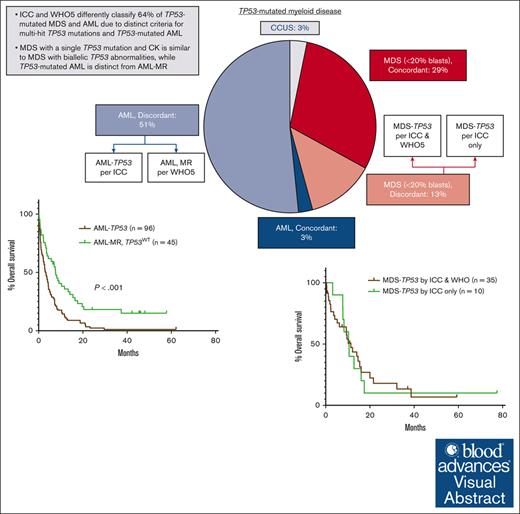
Key Points
The ICC and WHO5 differently classify 64% of TP53-mutated disease due to distinct criteria for multihit TP53 mutations and TP53-mutated AML.
MDS with a single TP53 mutation and CK is similar to MDS with biallelic TP53 abnormalities, whereas TP53-mutated AML is distinct from AML-MR.
Visual Abstract
The International Consensus (ICC) and World Health Organization fifth edition (WHO5) classifications introduced new, but differing, categories of myeloid disease defined by TP53 mutations. We reviewed a cohort of 188 cases of TP53-mutated myeloid disease to determine how diagnoses and outcomes differ between the 2 classifications. Overall, 120 (64%) cases were classified differently by the ICC and WHO5, including 24 of 80 (30%) cases with <20% blasts. These cases were discrepantly categorized primarily because of inclusion of complex karyotype (CK) as a surrogate for biallelic TP53 inactivation only in the ICC. However, there were no significant differences in clinicopathologic characteristics or overall survival between cases categorized as TP53-mutated disease by both classifications and those with a single TP53 mutation and CK, suggesting that CK reliably identifies TP53-mutated cases with biallelic TP53 inactivation. Most cases of acute myeloid leukemia (AML; 96/102 [94%]) were discrepantly diagnosed between the ICC and WHO5 because of the introduction of AML with mutated TP53 as a distinct category only in the ICC. Nearly all of these were instead diagnosed as AML, myelodysplasia related (AML-MR) by WHO5. However, when compared with a separate cohort of patients with AML-MR without TP53 mutations, patients with TP53-mutated AML showed a distinct genetic profile and significantly worse overall survival, supporting the inclusion of AML with mutated TP53 as a distinct disease category. Overall, our results show that a significant percentage of TP53-mutated myeloid disease is classified differently by the ICC and WHO5 and highlight areas to address in future classification systems.
Introduction
The TP53 tumor suppressor gene, located on chromosome 17p, encodes a transcription factor that functions as a master regulator of the cell, affecting proliferation, DNA stability and repair, cell cycle arrest, and apoptosis, among many other processes.1TP53 mutations are present in ∼10% of myeloid neoplasms as a whole,2-7 and are enriched in patients with secondary and therapy-related disease.8,9TP53 mutations are associated with particularly adverse outcomes in myeloid disease, and patients with acute myeloid leukemia (AML) with a TP53 mutation at an allele frequency of ≥10% are classified as adverse risk by the European LeukemiaNet guidelines.10 In both myelodysplastic syndrome (MDS) and AML, TP53 mutations are associated with therapeutic resistance, increased rates of relapse, and significantly shorter survival.2-5,11-17 In patients with MDS with <10% blasts, these poor outcomes have been shown to be associated specifically with biallelic TP53 inactivation, whereas such patients with MDS with monoallelic TP53 alteration showed similar survival outcomes to patients with wild-type TP53.4,11,14 Conversely, in cohorts of patients with ≥10% blasts, studies have shown that both monoallelic and biallelic TP53 mutations predict poor outcome, without significant differences between patients with MDS with excess blasts (10% to <20% blasts) and AML (≥20% blasts).2,3,11,15-18
The adverse effect of TP53 mutations in myeloid disease appears to be independent of other high risk disease features. While TP53-mutated MDS and AML show high rates of monosomal and complex karyotypes,3-5,11,19-21 the presence of a TP53 mutation is associated with adverse outcome compared with patients with complex karyotype without TP53 mutation.11,18 Similarly, although TP53-mutations are more common in therapy-related disease, studies comparing de novo and therapy-related TP53-mutated myeloid neoplasms have shown no significant difference in disease characteristics or outcomes between the 2 groups, suggesting that disease ontogeny is less predictive of outcome than TP53 mutant status.11,20
The effect of variant allele frequency (VAF) on outcome in TP53-mutated myeloid neoplasms remains unclear. In several studies, increased TP53 VAF has been correlated with therapeutic resistance, higher rates of transformation to AML, and decreased overall survival (OS).4,5,17,22 Additionally, patients with MDS with TP53 mutations at a VAF of <10% and lacking a complex karyotype showed similar outcomes to patients with wild-type TP53 in 1 study.5 However, other studies have shown no correlation between TP53 VAF and survival.11,14,15,23
