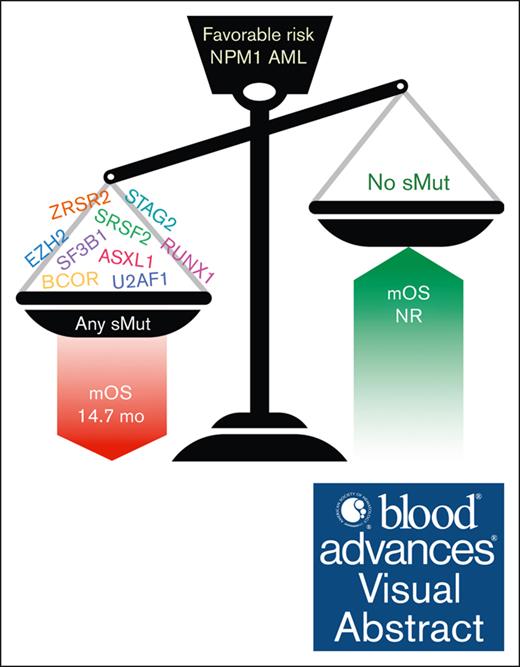
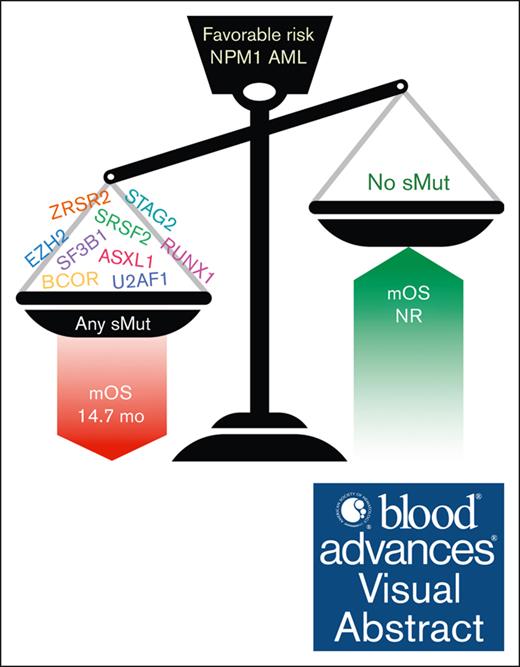
Patients with favorable-risk AML who had NPM1 mutation have poor prognosis if they also harbor mutations highly specific for sMut.
Patients with sMut had better outcomes if they achieved NPM1 MRD negativity.
Visual Abstract
Acute myeloid leukemia (AML) is a heterogeneous malignancy with outcomes largely predicted by genetic abnormalities. Mutations of NPM1 are common in AML, occurring in ∼30% of cases, and generally considered a favorable risk factor. Mutations highly specific for secondary AML (sMut) have been shown to confer poor prognosis, but the overall impact of these mutations in the setting of favorable-risk AML defined by mutant NPM1 remains unclear. In this multicenter study of patients with AML (n = 233) with NPM1 mutation at diagnosis, we observed that patients with sMut had worse overall survival (OS) than those without sMut (15.3 vs 43.7 months; P = .002). Importantly, this finding persisted in the European LeukemiaNet (ELN) 2017–defined favorable risk subset (14.7 months vs not reached; P < .0001). Among patients who achieved NPM1 measurable residual disease (MRD) negativity, longer OS was observed in the entire cohort (P = .015) as well as in both the sMut subset (MRD negative: median OS (mOS) 73.9 months vs MRD positive: 12.3 months; P = .0170) and sMut ELN 2017–favorable subset (MRD negative: mOS 27.3 vs MRD positive: 10.5 months; P = .009). Co-occurrence of sMut and mutant NPM1 confers a poor prognosis in AML.
