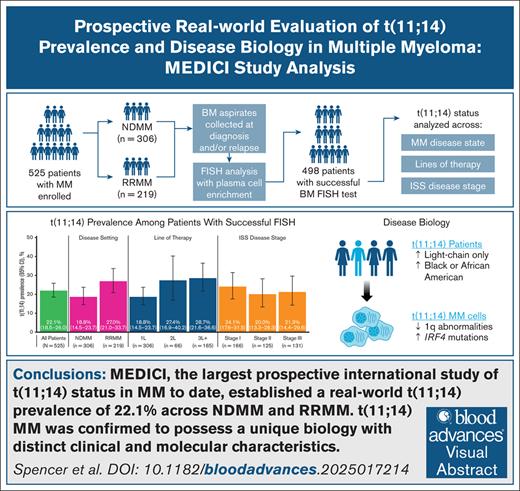
Key Points
This prospective international analysis demonstrated a real-world t(11;14) detection rate of 22.1% in patients with MM.
Increased frequency of IRF4 mutations was observed for t(11;14)-positive disease among patients with both NDMM and RRMM.
Visual Abstract
Genetic abnormalities in multiple myeloma (MM) influence treatment outcomes and may inform therapeutic decisions. The most common chromosomal translocation in MM is t(11;14); however, its role in disease progression is not well defined. We report results from MEDICI, a global, minimally invasive, prospective study evaluating t(11;14) status in newly diagnosed MM (NDMM) and relapsed/refractory MM (RRMM). MEDICI enrolled adult patients (≥18 years) with MM with bone marrow (BM) aspirates collected at diagnosis and/or disease relapse. The primary objective was to determine t(11;14) prevalence in patient BM samples using fluorescent in situ hybridization (FISH) with plasma cell enrichment. Samples from 525 patients (NDMM, n = 306; RRMM, n = 219) were analyzed for t(11;14), with 498 patients (95%) having successful BM FISH test. Prevalence of t(11;14) was 22.1% (110/498 patients; 95% confidence interval [CI], 18.5-26.0) in the overall patient population, 18.8% (56/298 patients; 95% CI, 14.5-23.7) in NDMM, and 27.0% (54/200 patients; 95% CI, 21.0-33.7) in RRMM. Patients with t(11;14)-positive MM were evenly distributed across disease stages (stage I, 24.1%; stage II, 20%; stage III, 21.3%); however, higher rates were observed in African American patients (RRMM odds ratio [OR], 7.27, 95% CI, 1.33-39.83; P = .022) and light-chain only disease (NDMM OR, 3.02, 95% CI, 1.33-6.87; P = .008). Chromosome 1q abnormalities occurred more often in the absence of t(11;14); however, higher frequency of mutation in the plasma cell differentiation regulator IRF4 was detected in t(11;14) presence. In conclusion, the results from MEDICI demonstrated reproducible t(11;14) detection with a real-world prevalence of 22.1% in MM. This trial was registered at www.ClinicalTrials.gov as #NCT04721002.
Introduction
Multiple myeloma (MM) is a hematologic malignancy characterized by clonal proliferation of bone marrow (BM) plasma cells (PCs) with a variety of well-defined genetic abnormalities.1,2 The most common translocation occurring in MM is of chromosomes 11 and 14, referred to as t(11;14), which has been estimated to be present in ∼15% to 20% of patients with MM.3,4 The impact of t(11;14) on prognosis is unclear. The 2013 Mayo Stratification of Myeloma and Risk-Adapted Therapy criteria considered t(11;14) a standard risk factor based on data available at that time (eg, the French IFM99 trial).3,5 However, more recent retrospective studies conducted after the availability of novel agents (eg, immunomodulatory agents) have associated t(11;14) with inferior overall survival (OS), suggesting that it confers a higher-than-standard risk,6-8 but other studies have not observed such an effect, and the risk associated with t(11;14) remains controversial.9,10 Understanding the prevalence and biology of t(11;14) in patients with MM in a global, real-world setting may provide insights into targeted therapies for this subpopulation.
Pathobiological features of t(11;14) MM are driven by the proto-oncogene CCDN1 on chromosome 11, resulting in overexpression of cyclin D1.11 Cells with this phenotype show elevated levels of the antiapoptotic protein BCL-2, on which they depend for survival.12 Clinical trials have investigated the use of BCL-2 inhibitors in MM, including venetoclax,13-16 lisaftoclax,17,18 sonrotoclax,19 and loxo-338.20
Although BM biopsy is performed for MM diagnosis and collection of biopsy materials is recommended for evaluation of cytogenetic abnormalities,21 evaluation of t(11;14) status is not currently required for risk stratification and methods used to assess t(11;14) lack standardization. PC enrichment is crucial for reliable identification of cytogenetic abnormalities by fluorescence in situ hybridization (FISH), particularly in samples with <20% PCs22,23; however, enrichment is not performed consistently, and enrichment methods differ in PC yield, resulting in inconsistencies in the results of t(11;14) assessments.24 Therefore, reliable data on the prevalence of t(11;14), particularly across patient subgroups based on clinical features and demographic characteristics, including stability of t(11;14) over the disease course, are limited. To address these gaps, the global MEDICI study (ClinicalTrials.gov identifier: NCT04721002) was performed to obtain data on t(11;14) as determined by FISH on CD138-enriched BM samples in a real-world setting. To our knowledge, this is the largest prospective study of t(11;14) prevalence among patients with MM.