In 2022, the International Consensus Classification (ICC)24 and World Health Organization fifth edition (WHO5)25 introduced new, but differing, categories of myeloid disease defined by the presence of TP53 mutations (Table 1). Both classification systems introduce MDS with biallelic TP53 alterations (MDS-TP53) as a distinct category and consider the following as evidence of biallelic or multihit TP53 mutation: 2 distinct TP53 mutations; or a single TP53 mutation with evidence of TP53 copy loss (17p deletion or VAF of >50%) or copy-neutral loss of heterozygosity. However, other diagnostic criteria for this category differ between the 2 classification systems. The ICC requires a VAF of >10% for TP53 mutations, whereas the WHO5 does not include any VAF criteria. Additionally, in patients with a single TP53 mutation, the ICC, but not WHO5, includes the presence of a complex karyotype as evidence of TP53 copy loss. Furthermore, the WHO5 includes MDS with biallelic TP53 inactivation as the only category for TP53-mutated disease, and includes MDS cases with 0% to 19% blasts in this category. The WHO5 does not include a distinct category of TP53-mutated AML for cases with ≥20% blasts. Conversely, the ICC divides TP53-mutated cases into 3 categories based on blast count. MDS with biallelic TP53 inactivation includes only cases with 0% to <10% blasts. Cases with 10% to 19% or ≥20% blasts are classified as MDS/AML (MDS/AML-TP53) or AML (AML-TP53) with mutated TP53, respectively. In contrast to cases with <10% blasts, these cases require only a single TP53 mutation (with a VAF of >10%) to be classified as TP53-mutated disease.
Diagnostic criteria for TP53-mutated myeloid disease
| BM blast % . | ICC diagnosis . | Genetic criteria . | WHO5 diagnosis . | Genetic criteria . |
|---|---|---|---|---|
| 0%-9% | MDS with mutated TP53 | Multihit TP53 mutation∗ (VAFs > 10%) or TP53 mutation and CK | MDS with biallelic TP53 inactivation | Biallelic TP53 alterations∗ |
| 10%-19% | MDS/AML with mutated TP53 | Any somatic TP53 mutation (VAF > 10%) | MDS with biallelic TP53 inactivation | Biallelic TP53 alterations∗ |
| ≥20% | AML with mutated TP53 | Any somatic TP53 mutation (VAF > 10%) |
| BM blast % . | ICC diagnosis . | Genetic criteria . | WHO5 diagnosis . | Genetic criteria . |
|---|---|---|---|---|
| 0%-9% | MDS with mutated TP53 | Multihit TP53 mutation∗ (VAFs > 10%) or TP53 mutation and CK | MDS with biallelic TP53 inactivation | Biallelic TP53 alterations∗ |
| 10%-19% | MDS/AML with mutated TP53 | Any somatic TP53 mutation (VAF > 10%) | MDS with biallelic TP53 inactivation | Biallelic TP53 alterations∗ |
| ≥20% | AML with mutated TP53 | Any somatic TP53 mutation (VAF > 10%) |
CK, complex karyotype.
Characterized by ≥2 TP53 mutations or a single TP53 mutation with (1) concurrent evidence of TP53 copy loss (17p deletion on cytogenetics; or VAF of >50%) or (2) copy-neutral loss of heterozygosity.
Here, we investigated how the divergent diagnostic criteria between the WHO5 and ICC affect clinical practice by reviewing a cohort of patients with TP53-mutated myeloid disease, determining how diagnoses differ between the 2 classification systems, and examining whether diagnostic discrepancies reflect differences in disease biology or patient outcome.
Methods
Patient cohort and clinical data
The pathology archives at Vanderbilt University Medical Center were searched from 2015 through 2023 for cases showing TP53 mutations at the time of diagnosis. Pathology reports and medical records were reviewed to confirm diagnoses and collect morphologic, cytogenetic, next-generation sequencing, and clinical outcome data. Patients were excluded if there was incomplete clinical or pathologic data or if they had a diagnosis or prior history of a myeloproliferative neoplasm (MPN), mixed phenotype acute leukemia, or lymphoma. A separate cohort of 45 patients with AML, myelodysplasia related (AML-MR) without TP53 mutations was also collected. These patients had a known history of MDS or MDS/MPN or defining gene mutations or cytogenetic abnormalities, as defined by WHO5 and ICC. When categorizing therapies that patients received, hypomethylating agent therapy included decitabine and azacitidine, alone or in combination; venetoclax-based therapy included venetoclax alone or in combination with hypomethylating agent or low dose cytarabine; and intensive induction chemotherapy included cytarabine plus daunorubicin or idarubicin, vosaroxin/cytarabine,26,27 or liposomal cytarabine/daunorubicin. OS was calculated from date of diagnosis until time of death or last follow-up. This study was approved by the Vanderbilt University institutional review board.