Introduction
Mutated nucleophosmin (NPM1) is one of the most common molecular abnormalities in acute myeloid leukemia (AML), occurring in ∼30% of cases regardless of cytogenetics.1 Wild-type NPM1 contains 12 exons and encodes for a ubiquitous multifunctional protein that shuttles between the nucleus and cytoplasm. Some of the major functions of NPM1 include ribosome biogenesis, p53-dependent stress response, and genomic stability maintenance.2 Aberrant cytoplasmic dislocation is the hallmark feature in NPM1-mutated AML, a discovery leading to the recognition of NPM1-mutated AML as a distinct entity within the 2016 World Health Organization classification.3 Mutated NPM1 is enriched in AML with normal cytogenetics and is associated with higher response rates to induction chemotherapy and better overall survival (OS) than many other AML subtypes.4,5 Besides CEBPA-bZip mutations, mutant NPM1 is the only other mutation that can define favorable-risk disease under the European LeukemiaNet (ELN) 2017 and 2022 criteria.6,7 It is well documented that certain gene-gene interactions may affect the prognostic impact of a single gene mutation. For example, in ELN 2022, co-occurrence of NPM1 and FLT3-ITD mutations defines intermediate-risk disease rather than favorable-risk disease (seen in cases with NPM1 mutation and wild-type FLT3). A recent study observed that outcomes of NPM1 and WT1–comutated AML in patients aged <60 years resembled outcomes in patients at adverse risk.8 Another study showed the combination of NPM1, DNMT3A, and IDH mutations led to an inferior OS.9 In ELN 2022, the presence of mutations highly specific for secondary AML (sAML), including ASXL1, BCOR, EZH2, RUNX1, SF3B1, SRSF2, STAG2, U2AF1, and/or ZRSR2 (sMut)–defined adverse risk.6,10 Older patients with de novo AML (dAML) with these genetic alterations share outcomes similar to those with conventional-defined sAML, with lower complete remission (CR) rate and decreased survival.10 To date, ELN 2022 has not yet classified AML with sMut that co-occurs with favorable-risk molecular features (such as mutant NPM1) as adverse.6 As such, we set out to study the prognostic impact of sMut co-occurring with mutant NPM1 in AML, including its prognostic effect among patients who achieve mutant NPM1–measurable residual disease (MRD) negativity.
Patients and methods
Patient selection
We performed a retrospective review of 233 patients with AML who had NPM1 mutation and were treated at the Moffitt Cancer Center (n = 160), Weill Cornell (n = 51), and Memorial Healthcare System (n = 22) between 2013 and 2022. Inclusion was restricted to patients who had next-generation sequencing (NGS) performed at diagnosis. Patients who participated in clinical trials at any point during their treatment course were included. Demographics, disease-specific variables, and clinical outcomes were collected in accordance with the institutional review board–approved protocol. The study was conducted in accordance with the Declaration of Helsinki. dAML is defined as AML developed in patients without clinical history of prior myeloid hematologic neoplasm or exposure to potentially leukemogenic agents. sAML refers to AML arising from an antecedent myeloid hematologic neoplasm (based on clinical history) such as myelodysplastic syndromes or myeloproliferative neoplasm. Therapy-related AML is defined as AML arising from prior cytotoxic, radiation, or immunosuppressive therapy for an unrelated disease. Risk stratification and response to therapy were defined using the ELN 2017 criteria, unless otherwise specified.7