Methods
Study design and patients
MEDICI (ClinicalTrials.gov identifier: NCT04721002) was a global, minimally invasive study. Eligible patients were aged ≥18 years with confirmed newly diagnosed MM (NDMM) or relapsed/refractory MM (RRMM), had BM aspirates and blood samples available at time of diagnosis and/or confirmation of relapse, and provided informed consent for the use of biological material for research purposes. Eligibility did not restrict line or type of treatment. Patients were enrolled between 7 July 2021 and 31 October 2023, across 20 countries. Duration of follow-up was not mandated by the study, but longitudinal samples were collected from previously enrolled patients who relapsed in the course of the study recruitment period. Obtained results did not affect therapeutic decisions. This study was conducted in accordance with ethical principles that have their origin in the current Declaration of Helsinki and was consistent with the International Conference Harmonization Good Clinical Practice. Any independent review board or ethic committee approval or notification was obtained before the initiation of the study as necessary per local regulation.
End points and assessments
The primary objective of the study was to estimate the prevalence of t(11;14)-positive NDMM and RRMM using interphase FISH analysis with BM PC enrichment. Key secondary objectives included evaluation of the change in intrapatient t(11;14) status in longitudinal BM samples collected at diagnosis and across lines of therapies. Other secondary objectives included prevalence of t(11;14) across lines of therapy and distribution of FISH fusion (F) categories among t(11;14)-positive samples. BM samples were also subjected to whole-exome sequencing for mutational analysis (supplemental Methods).
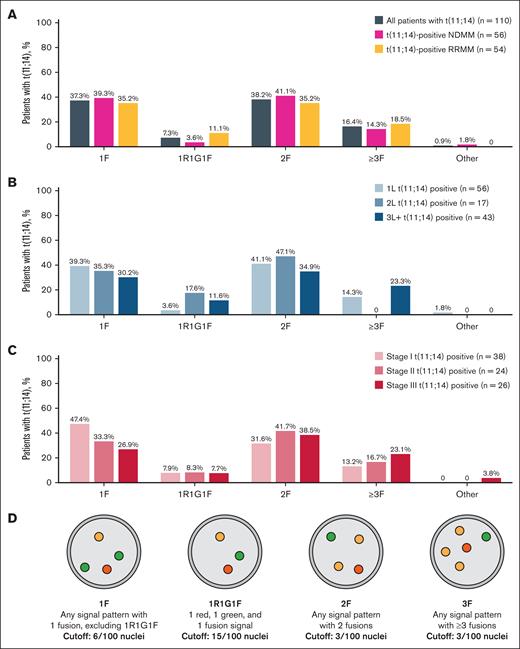
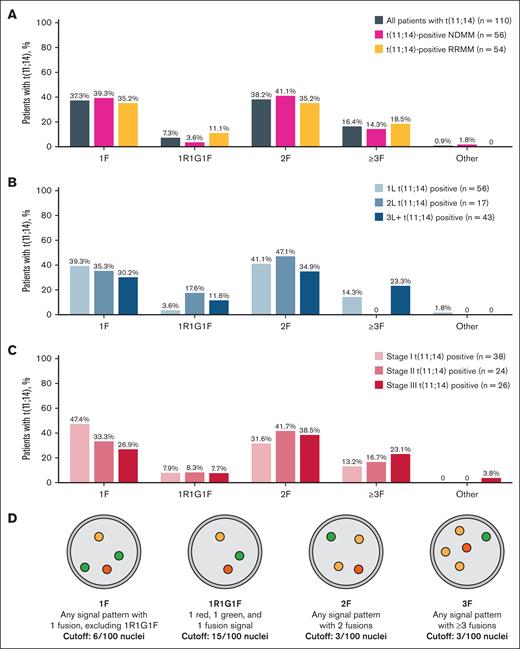
For assessment of t(11;14), BM PCs underwent bead-based enrichment using CD138+ selection with immunomagnetic particles (EasySep Human Bone Marrow CD138 Positive Selection Kit [IVD]; STEMCELL Technologies, Vancouver, BC, Canada) and locally evaluated using Vysis IntelliFISH CCND1/IGH XT FISH Probe Kit (Abbott, Abbott Park, IL). Samples were then locally analyzed for t(11;14) positivity using FISH F patterns (1F, 1R1G1F, 2F, ≥3F; Figure 2D). A positive sample contained ≥6 of 100 nuclei with 1F, ≥15 of 100 nuclei with 1R1G1F, ≥3 of 100 nuclei with 2F, or ≥3 of 100 nuclei with ≥3F. Demographic characteristics and treatments were collected from electronic health records.
Fusion phenotypes of t(11;14) samples. Fusion phenotypes by disease setting (A), line of therapy (B), and stage of disease (C), and visual schematic of fusion patterns (D).
Fusion phenotypes of t(11;14) samples. Fusion phenotypes by disease setting (A), line of therapy (B), and stage of disease (C), and visual schematic of fusion patterns (D).
Statistical analysis
A sample size of ≥500 patients was estimated to meet primary and secondary objectives. For the primary objective of prevalence of t(11;14), a sample size of ≥240 could provide precision of ≥5% based on the reported t(11;14) prevalence of ∼15% to 20%.3,4 For the secondary objective of intrapatient t(11;14) longitudinal stability, assuming 85% or 90% stability across lines, 30% progressing from first-line therapy (1L) and 50% in later-line therapies, and 50% sample availability, a sample size of 500 was projected to result in precision of 10% or better. Patient demographics, clinical characteristics, diagnosis, and medical history were reported using descriptive statistics.