Molecular and cytogenetic analysis
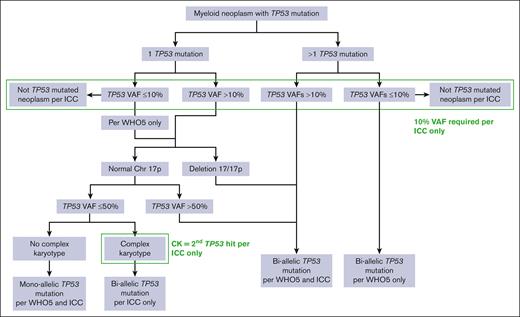
Targeted amplicon sequencing was performed on all cases using an Illumina MiSeq or Nextseq (Illumina, Inc, San Diego, CA) with a 37-gene (2015 to June 2021) or 50-gene (July 2021-2023) panel covering genes recurrently mutated in myeloid malignancies. The sequencing assays evolved over the study period, but single-nucleotide variants and small insertion/deletion/duplication mutations were reported down to a VAF threshold of 2% to 5%, depending on the assay and mutation type. A small number of recurrent hot spot variants were reported to a VAF threshold of 1%. Cases were classified as having biallelic TP53 abnormalities by WHO5 if there were ≥2 TP53 mutations, a single TP53 mutation with a VAF of >50%, or a single TP53 mutation with 17p loss on karyotype. Cases were also classified as having biallelic TP53 abnormalities by the ICC if any of the aforementioned were present and the TP53 VAF was >10%; or if a single TP53 mutation (VAF of >10%) and a complex karyotype (≥3 unrelated clonal chromosomal abnormalities) were present (Figure 1). Monosomal karyotype (MK) was identified as previously defined by the presence of at least 2 autosomal monosomies or 1 monosomy plus ≥1 structural aberrations.28
Determination of mutant TP53 allelic status. Algorithm used to determine TP53 allelic status by the ICC and WHO5 classifications.
Determination of mutant TP53 allelic status. Algorithm used to determine TP53 allelic status by the ICC and WHO5 classifications.
Statistical analysis
Fisher exact tests were used to compare categorical variables, and Mann-Whitney U tests were used for continuous variables. OS was estimated using the Kaplan-Meier method and compared between groups using log-rank tests. Survival analyses were performed using GraphPad Prism (10.0). A P value ≤.05 was considered statistically significant.
Results
Patient cohort
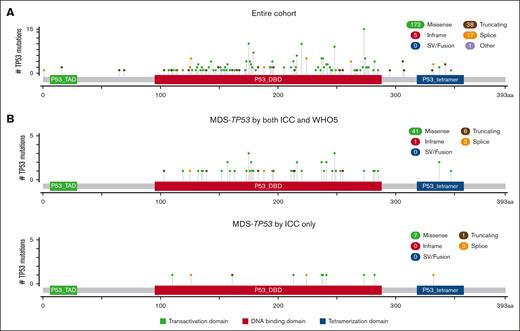
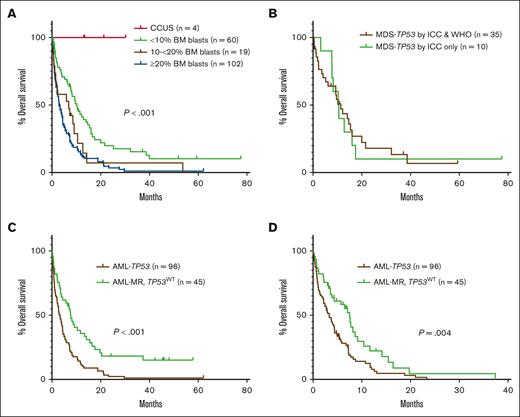
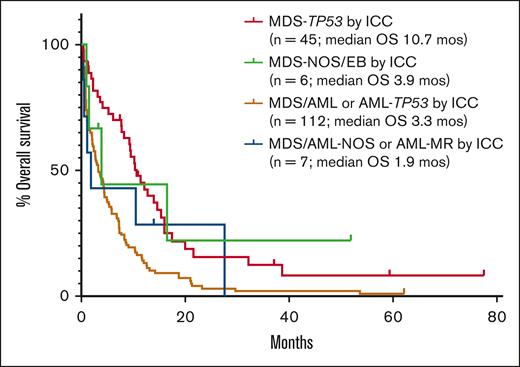
The cohort included 188 cases of TP53-mutated myeloid disease, including 6 cases of clonal cytopenia of undetermined significance (CCUS), 80 cases of MDS or MDS/AML (<20% blasts), and 102 cases of AML. Patients were divided into groups based on bone marrow (BM) blast percentage, because the diagnosis of MDS or MDS/AML varied between the WHO5 and ICC for some cases. The clinicopathologic features of the entire cohort are shown in Table 2. Overall, there was an increased depth of cytopenias with progression of BM blast count. Except for cases of CCUS, there were very high rates of complex and MKs across disease categories. A total of 234 TP53 mutations were identified (Figure 2A), most of which were missense mutations (173, 74%), predominantly located in the DNA binding domain. In 118 of 188 (63%) cases, there were no additional gene mutations present beyond the TP53 mutation(s). For patients with <20% blasts, 52 of 80 (65%) received hypomethylating agent therapy. Of patients with AML, ∼40% of patients received intensive induction chemotherapy, hypomethylating agent therapy, and venetoclax-based therapy at some point during their disease course. Overall, only 23 of 188 (12%) patients underwent stem cell transplant. All patients with CCUS were alive at last follow-up. Patients with <10% blasts showed significantly prolonged OS (median, 10.2 months) compared with patients with ≥10% blasts (P > .001), whereas there was no significant difference in OS between patients with 10% to <20% blasts (median, 6.9 months) and patients with ≥20% blasts (median, 3.0 months; Figure 3A). In total, 51 cases occurred in the setting of prior cytotoxic chemotherapy, including 33 of 80 cases of MDS or MDS/AML, and 18 of 102 cases of AML. There was no significant difference in OS between patients with and without a prior history of cytotoxic chemotherapy exposure for patients with <10% blasts, 10% to <20% blasts, or ≥20% blasts (supplemental Figure 1).