Molecular analysis
The majority of somatic mutations were assessed using TruSight Myeloid Sequencing Panel (Illumina, San Diego, CA) that targets 54 genes by NGS with a detection threshold of 5% to determine presence or absence of mutations based on institutional standards. These genes include ABL1, ASXL1, ATRX, BCOR, BCORL1, BRAF, CALR, CBL, CBLB, CBLC, CDKN2A, CEBPA, CSF3R, CUX1, DNMT3A, ETV6, EZH2, FBXW7, FLT3, GATA1, GATA2, GNAS, HRAS, IDH1, IDH2, IKZF1, JAK2, JAK3, KDM6A, KIT, KMT2A, KRAS, MPL, MYD88, NOTCH1, NPM1, NRAS, PDGFRA, PHF6, PTEN, PTPN11, RAD21, RUNX1, SETBP1, SF3B1, SMC1A, SMC3, SRSF2, STAG2, TET2, TP53, U2AF1, WT1, and ZRSR2. In patients sequenced after 2020 at Moffitt, somatic mutations were assessed using 98-gene Moffitt Myeloid Action Panel that targets 98 genes with a detection threshold of 3%. These genes include ABL1, ANKRD26, ASXL1, ASXL2, ATM, ATRX, BCOR, BCORL1, BRAF, BRCC3, CALR, CBL, CBLB, CBLC, CCND2, CDKN2A, CEBPA, CHEK2, CREBBP, CSF3R, CSMD1 , CSNK1A1, CTCF, CUX1, DDX41, DHX15,DNMT3A, ELANE, ETNK1, ETV6, EZH2, FANCA, FANCL, FLT3, GATA1, GATA2, GNAS, GNB1, HNRNPK, HRAS, IDH1, IDH2, IKZF1, JAK1, JAK2, JAK3, KDM6A, KIT, KMT2A, KRAS, LUC7L2, MECOM, MET, MLL2, MPL, MYC, NF1, NOTCH1, NOTCH2, NPM1, NRAS, PAX5, PDGFRA, PHF6, PIGA, PML, PPM1D, PTPN11, RAD21, RAF1, RB1, RBBP6, RPS19, RTEL1, RUNX1, SAMD9, SAMD9L, SBDS, SETBP1, SF3B1, SH2B3, SMC1A, SMC3, SOS1, SRP72, SRSF2, STAG1, STAG2, STAT3, STAT5B, TERC, TERT, TET2, TP53, U2AF1, WT1, ZBTB7A, and ZRSR2. A subset of molecular analysis was performed using TruSeq (Illumina, San Diego, CA), which includes 31 common myeloid targeted genes, in patients who were diagnosed before 2015. Older NGS tests only reported presence or absence of the mutation and its location without variant allele frequency (VAF) information. Weill Cornell’s NGS panel has a detection limit of 5% VAF. Memorial Healthcare System uses OnkoSight Advanced NGS Myeloid Panel (GenPath, Elmwood Park, NJ) that has a detection limit of 4%. In a subset of the total cohort, NPM1 MRD testing was performed using a propriety NGS assay by Invivoscribe. The analytic sensitivity of this assay is 1 × 10−5 mutant alleles per total alleles. In another subset of patients, NPM1 MRD was assessed using quantitative reverse transcription polymerase chain reaction. Using the delta-delta Ct method relative to a plasmid calibrator, NPM1 variant transcripts are quantified and reported as a normalized ratio to ABL1 transcripts present in the sample. The limit of detection of this assay is 1:100 000 cells (0.001%). These were performed at a reference laboratory, ARUP Laboratories in Salt Lake City, UT. Timing of MRD collection varied between patients and their treatment course.
Statistical analysis
Clinical variables were summarized using descriptive statistics. Fisher exact test and χ2 test were used to assess the association between categorical variables. The Wilcoxon rank-sum test was used to compare the 2 groups’ continuous variables. A 2-sided P value of <5% was considered statistically significant. The OS and relapse-free survival (RFS) were estimated by the Kaplan-Meier method and calculated from the date of diagnosis.
Multivariate analyses were conducted using Cox proportional hazard regression model. Statistical analysis was performed using R version 4.0.2 (R Foundation for Statistical Computing, Vienna, Austria).11
Results
Patient characteristics