The analysis populations were defined as population 1, consisting of all patients with ≥1 sample, and population 2 consisting of all population 1 patients with longitudinal samples available. The primary and FISH fusion category analyses were performed on population 1, with no hypothesis testing. Longitudinal analysis was carried out in analysis population 2. A logistic regression model was conducted to calculate odds ratios (ORs) for t(11;14) positivity vs negativity by baseline patient and disease characteristics (supplemental Methods). Multiple tests were conducted without applying a multiplicity correction. Statistical significance was determined by a P value of ≤.05; however, this was considered nominal. All calculations were performed using R software version 3.4.1 (R Foundation for Statistical Computing, Vienna, Austria).
Results
Patient demographics and baseline characteristics
At the data cutoff of 31 October 2023, 592 adults were screened to participate in the study, of whom 525 (NDMM, n = 306; RRMM, n = 219) were enrolled and analyzed, including 14 patients with data available for longitudinal analysis. BM aspirates were obtained from 526 of 538 patients (97.8%) with a confirmed diagnosis of NDMM (n = 308) or RRMM (n = 230). Almost all samples (525/526 aspirates [99.8%]) included a volume, median of 10 mL (range, 1.0-15.0), that was sufficient for the FISH testing. The plasma cell percentage before CD138+ selection was available on a subset of BM aspirates (392/525 aspirates [74.7%]), which ranged from 0.0% to 100.0% (median, 18.5%) by morphologic or flow cytometric analysis.
Patients had a median age of 66.0 years and were predominantly White (74.5%) and enrolled from Europe (61.9%; Table 1). More than half (55.4%) were men. Most patients had an Eastern Cooperative Oncology Group performance status of ≤2 (83.2%), with disease stage I (31.6%) being slightly more common than later stage disease (stage II, 23.8%; stage III, 25.0%). Patients with RRMM had a median of 2 (range, 1-8) prior lines of therapy. Although presence of chromosome 17p deletion [del(17p)] or chromosome 1q gain [gain(1q)] was unknown for half of the patients [del(17p), 45.5%; gain(1q), 50.5%], most tested samples did not have these cytogenetic abnormalities (17p not deleted, 219/286 [77%]; 1q no gain, 175/260 [67%]). Patients with NDMM and RRMM exhibited broadly similar demographics and baseline characteristics.
FISH testing performance
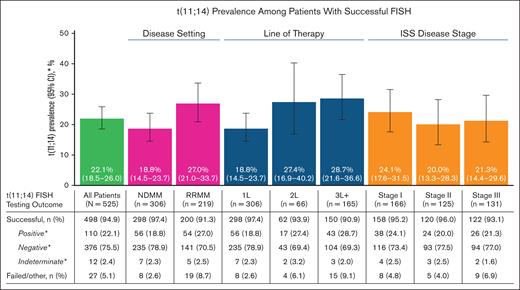
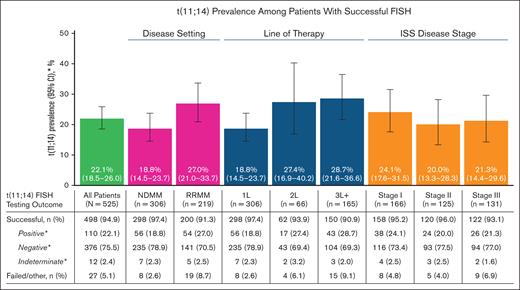
After PC enrichment, FISH testing for t(11;14) was successful for 498 samples (94.9% success rate) and failed in 27 samples (5.1% failure rate) of the 525 total samples available (Figure 1). The success rate was the highest in NDMM (97.4%) and decreased with each successive line of treatment, reaching a minimum of 90.9% in the third-line or greater (3L+) therapy. Samples across different disease stages had similar success rates (stage I, 95.2%; stage II, 96.0%; stage III, 93.1%).
Prevalence of t(11;14) by disease severity and FISH testing outcomes. ∗Percentages for the categories of positive, negative, and indeterminate were based on the number of patients with successful t(11;14) FISH testing and excluded patients with failed/other test results. ISS, International Staging System.
Prevalence of t(11;14) by disease severity and FISH testing outcomes. ∗Percentages for the categories of positive, negative, and indeterminate were based on the number of patients with successful t(11;14) FISH testing and excluded patients with failed/other test results. ISS, International Staging System.
Prevalence and fusion patterns of t(11;14)
Of the 498 successfully analyzed samples, 22.1% (110/498) were positive for t(11;14), 75.5% (376/498) were negative for t(11;14), and 2.4% (12/498) had indeterminate results (Figure 1). The presence of t(11;14) was numerically higher in RRMM (27.0%) than in NDMM (18.8%) with no further increase in later lines of treatment (second-line therapy [2L], 27.4%; 3L+, 28.7%). Similar t(11;14) prevalence was observed across different disease stages (stage I, 24.1%; stage II, 20.0%; stage III, 21.3%).
Among the 110 t(11;14)-positive samples, the median percent of abnormal nuclei was 77.0% (range, 7.0%-99.0%). Samples from patients with NDMM had a higher percentage of abnormal nuclei than samples from patients with RRMM (median, 79.5% vs 69.0%), although this percentage did not further decrease with additional lines of treatment (median, 2L, 60.0%; 3L+, 69.5%). There was a trend toward an increased percentage of abnormal nuclei with later disease stage (median: stage I, 71.5%; stage II, 79.0%; stage III, 85.0%).