Clinicopathologic features of the cohort
| Characteristic . | Total cohort (N = 188) . | CCUS (n = 6) . | MDS and MDS/AML (n = 80) . | AML (n = 102) . | |
|---|---|---|---|---|---|
| 0% to <10% blasts (n = 61) . | 10% to <20% blasts (n = 19) . | ≥20% blasts . | |||
| Age at dx (y), median (range) | 68.5 (28.0-88.0) | 69.5 (56.5-79.1) | 68.5 (38.0-84.0) | 71.1 (38.5-81.0) | 68.3 (28.0-88.0) |
| Sex, male, n (%) | 98 (52) | 3 (50) | 27 (44) | 13 (68) | 55 (54) |
| CBC at dx, median (range) | |||||
| ANC, ×103/μL | 0.8 (0-16.1) | 2.7 (0.5-4.6) | 1.4 (0.1-11.5) | 1.2 (0.3-2.5) | 0.6 (0.0-16.1) |
| Hb, g/dL | 8.4 (5.5-14.1) | 10.6 (7.3-13.2) | 8.8 (5.6-13.8) | 8.4 (6.6-12.3) | 8.3 (5.5-14.1) |
| Plt, ×103/μL | 48 (7-347) | 123 (81-232) | 62 (7-347) | 51 (8-114) | 36 (7-228) |
| BM blast %, median (range) | 23 (0-100) | 2 (0.5-2.5) | 3 (0-9.5) | 15 (10-17.5) | 35 (20.8-100) |
| Karyotype | |||||
| Complex, n (%) | 161 (86) | 0 (0) | 47 (77) | 19 (100) | 95 (93) |
| Monosomal, n (%) | 154 (82) | 0 (0) | 46 (75) | 17 (90) | 91 (89) |
| Del 17/17p, n (%) | 56 (30) | 0 (0) | 12 (20) | 4 (21) | 40 (40) |
| Mutations by NGS | |||||
| 1 TP53 mutation, n (%) | 145 (77) | 6 (100) | 43 (70) | 15 (79) | 80 (78) |
| ≥2 TP53 mutations, n (%) | 43 (23) | 0 (0) | 18 (30) | 4 (21) | 21 (21) |
| Highest TP53 VAF, median (range) | 41 (1.1-97) | 8.9 (4.5-14.0) | 35 (4-97) | 41 (4-89) | 46.5 (1.1-93.0) |
| 0 additional mutations, n (%) | 118 (63) | 4 (67) | 42 (67) | 16 (84) | 56 (55) |
| 1 additional mutation, n (%) | 38 (20) | 2 (33) | 11 (18) | 1 (5) | 24 (24) |
| ≥2 additional mutations, n (%) | 32 (17) | 0 (0) | 8 (13) | 2 (11) | 22 (22) |
| Therapy | |||||
| Intensive induction, n (%) | 40 (21) | 0 (0) | 1 (2) | 1 (5) | 38 (37) |
| HMA, n (%) | 106 (56) | 0 (0) | 39 (64) | 15 (79) | 52 (51) |
| Venetoclax, n (%) | 70 (37) | 0 (0) | 12 (20) | 7 (37) | 51 (50) |
| Stem cell transplant, n (%) | 23 (12) | 0 (0) | 11 (18) | 2 (11) | 10 (10) |
| Characteristic . | Total cohort (N = 188) . | CCUS (n = 6) . | MDS and MDS/AML (n = 80) . | AML (n = 102) . | |
|---|---|---|---|---|---|
| 0% to <10% blasts (n = 61) . | 10% to <20% blasts (n = 19) . | ≥20% blasts . | |||
| Age at dx (y), median (range) | 68.5 (28.0-88.0) | 69.5 (56.5-79.1) | 68.5 (38.0-84.0) | 71.1 (38.5-81.0) | 68.3 (28.0-88.0) |
| Sex, male, n (%) | 98 (52) | 3 (50) | 27 (44) | 13 (68) | 55 (54) |
| CBC at dx, median (range) | |||||
| ANC, ×103/μL | 0.8 (0-16.1) | 2.7 (0.5-4.6) | 1.4 (0.1-11.5) | 1.2 (0.3-2.5) | 0.6 (0.0-16.1) |
| Hb, g/dL | 8.4 (5.5-14.1) | 10.6 (7.3-13.2) | 8.8 (5.6-13.8) | 8.4 (6.6-12.3) | 8.3 (5.5-14.1) |
| Plt, ×103/μL | 48 (7-347) | 123 (81-232) | 62 (7-347) | 51 (8-114) | 36 (7-228) |
| BM blast %, median (range) | 23 (0-100) | 2 (0.5-2.5) | 3 (0-9.5) | 15 (10-17.5) | 35 (20.8-100) |
| Karyotype | |||||
| Complex, n (%) | 161 (86) | 0 (0) | 47 (77) | 19 (100) | 95 (93) |
| Monosomal, n (%) | 154 (82) | 0 (0) | 46 (75) | 17 (90) | 91 (89) |