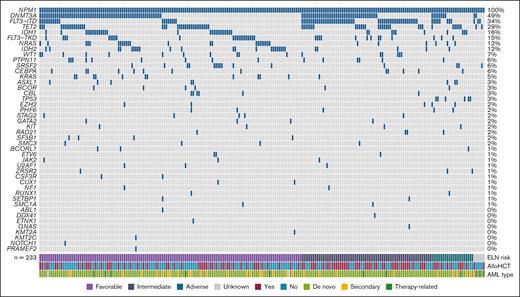
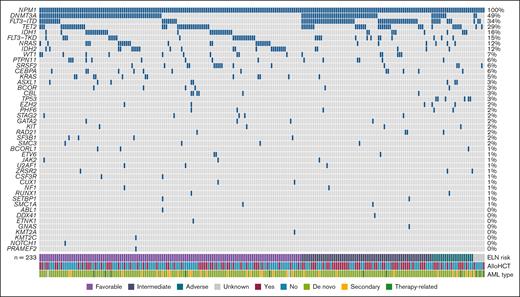
A total of 233 patients with AML (111 male and 122 female) with NPM1 mutation were included in this analysis (Table 1). The median age at the time of diagnosis was 63 years (range, 22-86). Clinically, patients with dAML formed the majority at 81.1% (189/233) followed by 13.3% (31/233) sAML, and 4.7% (11/233) therapy-related AML. According to the ELN 2017 criteria, 137 patients (58.8%) had favorable-risk, 68 (29.2%) had intermediate-risk, and 22 (9.4%) had adverse-risk disease. A total of 43 patients (18.5%) had sMut (any ≥1 of the following: ASXL1, BCOR, EZH2, RUNX1, SF3B1, SRSF2, STAG2, U2AF1, and ZRSR2), of whom 30 (69.8%) had ELN 2017–defined favorable-risk disease. When reclassified based on ELN 2022, a total of 19 patients had risk escalation from favorable to intermediate, and 2 patients with previously unknown risk were now intermediate. This is because the updated ELN no longer takes allelic ratio of FLT3-ITD into account. The most frequent comutations in our cohort included DNMT3A (114/233 [48.9%]), FLT3-ITD (79/233 [33.9%]), TET2 (67/233 [28.7%]), IDH1 (37/233 [15.8%]), FLT3-TKD (35/233 [15%]), NRAS (29/233 [12.4%]), and IDH2 (27/233 [11.6%]) (Figure 1). Among patients with sMut, the most common sMuts were SRSF2 (15/43 [34.9%]) and ASXL1 (8/43 [18.6%]). Among patients without sMut, common comutations included DNMT3A (99/190 [52.1%]), FLT3-ITD (68/190 [35.8%]), TET2 (50/190 [26.3%]), IDH1 (28/190 [14.7%]), FLT3-TKD (31/190 [16.3%]), NRAS (26/190 [13.7%]), and IDH2 (21/190 [11.1%]). Of note, there were 6 patients with TP53 mutations, and none of them were categorized as having sMut based on their comutations. The average number of mutations in the sMut and no sMut cohorts was 4.1 and 3.3, respectively (Table 1). NPM1 VAF was available in 151 patients (64.8%) with a median of 26.2% (range, 2.37%-62%). Among patients with sMut with available NPM1 VAF (n = 31), median NPM1 VAF was 19% (range, 3%-49.6%). The majority of patients (177/233 [76%]) received frontline intensive chemotherapy, and 46.8% (109/233) proceeded to allogeneic hematopoietic cell transplantation (allo-HCT). Fewer patients with sMut received frontline intensive chemotherapy than those without (26/43 [60.5%] vs 151/190 [79.5%]; P = .016), and more received nonintensive regimens (37.2% [16/43] vs 17.9% [34/190]; P = .008). A total of 15 out of 43 (∼35%) of the sMut cohort received transplant compared with 94 of 190 (∼50%) in those without sMut (P = .093).
Molecular landscape of AML with NPM1 mutation.NPM1 is frequently comutated with DNMT3A (48.9%), FLT3-ITD (33.9%), and TET2 (28.7%).
Molecular landscape of AML with NPM1 mutation.NPM1 is frequently comutated with DNMT3A (48.9%), FLT3-ITD (33.9%), and TET2 (28.7%).
Responses by secondary mutations status
Patients with sMut achieved a CR/CR with incomplete hematologic recovery (CRi) rate of 69.7% (30/43) compared with 85.3% (162/190) in those without sMut (P = .025) (Table 2). Relapse occurred in 34.9% (15/43) and 36.8% (70/190) of sMut and no sMut cohorts, respectively (P = .862). There was a higher rate of primary refractory disease in patients with sMut (13/43 [30.2%] vs 20/190 [10.5%]; P = .003). Posttreatment MRD testing for mutant NPM1 was performed in 95 of 233 patients (40.8%) and was negative in 57 of 95 (60%). The proportion of posttreatment MRD negativity between sMut (9/43 [20.9%]) and non-sMut (48/190 [25.3%]) was not significantly different (P = .695) (Table 2).