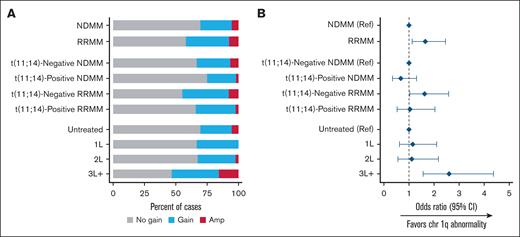
Most t(11;14)-positive samples displayed 1F (37.3%) or 2F (38.2%) fusion patterns, with ≥3F (16.4%) and 1R1G1F (7.3%) being less common (Figure 2A). The distribution of fusion patterns was generally similar between patients with NDMM vs RRMM. The proportion of 1F fusions decreased from 1L (39.3%) to the later lines of treatment (2L, 35.3%; 3L+, 30.2%; Figure 2B). Other fusion patterns varied between treatment line settings with no clear trends. The presence of the 1F pattern decreased with higher disease stage (stage I, 47.4%; stage II, 33.3%; stage III, 26.9%), whereas the ≥3F pattern increased (stage I, 13.2%; stage II, 16.7%; stage III, 23.1%; Figure 2C). 1R1G1F had equal prevalence of ∼8% across all disease stages, and proportion of 2F pattern varied.
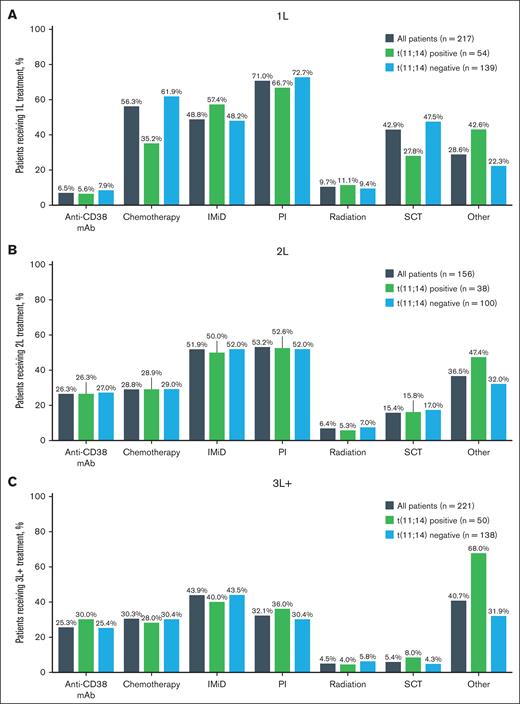
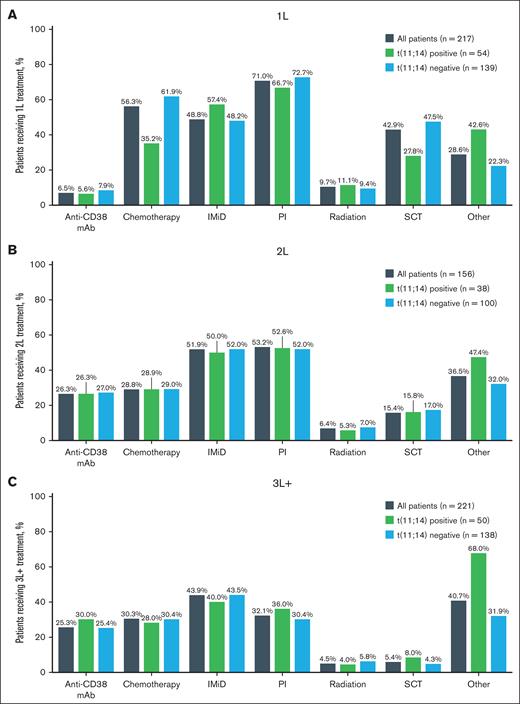
Treatment patterns by t(11;14) status
Prior types of treatment were analyzed among patients with RRMM. There was no difference between t(11;14)-positive and t(11;14)-negative subgroups regarding what type of treatment patients underwent most often in each line of therapy (Figure 3). In the 1L setting, the most common type of therapy was proteasome inhibitor based: 66.7% in t(11;14)-positive patients and 72.7% on t(11;14)-negative patients (Figure 3A). Half of the patient population received proteasome inhibitor [(t(11;14) positive, 52.6%; t(11;14) negative, 52.0%] and/or immunomodulatory drug (IMiD) treatment [t(11;14) positive, 50.0%; t(11;14) negative, 52.0%] as 2L treatment (Figure 3B). IMiD was the most used treatment option [t(11;14) positive, 40.0%; t(11;14) negative, 43.5%] in the 3L+ setting (Figure 3C).
Prior treatments received by line of therapy. Prior treatment received by t(11;14) status in 1L (A), 2L (B), and 3L+ (C) settings. Patients were counted once per type of prior therapy received. mAb, monoclonal antibody; PI, proteasome inhibitor; SCT, stem cell transplant.
Prior treatments received by line of therapy. Prior treatment received by t(11;14) status in 1L (A), 2L (B), and 3L+ (C) settings. Patients were counted once per type of prior therapy received. mAb, monoclonal antibody; PI, proteasome inhibitor; SCT, stem cell transplant.
Stability of t(11;14)
Of the 14 patients included in the longitudinal population, 12 had paired BM samples tested for t(11;14) at enrollment and relapse. Furthermore, 11 patients had successful paired FISH tests and were included in the stability analysis. Among these, 5 were t(11;14) positive and 6 were negative. All 11 demonstrated the same t(11;14) status at enrollment and relapse. Among 5 patients with t(11;14)-positive samples, fusion patterns similarly remained unchanged: 1 patient had a 1R1G1F fusion pattern at diagnosis and at later sample analysis, whereas the other 4 patients consistently had 2F fusion patterns.