| Del 17/17p, n (%) | 56 (30) | 0 (0) | 12 (20) | 4 (21) | 40 (40) |
| Mutations by NGS | |||||
| 1 TP53 mutation, n (%) | 145 (77) | 6 (100) | 43 (70) | 15 (79) | 80 (78) |
| ≥2 TP53 mutations, n (%) | 43 (23) | 0 (0) | 18 (30) | 4 (21) | 21 (21) |
| Highest TP53 VAF, median (range) | 41 (1.1-97) | 8.9 (4.5-14.0) | 35 (4-97) | 41 (4-89) | 46.5 (1.1-93.0) |
| 0 additional mutations, n (%) | 118 (63) | 4 (67) | 42 (67) | 16 (84) | 56 (55) |
| 1 additional mutation, n (%) | 38 (20) | 2 (33) | 11 (18) | 1 (5) | 24 (24) |
| ≥2 additional mutations, n (%) | 32 (17) | 0 (0) | 8 (13) | 2 (11) | 22 (22) |
| Therapy | |||||
| Intensive induction, n (%) | 40 (21) | 0 (0) | 1 (2) | 1 (5) | 38 (37) |
| HMA, n (%) | 106 (56) | 0 (0) | 39 (64) | 15 (79) | 52 (51) |
| Venetoclax, n (%) | 70 (37) | 0 (0) | 12 (20) | 7 (37) | 51 (50) |
| Stem cell transplant, n (%) | 23 (12) | 0 (0) | 11 (18) | 2 (11) | 10 (10) |
ANC, absolute neutrophil count; CBC, complete blood count; dx, diagnosis; Hb, hemoglobin; HMA, hypomethylating agent; NGS, next-generation sequencing; Plt, platelets.
Distribution of TP53 mutations. Lolliplot figures showing the types and distribution of TP53 mutations in the entire cohort (A) and in cases of MDS with <10% blasts classified as MDS-TP53 by both classifications or by the ICC alone (B). SV, structural variant. Figure panels were generated using cBioPortal, as previously described.29,30
Distribution of TP53 mutations. Lolliplot figures showing the types and distribution of TP53 mutations in the entire cohort (A) and in cases of MDS with <10% blasts classified as MDS-TP53 by both classifications or by the ICC alone (B). SV, structural variant. Figure panels were generated using cBioPortal, as previously described.29,30
OS in TP53-mutated myeloid disease. (A) OS of the entire cohort based on diagnosis and blast percentage. (B) OS for cases of MDS with <10% blasts classified as MDS-TP53 by both classifications vs cases of MDS with <10% blasts classified as MDS-TP53 by the ICC only (P=NS). (C-D) OS in patients with AML-TP53 compared to patients with AML-MR with wild-type TP53, both overall (C) and censored at the time of stem cell transplant (D).
OS in TP53-mutated myeloid disease. (A) OS of the entire cohort based on diagnosis and blast percentage. (B) OS for cases of MDS with <10% blasts classified as MDS-TP53 by both classifications vs cases of MDS with <10% blasts classified as MDS-TP53 by the ICC only (P=NS). (C-D) OS in patients with AML-TP53 compared to patients with AML-MR with wild-type TP53, both overall (C) and censored at the time of stem cell transplant (D).
TP53-mutated disease with <10% BM blasts
Patients with CCUS harbored a single TP53 mutation, lacked complex karyotypes, with normal karyotypes in 5 of 6 (83%) cases, and had no additional comutations in 4 of 6 (67%) of cases. The CCUS cohort demonstrated a median TP53 VAF of 8.9% (range, 4.5-14.0), with 3 of 6 patients notably showing VAFs of >10%.