Impact of sMut on survival
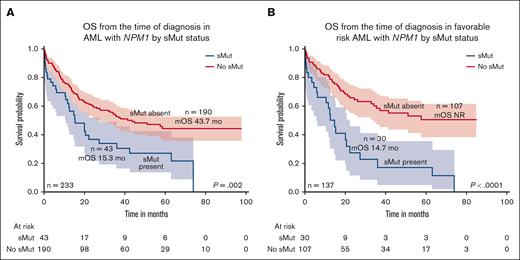
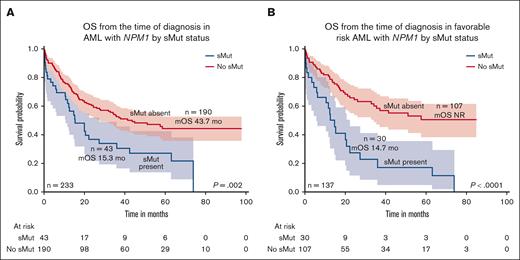
The median OS of the entire cohort was 35.5 months with a median follow-up of 52.1 months. Those who harbored sMut (n = 43) had worse OS than those without sMut (n = 190) (15.3 months vs 43.7 months; P = .002) (Figure 2A). Median RFS was 22.1 months in sMut (n = 43) and 37.5 months in those without sMut (n = 190) (P = .335). Focusing only on patients with ELN 2017 favorable-risk disease (n = 137), OS was 14.7 months vs not reached for those with sMut (n = 30) and those without sMut (n = 107), respectively (P < .0001) (Figure 2B). In this same subset of patients with ELN 2017 favorable risk, RFS was 14.7 months in sMut (n = 30) and 37.5 months in no sMut (n = 107) (P = .124). Similar findings were observed when patients were reclassified according to ELN 2022. Those with ELN 2022 favorable risk (n = 119) had significantly shorter OS if they had sMut (n = 26) than those with no sMut (n = 93) (14.7 months vs not reached; P < .0001). In addition, omitting FLT3-ITD from the sMut cohort or the entire cohort did not change the adverse prognostic impact of sMut in all risks or favorable risk only. Univariate analysis demonstrated that age ≥60 years at diagnosis, nonintensive frontline treatment (compared with intensive treatment), no treatment, and sMut negatively affected OS (age ≥ 60 years; hazard ratio [HR], 2.63; 95% confidence interval [CI], 1.47-4.69; P = .001; nonintensive frontline treatment: HR, 3.67; 95% CI, 2.16-6.24; P < .001; no treatment: HR, 57.66; 95% CI, 14.48-229.5; P < .001; sMut: HR, 2.97; 95% CI, 1.77-4.96; P < .001). Patients who received allo-HCT had better outcomes than those who did not (HR, 0.31; 95% CI, 0.18-0.53; P < .001). In multivariate analysis, sMut retained independent negative prognostic significance on OS (HR, 2.95; 95% CI, 1.56-5.59; P < .001) along with those who did not get intensive treatment or allo-HCT (Table 3). Among patients with sMut with available NPM1 VAF data (n = 31), those with NPM1 VAF ≤20% (n = 16) had a shorter median OS of 7.9 months compared with 42.4 months (P = .014) in those with VAF >20% (n = 15). Among patients (n = 120) without sMut, the median NPM1 VAF was 27.4% (range, 2.37%-62%), and there was no significant difference in OS between those with VAF ≤20% (n = 44) and those with VAF >20% (n = 76; P = .39).
OS by sMut status of the entire cohort and favorable risk subset. (A) Median OS was shorter in patients with NPM1-mutated AML with sMut than those without sMut (15.3 months vs 43.7 months; P = .002). (B) When restricted to only patients with favorable risk, median OS was 14.7 months vs not reached (P < .0001).
OS by sMut status of the entire cohort and favorable risk subset. (A) Median OS was shorter in patients with NPM1-mutated AML with sMut than those without sMut (15.3 months vs 43.7 months; P = .002). (B) When restricted to only patients with favorable risk, median OS was 14.7 months vs not reached (P < .0001).
Impact of NPM1 MRD on outcomes
Patients who achieved MRD negativity (n = 57) had longer OS than those with MRD positivity (n = 38) (MRD negative median OS (mOS), 73.9 months vs MRD positive mOS, 34.5 months; P = .015). Similarly, MRD negativity predicted longer OS in the overall sMut subset, although patient numbers were low (n = 13 [9 MRD negative and 4 MRD positive]; mOS, 73.9 vs 12.3 months; P = .017). Among patients with favorable-risk sMut, MRD negativity also predicted longer survival (n=8 [6 MRD negative and 2 MRD positive]; mOS, 27.3 vs 10.5 months; P = .009).