Before enrollment in the MEDICI study, 98 of 219 patients with RRMM (44.7%) had previously undergone FISH testing, with 67 of 98 (58.8%) incorporating PC enrichment similar to the MEDICI method. Prior successful FISH testing results were available for 41, 5, and 26 patients at 1L, 2L, and 3L+ treatment, respectively. Moreover, >90% of patients demonstrated concordant results at each line of treatment between the prior and MEDICI FISH testing (1L, 92.7% [38/41]; 2L, 100% [5/5]; 3L, 92.3% [24/26]).
Correlation of t(11;14) positivity with patient demographics and clinical characteristics
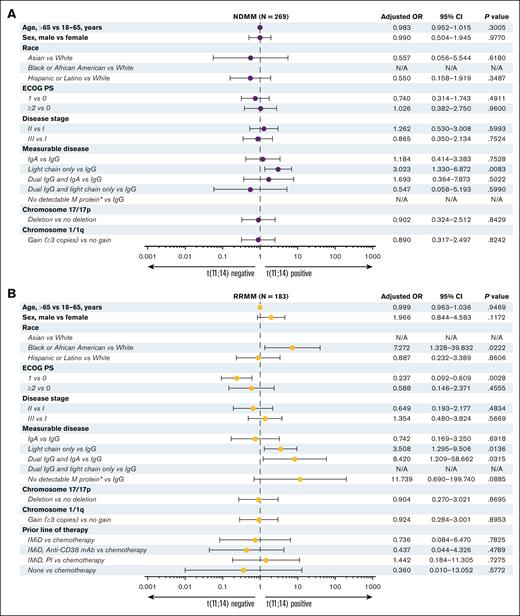
In NDMM, t(11;14) positivity or negativity was not correlated with age, sex, race, Eastern Cooperative Oncology Group performance status, disease stage, or the presence of cytogenetic abnormalities del(17p) or gain(1q) (Figure 4A). Conversely, there was a significantly higher likelihood of t(11;14) in patients with light-chain only disease when compared with immunoglobulin G disease (IgG; OR, 3.02; 95% confidence interval [CI], 1.33-6.87; P = .008).
Adjusted OR for t(11;14)-positive or -negative status among the patients at enrollment. Patients with NDMM (A) or RRMM (B). ∗Includes nonsecretory MM. N/A, not applicable; PI, proteasome inhibitor.
Adjusted OR for t(11;14)-positive or -negative status among the patients at enrollment. Patients with NDMM (A) or RRMM (B). ∗Includes nonsecretory MM. N/A, not applicable; PI, proteasome inhibitor.
Among the patients with RRMM (Figure 4B), Black or African American patients were significantly more likely to have t(11;14) than White patients (OR, 7.27; 95% CI, 1.33-39.83; P = .022). Similar to NDMM, patients with light-chain only disease vs IgG disease demonstrated a greater likelihood of t(11;14) positivity (OR, 3.51; 95% CI, 1.30-9.51; P = .014). Moreover, in contrast to NDMM, in RRMM, there was a statistically higher rate of t(11;14) observed in dual IgG disease and IgA disease elevation vs IgG disease alone (OR, 8.42; 95% CI, 1.21-58.66; P = .03). A trend toward t(11;14) positivity was observed for nonsecretory MM disease vs IgG disease (OR, 11.74; 95% CI, 0.69-199.74; P = .089), albeit the sample size of patients with RRMM with no detectable M protein or nonsecretory MM was very small (all patients, n = 6; t(11;14) positive, n = 2; t(11;14) negative, n = 2; Table 1). No significant difference in t(11;14) status was observed based on cytogenetic abnormalities [del(17p) or gain(1q)] or type of previous therapy.
Whole exome sequencing analysis of chromosome 1q abnormalities and MM driver gene mutations
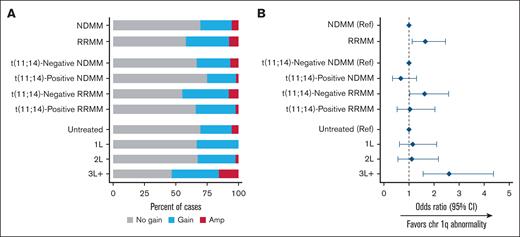
The prevalence of chromosome 1q abnormalities, gain(1q) and amp(1q), was, as would be expected, higher in patients with RRMM vs NDMM and in patients with ≥3 lines of treatment vs those with ≤2 lines of treatment (Figure 5). Patients with t(11;14)-negative MM, both NDMM and RRMM, had higher likelihood of 1q+ vs those with t(11;14)-positive status from the corresponding MM subgroup.
Gain(1q) and amp(1q) prevalence. Rates (A) and odds (B) of chr 1q abnormalities by population. Amp, amplification; chr 1q, chromosome 1q; Ref, reference.
Gain(1q) and amp(1q) prevalence. Rates (A) and odds (B) of chr 1q abnormalities by population. Amp, amplification; chr 1q, chromosome 1q; Ref, reference.