The cohort included 61 cases of MDS with <10% blasts and TP53 mutation(s). A total of 12 cases (20%) harbored monoallelic TP53 abnormalities per both ICC and WHO5 and were classified as MDS with deletion 5q (n = 3) or MDS, not otherwise specified (NOS)/MDS with low blasts (n = 9), whereas 35 cases (57%) met criteria for a diagnosis of MDS-TP53 by both ICC and WHO5 criteria. In 14 cases (23%), the diagnosis differed between the 2 classification systems. Specifically, 10 cases showed a TP53 mutation in addition to a complex karyotype, without a specific 17p abnormality on karyotype; these were classified as MDS-TP53 only by the ICC. In contrast, 4 cases harbored biallelic TP53 abnormalities but with VAFs of <10%. These cases were classified as MDS-TP53 only by WHO5.
We compared the cases classified as MDS-TP53 by both ICC and WHO5 (n = 35) to the cases classified as MDS-TP53 by the ICC only, based on the presence of a complex karyotype (n = 10). Cases classified as MDS-TP53 by both systems showed a significantly higher median TP53 VAF, because any case with a VAF of >50% was automatically included in this category. Otherwise, there were no significant differences in the clinicopathologic features between the 2 groups (Table 3), and the 2 groups also showed a similar distribution of TP53 mutations (Figure 2B). Additionally, there was no difference in OS between cases diagnosed as MDS-TP53 by both ICC and WHO5 (median OS, 11.4 months) and cases classified as MDS-TP53 by the ICC only (median OS, 10.5 months; Figure 3B).
Clinicopathologic features of TP53-mutated myeloid disease with less than 10% blasts
| Characteristic . | MDS-TP53 by ICC and WHO (n = 35) . | MDS-TP53 by ICC only (n = 10) . | P value . |
|---|---|---|---|
| Age at dx (y), median (range) | 68.5 (36-83.2) | 70.2 (60.6-84) | NS |
| Sex, male, n (%) | 14 (40) | 5 (50) | NS |
| CBC at dx, median (range) | |||
| ANC, ×103/μL | 1.4 (0.2-11.5) | 0.48 (0.4-2.0) | NS |
| Hb, g/dL | 8.5 (7-12.4) | 8.8 (5.6-13.8) | NS |
| Plt, ×103/μL | 54 (9-347) | 75 (7-226) | NS |
| BM blast %, median (range) | 3.0 (0-9.5) | 5.5 (0.5-9.0) | NS |
| CK, n (%) | 32 (94) | 10 (100) | NS |
| MK, n (%) | 30 (88) | 9 (90) | NS |
| TP53 VAF, median (range) | 45.7 (14.4-97) | 30 (11.8-45) | .02 |
| 0 additional mutations, n (%) | 24 (69) | 7 (70) | NS |
| HMA therapy, n (%) | 23 (66) | 7 (70) | NS |
| Stem cell transplant, n (%) | 6 (17) | 3 (30) | NS |
| Characteristic . | MDS-TP53 by ICC and WHO (n = 35) . | MDS-TP53 by ICC only (n = 10) . | P value . |
|---|---|---|---|
| Age at dx (y), median (range) | 68.5 (36-83.2) | 70.2 (60.6-84) | NS |
| Sex, male, n (%) | 14 (40) | 5 (50) | NS |
| CBC at dx, median (range) | |||
| ANC, ×103/μL | 1.4 (0.2-11.5) | 0.48 (0.4-2.0) | NS |
| Hb, g/dL | 8.5 (7-12.4) | 8.8 (5.6-13.8) | NS |
| Plt, ×103/μL | 54 (9-347) | 75 (7-226) | NS |
| BM blast %, median (range) | 3.0 (0-9.5) | 5.5 (0.5-9.0) | NS |
| CK, n (%) | 32 (94) | 10 (100) | NS |
| MK, n (%) | 30 (88) | 9 (90) | NS |
| TP53 VAF, median (range) | 45.7 (14.4-97) | 30 (11.8-45) | .02 |
| 0 additional mutations, n (%) | 24 (69) | 7 (70) | NS |
| HMA therapy, n (%) | 23 (66) | 7 (70) | NS |
| Stem cell transplant, n (%) | 6 (17) | 3 (30) | NS |
Abbreviations are explained in Tables 1 and 2.
TP53-mutated disease with 10% to <20% BM blasts
Of 19 cases with 10% to <20% blasts, 1 case (5%) had a single TP53 mutation at a VAF of 4.6% and was classified as MDS/AML-NOS by ICC and MDS with increased blasts by WHO5. There were 8 cases (42%) classified as MDS/AML-TP53 or MDS-TP53 by ICC and WHO5, respectively. These cases all had biallelic TP53 abnormalities per ICC and WHO5 criteria, although only a monoallelic TP53 abnormality is required for a diagnosis of MDS/AML-TP53 per ICC. Discrepant diagnoses were seen in 10 cases (53%). These included 8 cases with a single TP53 mutation, all of which harbored a complex karyotype without 17p loss on karyotype, which were classified as MDS/AML-TP53 by ICC and as MDS with increased blasts by WHO5. Finally, 2 cases harbored biallelic TP53 abnormalities but with VAFs of <10% and were classified as MDS-TP53 only by WHO5.
