L.T., P.R., A.M., H.T., S.R., R.Z., H.L.P., F.B., J.H., B.L., G.S., P.M., N.C.-W. contributed to the methodology; Y.W.Z. and N.C.-W. supervised and wrote the original draft; and all the authors participated in editing the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Nina Cabezas-Wallscheid, Max Planck Institute of Immunobiology and Epigenetics, Stuebeweg 51, 79108 Freiburg, Germany; email: cabezas@ie-freiburg.mpg.de; and Yu Wei Zhang, Max Planck Institute of Immunobiology and Epigenetics, Stuebeweg 51, 79108 Freiburg, Germany; email: zhang@ie-freiburg.mpg.de.
References
Author notes
∗T.V.H. and J.M. contributed equally to this study.
The raw data are deposited in ArrayExpress (accession number E-MTAB-11690) at http://www.ebi.ac.uk/arrayexpress.
The full-text version of this article contains a data supplement.