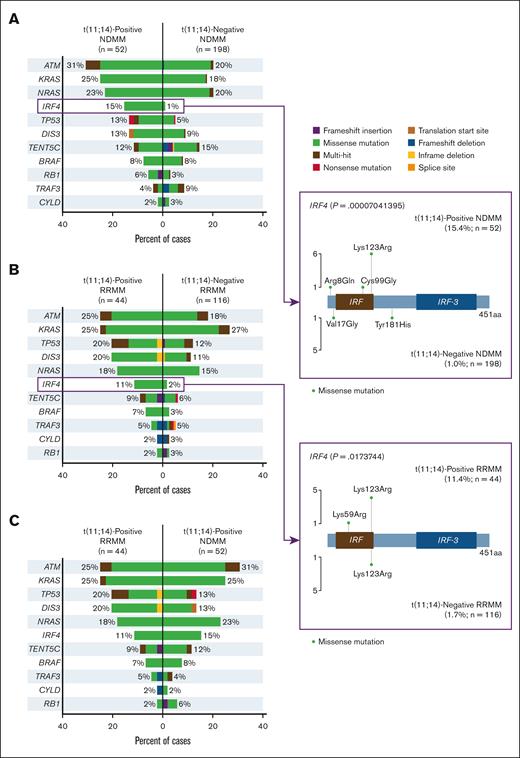
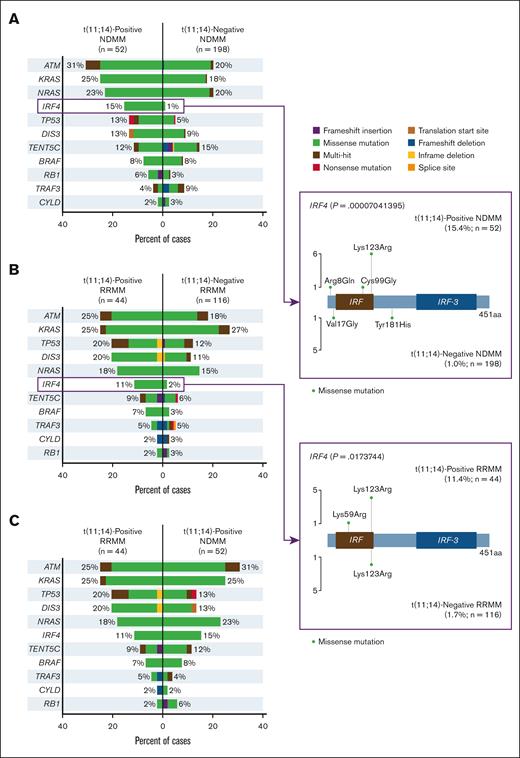
Analysis of MM driver gene mutations revealed mutation frequencies for ATM, KRAS, and NRAS among patients with t(11;14)-positive vs -negative NDMM of 31% vs 20%, 25% vs 18%, and 23% vs 20%, respectively (Figure 6A). Among patients with t(11;14)-positive and -negative RRMM, the highest mutation frequencies were for ATM, KRAS, TP53, DIS3, and NRAS, at 25% vs 18%, 25% vs 27%, 20% vs 12%, 20% vs 11%, and 18% vs 15%, respectively (Figure 6B). IRF4 mutation frequency was significantly higher among patients with t(11;14)-positive vs -negative NDMM, at 15% and 1%, respectively (P < .0001) and among patients with t(11;14)-positive vs -negative RRMM, 11% and 2%, respectively (P = .017). Among patients positive for t(11;14), mutation frequency for ATM was numerically higher in NDMM vs RRMM (31% vs 25%) and was the same for KRAS (both 25%; Figure 6C).
Mutational frequencies in MM driver genes.P values not adjusted for multiplicity. aa, amino acid.
Mutational frequencies in MM driver genes.P values not adjusted for multiplicity. aa, amino acid.
Discussion
Translocation t(11;14) dysregulating cyclin D is thought to represent a primary oncogenetic event25 and is the most common translocation in MM, with an estimated prevalence of ∼15% to 20%.3,4 However, the real-world prevalence of t(11;14) and its relationship to disease severity is unclear. Prior work has predominantly reported t(11;14) status in NDMM because re-evaluation of mutational status is not widely undertaken at relapse,26 with few observational studies having investigated the prevalence of t(11;14) among patients with RRMM, with one of these studies reporting t(11;14) in 14 of 77 (18%) patients.27 In the MEDICI study, PC enrichment was leveraged in an attempt to achieve a high success rate for t(11;14) FISH among an international patient population, including >200 patients with RRMM.
In our analysis, the prevalence of t(11;14) was 22.1%, with a similar prevalence across disease stages (Figure 1), which is slightly higher than previously reported.4 This difference might be attributed to the PC enrichment uniformly used in the MEDICI study, which has previously been found to improve detection of cytogenetic abnormalities in MM.24 Although the absence of universal use of PC enrichment in the prior studies could have resulted in underreporting of t(11;14) prevalence.
Notably, the prevalence of t(11;14) was numerically higher (27.0%) in RRMM than in NDMM (18.8%). A possible reason for this difference is that patients with higher risk disease corresponding with t(11;14)-negative status were more likely to succumb to their disease earlier than patients with t(11;14) and lower risk disease. Therefore, patients without t(11;14) could be underrepresented in RRMM population compared with the NDMM group resulting in an apparent increase in the proportion of t(11;14)-positive patients at later lines of treatment. As previously reported, t(11;14) occurs early in tumorigenesis and remains stable throughout disease evolution, including from monoclonal gammopathy of undetermined significance/smoldering MM transition to MM, and across lines of MM treatment to relapse.28 Therefore, transition from t(11;14)-negative to t(11;14)-positive status at first relapse would be unlikely. Similar to the prior report, intrapatient longitudinal analysis data reported here also revealed stability of t(11;14) status, although the small sample size limits interpretation of these data.28 The overwhelming majority of patients with available historical FISH testing results also showed the same t(11;14) status before and after treatment.