We compared cases classified as MDS/AML-TP53 or MDS-TP53 by both classification systems (n = 8) to cases classified as MDS/AML-TP53 by ICC but not WHO5 (n = 8). Similar to cases with <10% blasts, there were no significant differences in clinicopathologic features (Table 4) or OS between the 2 groups (median OS of 4 months and 9.1 months for the concordant and discordant groups, respectively). Although not a statistically significant difference, there were 2 patients with disease classified as MDS/AML-TP53 by ICC only who underwent stem cell transplant, whereas none in the concordantly diagnosed group underwent stem cell transplant. Therefore, we also examined OS when censoring patients at the time of stem cell transplant and again saw no significant difference in OS between the 2 groups (data not shown).
Clinicopathologic features of TP53-mutated myeloid disease with blasts of 10% to less than 20%
| Characteristic . | M/A or MDS-TP53 by ICC and WHO (n = 8) . | M/A-TP53 by ICC only (n = 8) . | P value . |
|---|---|---|---|
| Age at dx (y), median (range) | 64.1 (51.4-74) | 67.5 (38.5-77.1) | NS |
| Sex, male, n (%) | 6 (75) | 4 (50) | NS |
| CBC at dx, median (range) | |||
| ANC, ×103/μL | 1.0 (0.4-1.5) | 1.2 (0.3-1.9) | NS |
| Hb, g/dL | 8.7 (6.8-12.3) | 8.4 (6.6-11.3) | NS |
| Plt, ×103/μL | 56 (22-114) | 41 (10-88) | NS |
| BM blast %, median (range) | 15 (11-17.5) | 15 (10-17.5) | NS |
| CK, n (%) | 8 (100) | 8 (100) | NS |
| MK, n (%) | 7 (100) | 7 (100) | NS |
| TP53 VAF, median (range) | 48.5 (21-89) | 42.3 (19-48.3) | NS |
| 0 additional mutations, n (%) | 7 (88) | 7 (88) | NS |
| HMA therapy, n (%) | 6 (75) | 6 (75) | NS |
| SCT, n (%) | 0 (0) | 2 (25) | NS |
| Characteristic . | M/A or MDS-TP53 by ICC and WHO (n = 8) . | M/A-TP53 by ICC only (n = 8) . | P value . |
|---|---|---|---|
| Age at dx (y), median (range) | 64.1 (51.4-74) | 67.5 (38.5-77.1) | NS |
| Sex, male, n (%) | 6 (75) | 4 (50) | NS |
| CBC at dx, median (range) | |||
| ANC, ×103/μL | 1.0 (0.4-1.5) | 1.2 (0.3-1.9) | NS |
| Hb, g/dL | 8.7 (6.8-12.3) | 8.4 (6.6-11.3) | NS |
| Plt, ×103/μL | 56 (22-114) | 41 (10-88) | NS |
| BM blast %, median (range) | 15 (11-17.5) | 15 (10-17.5) | NS |
| CK, n (%) | 8 (100) | 8 (100) | NS |
| MK, n (%) | 7 (100) | 7 (100) | NS |
| TP53 VAF, median (range) | 48.5 (21-89) | 42.3 (19-48.3) | NS |
| 0 additional mutations, n (%) | 7 (88) | 7 (88) | NS |
| HMA therapy, n (%) | 6 (75) | 6 (75) | NS |
| SCT, n (%) | 0 (0) | 2 (25) | NS |
Abbreviations are explained in Tables 1 and 2.
M/A, MDS/AML; SCT, stem cell transplant.
TP53-mutated disease with ≥20% BM blasts
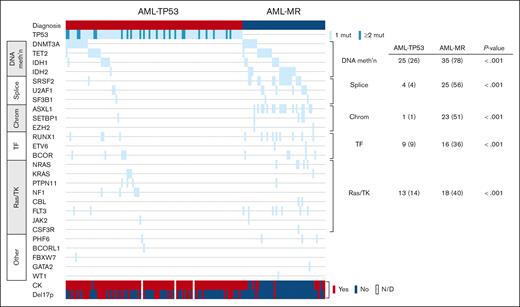
Among the cases with ≥20% blasts (n = 102), 2 were classified as AML with NPM1 mutation by both ICC and WHO5. Additionally, 4 cases harbored TP53 mutations at a VAF of <10% and were classified by WHO5 as AML-MR, based on defining cytogenetic abnormalities or gene mutations, and per ICC as AML with MR gene mutations (n = 2) or AML with MR cytogenetic abnormalities (n = 2). The remaining 96 cases were classified as AML-TP53 by ICC. Per the WHO5, 1 of 96 was classified as AML defined by differentiation; the rest met criteria for AML-MR by WHO5, based on cytogenetic abnormalities (n = 92) or a prior history of MDS (n = 3). However, when compared with a separate cohort of AML-MR with wild-type TP53 (n = 45), cases classified as AML-TP53 by ICC showed significantly higher rates of complex and MKs and harbored significantly fewer uniquely mutated genes (Table 5; Figure 4). Patients with AML-TP53 were also less likely to undergo stem cell transplant. Finally, patients with AML-TP53 demonstrated worse OS than patients with AML-MR lacking TP53 mutations, both as a whole (median OS, 3.3 months vs 7.7 months, respectively) and when censoring at stem cell transplant (median OS, 3.3 months vs 7.5 months, respectively; Figure 3C-D). Furthermore, specifically among patients with a complex karyotype, patients with AML-TP53 showed significantly worse OS (n = 92; median OS, 3.3 months) than patients with AML-MR (n = 11; median OS, 6.5 months; P = .046). Finally, we evaluated whether clone size (TP53 VAF) had any impact on OS in our AML-TP53 cohort. Using VAF thresholds of 30%, 40%, 50%, and 60%, we found no impact on OS at any threshold (data not shown).