Effects of age on response and survival in patients with NPM1
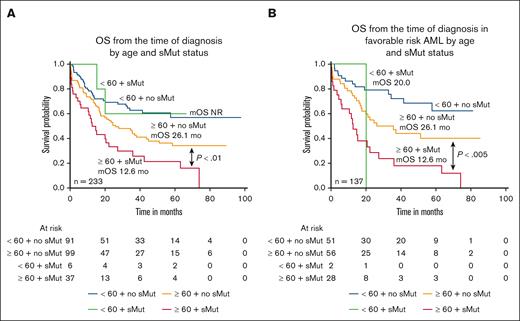
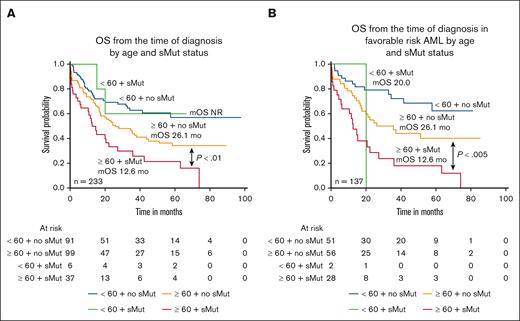
Lower CR/CRi rates were observed in patients with sMut within both age cohorts (age < 60 years CR/CRi, 5/6 [83.3%] with sMut vs 82/91 [90.1%] without sMut; age ≥ 60 years CR/CRi, 25/37 [67.6%] vs 80/99 [80.8%]) (Table 4). Among patients aged ≥60 years, those who harbored sMut (n = 37) fared significantly worse than patients without sMut (n = 99) (12.6 months vs 26.1 months; P < .01) (Figure 3A). In patients aged <60 years, there was no significant difference in OS between those with sMut (n = 6) and those without non-sMut (n = 91), although the numbers were quite small. Median OS was not reached for either group. Similarly, in the ELN 2017 subset of patients with favorable risk, OS was significantly shorter in patients age ≥60 years with sMut (n = 28) than in those without (n = 56) (P < .005), and this was not observed among patients aged <60 years (P = .31) (Figure 3B). When analyses were repeated using the age cutoff of 70 years for the entire cohort, among those aged <70 years (n = 160), the median OS were not reached and 35 months for those without (n = 137) and those with sMut (n = 23), respectively (P=.1471). Among patients aged ≥70 years (n = 73), the median OS were 21.1 and 10 months for those without (n = 53) and those with sMut (n = 20), respectively (P = .0115). In the favorable-risk subset, sMut status significantly affected both age cohorts (age <70 years [n = 92]: no sMut [n = 75] median OS, not reached vs sMut [n = 17] median OS, 22.2 months; P = .0059; age ≥70 years [n = 45] no sMut [n = 32] median OS, 24.4 months vs sMut [n = 13] median OS, 10 months; P = .0036)
OS by age and sMut status of the entire cohort and favorable risk subset. (A) Median OS was shorter in older patients (age ≥60) with NPM1-mutated AML with sMut than those without sMut. (12.6 months vs 26.1 months; P < .01). (B) When restricted to only patients with favorable risk, median OS was again significantly shorter in the older, sMut subset.
OS by age and sMut status of the entire cohort and favorable risk subset. (A) Median OS was shorter in older patients (age ≥60) with NPM1-mutated AML with sMut than those without sMut. (12.6 months vs 26.1 months; P < .01). (B) When restricted to only patients with favorable risk, median OS was again significantly shorter in the older, sMut subset.