Despite the stability of t(11;14) status, MM is highly heterogenous, and variability was observed between t(11;14)-positive and -negative disease in this study. Analysis of patients tested for gain(1q)/amp(1q) demonstrated that the prevalence was lower among patients with NDMM than those with RRMM, as would be expected. Interestingly, chromosome 1q abnormalities were lowest among patients with t(11;14)-positive NDMM and highest among those with t(11;14)-negative RRMM or in the 3L+ setting (Figure 5). These findings differ somewhat from prior data that revealed no difference in gain(1q) between t(11;14)-positive vs -negative MM.27 Although not considered high risk according to the current International Myeloma Working Group classification, gain(1q) has been associated with reduced OS and is a poor prognostic marker for patients treated with novel agents.29,30
Outcomes for patients with t(11;14)-positive MM differ based on study treatment and biological characteristics of the disease. Initial studies were small and suggested that outcomes for t(11;14)-positive disease were unfavorable.31,32 Larger, retrospective studies with better characterization of t(11;14) subsequently indicated that outcomes with prenovel era treatments were not inferior to other subgroups.33-35 Treatment with novel targeted therapies such as proteasome inhibitors and IMiDs have improved survival in MM,36,37 but varying outcomes suggest that underlying biological factors also play a role. For example, patients with t(11;14)-positive disease without CD20 were reported to have worse outcomes compared with patients with t(11;14)-positive disease with CD20.27 Prior reports noted the presence of t(11;14) to be associated with higher incidence of light-chain MM,38 which has poor prognosis. In both the NDMM and RRMM populations in our study, light-chain only disease was more common in patients with t(11;14) positivity (Figure 4), which aligns with prior reports. Although t(11;14) status is not currently driving treatment decisions, these results suggest that patients with t(11;14)-positive MM represent a unique, albeit heterogenous, subgroup that may not be achieving optimal treatment outcomes as presently being treated.
Analysis of MM driver mutations revealed the highest frequency of alterations in ATM, KRAS, and NRAS for NDMM and ATM, KRAS, and TP53 for RRMM (Figure 6). TP53 mutation is a known negative prognostic factor in multiple cancers including MM.39 Our study revealed numerically higher frequency of TP53 mutation in t(11;14)-positive subgroup compared with the patients with t(11;14)-negative status in both NDMM and RRMM. The frequency of DIS3 mutation in this study was also numerically higher among patients with t(11;14)-positive vs -negative disease in both NDMM and RRMM, in agreement with previously reported data.28DIS3 has been implicated in centrosome duplication cycle control in MM, and its loss-of-function was found to impair cell-cycle progression in MM cells in vitro.40BRAF alterations were also numerically higher in t(11;14)-positive vs -negative RRMM in this study but were previously reported at lower frequency in t(11;14)-positive vs -negative disease.28 Our study reported a significantly higher frequency of mutations in the plasma cell differentiation regulator IRF4 for t(11;14)-positive vs -negative disease for both NDMM and RRMM. In addition, IRF4 has been found to regulate plasma cell differentiation and to induce MYC expression in MM, which promotes MM cell survival.41 Downregulation of IRF4 is being investigated as a treatment approach for MM. Lys123Arg, a missense IRF4 mutation observed in our study, was previously detected in other patients with MM, although its effect on disease progression and response to treatment is not well understood.42
In the randomized, phase 3 BELLINI trial (NCT02755597), venetoclax in combination with bortezomib and dexamethasone provided significantly improved progression-free survival and response rates but unfavorable OS compared with placebo combined with bortezomib and dexamethasone in the overall population of patients with RRMM; however, the risk-benefit ratio was favorable among patients with RRMM harboring t(11;14).14 Treatment with venetoclax monotherapy has demonstrated evidence of efficacy among patients with t(11;14),13,43 and clinical studies have explored venetoclax combination therapy in this patient population. Preliminary results from the phase 1 study of venetoclax plus daratumumab and dexamethasone with or without bortezomib in RRMM (M15-654 study; NCT03314181) revealed high and durable responses in patients with t(11;14).16 In the phase 2 study of venetoclax with carfilzomib and dexamethasone in RRMM (M15-538 study; NCT02899052), the ORR in patients with RRMM was higher in those with t(11;14) vs those without t(11;14), at 92% and 75%, respectively.15 Given the high heterogeneity of t(11;14)-positive MM, there may be opportunities for t(11;14)-directed combination therapy with venetoclax.
Recent large-scale reports focused on t(11;14) have been limited to patients with NDMM and patients located in the United States.44,45 To our knowledge, this study is the largest prospective international study to date to estimate t(11;14) prevalence and represents the largest cohort of patients with RRMM tested for t(11;14) by FISH with PC enrichment. MEDICI demonstrated reproducibility of determining t(11;14) status using this assay in a real-world setting. However, further studies are required to better understand t(11;14) MM and its impact on response to treatment and clinical outcomes.
Acknowledgments
AbbVie and the authors thank all the trial investigators and the patients who participated in this clinical trial. Medical writing support was provided by Svetlana Pitts of Bio Connections, LLC, and funded by AbbVie.
This study was sponsored and funded by AbbVie. AbbVie funded ICON plc for this study.
AbbVie participated in the study design, research, analysis, data collection, interpretation of data, review, and approval of the publication. No honoraria or payments were made for authorship.
Authorship
Contribution: D.F., J.A.R., and C.H. conceptualized and designed study; O.H., R.T., and K.L. provided administrative support; A.S., M.G., D.C., A.L., N.C.G., F.E., D.B.E.F., R.H., F.S., G.K., R.B., S.B.K., M.P.C., H.S., J.M.-L., E.d.Q.C., D.L.C.d.F., M.O., E.A., Ö.G.S., V.M., and C.-L.T. provided study materials or patients; A.S., M.G., D.C., A.L., N.C.G., F.E., D.B.E.F., R.H., F.S., G.K., R.B., S.B.K., M.P.C., H.S., J.M.-L., E.d.Q.C., D.L.C.d.F., M.O., E.A., Ö.G.S., V.M., C.-L.T., X.L., L.M., P.J., E.L.R., S.K., and A.N. collected and assembled the data; A.S., D.F., R.T.-M., X.L., L.M., P.J., E.L.R., S.K., A.N., J.A.R., C.H., and O.H. analyzed and interpreted data; and all authors had access to relevant data, participated in the drafting, review, and approval of this publication, and were accountable for all aspects of the work.