Discussion
NPM1 mutation is generally considered a favorable prognostic marker in AML.6 Only selected co-occurring mutations such as FLT3-ITD or TP53 escalate the risk under ELN 2017. ASXL1 and RUNX1, 2 of the 9 mutations highly specific for sAML, are the only sMut in ELN 2017 that defines adverse-risk disease.7 More recent reports, including the seminal study by Lindsley et al, provided strong evidence of a poor prognosis conferred by sMut in AML.4,10 Based on this, 1 major update in ELN 2022 is the inclusion of all sMut being categorized as adverse risk. However, it was recommended that sMut should not currently be used as an adverse prognostic marker in the setting of favorable-risk AML subtypes, pending further evidence.6
In this multicenter study of 233 patients with mutant NPM1 at diagnosis, we demonstrated that the presence of co-occurring sMut was strongly associated with adverse prognosis, including within the subset of patients with ELN 2017–defined favorable-risk disease. The facts that we observed an incidence of co-occurring sMut of ∼20% in NPM1 mutant AML and that most of these patients had ELN 2017 favorable-risk AML suggest that a substantial proportion of traditionally defined patients with favorable-risk AML indeed have poor outcomes based on the presence of these secondary-like mutations, negating the potentially beneficial impact of mutant NPM1. Within the sMut cohort, the rate of primary refractory disease was higher than in the non-sMut cohort, likely contributing to worse OS in this cohort, a finding consistent with prior studies showing a correlation between sMut and lower CR rates.10
We next looked at NPM1 VAF at diagnosis among patients with sMut and those with non-sMut together and separately to see whether it alters survival. Using NPM1 VAF threshold of 20%, there was no difference in OS among patients without sMut (P = .39). Surprisingly, in the sMut group, lower VAF (n = 16) had worse outcome than higher VAF (n = 15) (VAF ≤20% mOS, 7.9 months vs VAF >20% mOS, 42.4 months; P = .014). A prior study showed higher NPM1 VAF (> 40%) conferred worse outcomes in de novo AML.12 It is unclear whether the lower NPM1 VAF in this population may have contributed to the worse outcomes observed. It is possible in the setting of sMut, low NPM1 VAF represents a smaller prognostic role of this favorable-risk mutation and a larger adverse impact from co-occurring mutations. However, our sample size is small precluding more extensive analysis. Further studies are needed to address this question more definitively.
Older age is also known to have an important prognostic role in patients with NPM1-mutated AML. For example, among patients aged ≥55 years treated with intensive chemotherapy who were enrolled onto Southwest Oncology Group and UK National Cancer Research Institute/Medical Research Council clinical trials, NPM1-positive/FLT3-ITD–negative genotype was a favorable prognostic factor in patients aged ≤65 years but had no significant prognostic impact in those aged >65 years.13 In this study of patients with NPM1, those who carried sMut were generally older than patients without sMut, indicating age as a contributor to the worse outcomes among patients with sMut but clearly not the only driver. Consequently, they are also less likely to receive frontline intensive chemotherapy and subsequent allo-HCT. Interestingly, the negative prognostic impact of sMut in patients with mutant NPM1 was not observed in patients aged <60 years, raising the possibility that sMut is a less important prognostic factor in younger patients, although the numbers of younger patients with sMut in our study were very low, precluding any definitive conclusions in younger patients exclusively.
MRD in AML is becoming increasingly important for the assessment of response, monitoring, and prognosis.14 Different methods of measuring MRD exist, and its detection predicts increased risk of relapse and decreased survival.15,16 Ivey et al assessed a large cohort of patients with NPM1-mutated AML with MRD data available (n = 194) after 2 cycles of chemotherapy using quantitative polymerase chain reaction with a median sensitivity level of 10–5. They found those with undetectable MRD had a significantly lower cumulative incidence of relapse (3-year, 30% vs 80%) and higher OS (3-year, 75% vs 24%) than those with detectable MRD.16 In this study, patients who achieved NPM1 MRD negativity had significantly longer OS within the entire cohort as well as within the sMut group itself (mOS, 73.9 vs 12.3 months; P = .017). Unfortunately, the number of patients in our data set who had postremission MRD testing was low, because widescale implementation of such testing is relatively recent. A lack of standardization as to the precise timing of MRD testing in our patients also might have affected any interpretation of the ultimate impact of this testing among patients with sMut. Nonetheless, the observation that NPM1 MRD negativity in patients with sMut suggests the possibility of identifying subsets of patients with favorable outcomes in an otherwise poor-risk disease. Larger scale studies with more standardized MRD testing (and timing of testing) to confirm these observations are warranted.