Conflict-of-interest disclosure: A.S. has received research funding and honoraria from AbbVie. M.G. has served in a consultancy/advisory role for Amgen, Takeda, Karyopharm, and GlaxoSmithKline (GSK); and has received honoraria from Amgen, Janssen Cilag, Genesis Pharma, Karyopharm, Takeda, Sanofi, and GSK. D.C. has served in a consultancy/advisory role for Accord and Genesis Pharma. N.C.G. has received honoraria from Janssen, Amgen, and Sanofi. F.E. has received honoraria from Amgen, Janssen, and Sanofi; and has served in a consultancy/advisory role for Takeda, Amgen, Janssen, Sanofi, Bristol Myers Squibb (BMS)/Celgene, BeiGene, and GSK. D.B.E.F. has served in a consultancy/advisory role for Takeda, Janssen, and Sanofi; has received honoraria from Janssen, Sanofi, and BMS/Celgene; and has received research funding from AbbVie, Roche, Janssen, and GSK. R.H. has served in a consultancy/advisory role for Janssen, Amgen, Celgene, AbbVie, BMS, Novartis, PharmaMar, and Takeda; and has received honoraria from Janssen, Amgen, Celgene, BMS, PharmaMar, and Takeda. F.S. has served in a consultancy/advisory role for AbbVie; and has received honoraria from Janssen and Novartis. G.K. has received honoraria from Janssen, Pfizer, BMS, Sanofi, Kedrion, Cellectar, and Arcellx; and has received institutional research funding from BMS, Arcellx, Janssen, and AbbVie. S.B.K. has received honoraria from and served in a consultancy/advisory role for Amgen, Janssen, AbbVie, and Takeda. M.P.C. has received honoraria from Takeda, Janssen, Pfizer, Gilead, AstraZeneca, and Amgen. H.S. has served as a speaker for Incyte, Blueprint Medicines, Astellas, and Sanofi; and has participated in clinical research funded by AbbVie. J.M.-L. has served in a consultancy/advisory role for Novartis, Janssen, BMS, Sanofi, GSK, Gilead, Roche, and Astellas; and has received research funding from BMS, Janssen, and Roche. E.d.Q.C. has served in a consultancy role for Janssen, BMS, Sanofi, GSK, Takeda, and Amgen; and has received research funding from Janssen. D.L.C.d.F. has served as a speaker for Janssen, AstraZeneca, Zodiac, Roche, Novartis, Takeda, Amgen, Roche, Libbs, BMS, Celgene, Pfizer, AbbVie, Sanofi, Pint Pharma, and Knight Therapeutics/United Medical; has served as an advisory board member for Kite/Gilead, Knight Therapeutics/United Medical, Janssen, Novartis, Amgen, Takeda, Roche, BMS, AbbVie, Libbs, Celgene, and Eurofarma; has served as a steering committee member for Janssen; and has served on an independent data monitoring committee for Libbs/mAbxience. M.O. has received honoraria from AbbVie, Amgen, BMS, Celgene, GSK, Janssen, Roche, Sanofi, and Takeda; and has served in a consultancy/advisory role for AbbVie, Amgen, BMS, Celgene, GSK, Janssen, Roche, Sanofi, and Takeda. E.A. has served in a consultancy/advisory role for Janssen, Pfizer, Sanofi, and BeiGene. Ö.G.S. has served in a consultancy/advisory role for Takeda, Janssen, AbbVie, and Sanofi; and has received honoraria from Janssen, Sanofi, BMS, Amgen, and Astellas Pharma. V.M. has served in a consultancy/advisory role for Amgen, BMS, GSK, Janssen, and Sanofi; and has received honoraria from Amgen, BMS, Janssen, and Sanofi. O.H. is employed with ICON plc. R.T., K.L., D.F., R.T.-M., X.L., L.M., P.J., E.L.R., S.K., A.N, J.A.R., and C.H. are employed with AbbVie and may hold stock or other options. The remaining authors declare no competing financial interests.
Correspondence: Andrew Spencer, Malignant Haematology, Transplantation, and Cellular Therapies Service, Alfred Health-Monash University, 55 Commercial Rd, Melbourne, VIC 3004, Australia; email: andrew.spencer@monash.edu.
References
Author notes
J.A.R. and C.H. are joint senior authors.
Presented, in part, at the International Myeloma Society Annual Meeting, Rio De Janeiro, Brazil, 25 to 28 September 2024.
AbbVie is committed to responsible data sharing regarding the clinical trials they sponsor. This includes access to anonymized, individual, and trial-level data (analysis data sets), as well as other information (eg, protocols, clinical study reports, or analysis plans), as long as the trials are not part of an ongoing or planned regulatory submission. This includes requests for clinical trial data for unlicensed products and indications.
These clinical trial data can be requested by any qualified researchers who engage in rigorous, independent, scientific research and will be provided after review and approval of a research proposal, statistical analysis plan, and execution of a data-sharing agreement. Data requests can be submitted at any time after approval in the United States and Europe and after acceptance of this article for publication. The data will be accessible for 12 months, with possible extensions considered. For more information on the process or to submit a request, visit the following link: https://vivli.org/ourmember/abbvie/, then select “Home.”
The full-text version of this article contains a data supplement.