In summary, our findings demonstrate that the co-occurrence of sMuts is not uncommon in NPM1-mutant AML, conferring a significantly detrimental effect on survival of such patients that is not well recognized by the current classification and prognostic systems. Our data not only support the recent ELN 2022 recommendations of sMut as an adverse prognostic indicator but strongly suggest that the risk classification of AML be further refined to consider the presence of sMuts as adverse risk in the presence of mutant NPM1. Such refinement would likely have significant implications on the management of AML along with clinical trial design, and additional studies should be undertaken to further strengthen and confirm these considerations to ultimately assist in the best utilization of current therapy and development of future therapies for AML.
Acknowledgment
The authors thank the patients and the supporting staff at each of the participating institution.
Authorship
Contribution: J.L. and O.C. designed the study; O.C., N.A., H.T., A.E., S.B., J.G., C.H., Y.D., L.Z., M.H., J.S., S.Y., A.K., E.P., P.D., D.S., K.S., R.K., and J.L. collected and analyzed the data; O.C. wrote the first draft; and all authors critically reviewed the first draft, provided edits, and approved the final version of the manuscript.
Conflict-of-interest disclosure: O.C. reports honoraria for consulting from Bristol Myers Squibb (BMS) and research funding from Jazz Pharmaceuticals. M.H. reports consulting and/or other fees from Adaptive Biotechnologies, Amgen, Aptitude Health, Blueprint Oncology, Celgene, Decibio, Diaceutics, Guidepoint, Seattle Genetics, Stemline, Tegus, Janssen, and BMS. C.T. is a current equity holder in private company, AbbVie. A.K. reports consulting and/or other fees from AbbVie, Blueprint Medicines, Celgene, Constellation, CTI Biopharma, Incyte, Novartis, Prelude, Protagonist, and Sierra Oncology; and grant funding from BMS, Constellation, Novartis, Prelude, Protagonist, and Sierra Oncology. E.P. reports consulting and/or other fees from Blueprint Medicines, Novartis, and Taiho Oncology. G.R. reports consulting and/or other fees from Actinium, AbbVie, Agios, Amgen, Astellas, AstraZeneca, BMS, Blueprint Medicines, bluebird bio, GlaxoSmithKline, Janssen, Jasper, Jazz, MEI Pharma, Mesoblast, Novartis, Pfizer, Syndax, and Takeda. P.D. reports consulting and/or other fees from Janssen, Servier, AbbVie, and BMS. D.S. reports consulting and/or other fees from Aprea Therapeutics, Avencell Europe GmbH, bluebird bio, BMS, Gilead Sciences, Incyte, Intellia Therapeutics, Janssen Global Services, Jazz Pharmaceuticals, Kite Pharma, Molecular Partners AG, Novartis, Servier Pharmaceuticals, Shattuck Labs, Syndax Pharmaceuticals, Syros Pharmaceuticals, and Takeda Pharmaceutical. K.S. reports consulting and/or other fees from Arog, Astellas Pharma, BerGenBio, BMS, Curis, Gilead Sciences, Mablytics, Novartis, Pfizer Pharmaceuticals, and Incyte. R.K. reports consulting and/or other fees from AbbVie, Acceleron Pharma, Agios Pharmaceuticals, Alexion Pharmaceuticals, BMS, Celgene, CTI Biopharma, Daiichi Sankyo, Geron, Janssen Biotech, Jazz Pharmaceuticals, Novartis Pharma, Pfizer, PharmaEssentia, Servier Pharmaceuticals LLC, Taiho Oncology, and Takeda Oncology. J.L. reports consulting and/or other fees from AbbVie, Boxer Capital, BMS, Jasper Therapeutics, Jazz Pharmaceuticals, Novartis Pharma, and Servier Pharmaceuticals. The remaining authors declare no competing financial interests.
Correspondence: Onyee Chan, Department of Malignant Hematology, Moffitt Cancer Center, 12902 Magnolia Dr, Tampa, FL 33612; email: onyee.chan@moffitt.org.
References
Author notes
Publication-related data are available upon reasonable request from the corresponding author, Onyee Chan (onyee.chan@moffitt.org).