Clinicopathologic features of TP53-mutated AML and AML-MR with wild-type TP53
| Characteristic . | AML-TP53 by ICC (n = 96) . | AML-MR (n = 45) . | P value . |
|---|---|---|---|
| Age at dx (y), median (range) | 68.8 (28-88) | 68.9 (41-91) | NS |
| CK, n (%) | 92 (99) | 12 (27) | <.0001 |
| MK, n (%) | 89 (97) | 12 (27) | <.0001 |
| No. of unique genes mutated, median (range) | 1 (1-4) | 3 (0-6) | <.0001 |
| Intensive induction, n (%) | 35 (36) | 24 (53) | NS |
| SCT, n (%) | 9 (10) | 10 (23) | NS |
| Characteristic . | AML-TP53 by ICC (n = 96) . | AML-MR (n = 45) . | P value . |
|---|---|---|---|
| Age at dx (y), median (range) | 68.8 (28-88) | 68.9 (41-91) | NS |
| CK, n (%) | 92 (99) | 12 (27) | <.0001 |
| MK, n (%) | 89 (97) | 12 (27) | <.0001 |
| No. of unique genes mutated, median (range) | 1 (1-4) | 3 (0-6) | <.0001 |
| Intensive induction, n (%) | 35 (36) | 24 (53) | NS |
| SCT, n (%) | 9 (10) | 10 (23) | NS |
Abbreviations are explained in Tables 1 and 2.
Distribution of mutations in AML-TP53 and AML-MR. Mutations and cytogenetic abnormalities are shown for patients with AML-TP53 and AML-MR with wild-type TP53, with each row representing a gene and each column representing a patient. The number (percent) of patients in each group harboring different types of mutations is shown at the right. chrom, chromatin modifier; mut, mutation; TF, transcription factor; TK, tyrosine kinase. Figure generated using cBioPortal, as previously described.29,30
Distribution of mutations in AML-TP53 and AML-MR. Mutations and cytogenetic abnormalities are shown for patients with AML-TP53 and AML-MR with wild-type TP53, with each row representing a gene and each column representing a patient. The number (percent) of patients in each group harboring different types of mutations is shown at the right. chrom, chromatin modifier; mut, mutation; TF, transcription factor; TK, tyrosine kinase. Figure generated using cBioPortal, as previously described.29,30
TP53-mutated disease with TP53 VAF of <10%
With the exception of 1 patient, TP53 variants were detected at VAFs of ≥4%. The 1 patient with a TP53 mutation detected at a VAF of 1.1% developed AML after a previous diagnosis of TP53-mutated MDS. At the time of AML diagnosis, the karyotype showed loss of both copies of 17p; however, manual review of the sequencing data identified the known TP53 variant. Excluding cases of CCUS, there were 19 of 182 (10%) cases with TP53 mutations at VAFs below the ICC VAF requirement of 10%, including 11 cases with <10% blasts and 8 cases with >10% blasts. Of the cases with <10% blasts, 5 cases lacked a second TP53 abnormality and so did not meet criteria for biallelic TP53 abnormalities. The other 6 cases showed a complex karyotype or 2 TP53 mutations but were not classified as MDS-TP53 by ICC because of the 10% VAF requirement. Of the 8 cases with >10% blasts, 1 was classified AML with NPM1 mutation. The remaining 7 harbored ≥1 TP53 mutations at VAFs ranging from 1.1% to 9.5% but were classified as MDS/AML-NOS or AML-MR by ICC. However, the 13 cases in which TP53 VAF precluded classification as TP53-mutated disease by ICC showed a high rate of complex karyotype (10/13 [77%]), and most (10/13 [77%]) had no additional mutations other than the TP53 mutation(s), similar to other cases of TP53-mutated disease. Additionally, these cases demonstrated equal or worse OS than cases with TP53 VAFs of >10% that were classified as TP53-mutated disease by ICC, both overall (Figure 5) and when censoring at stem cell transplant (not shown).
OS in cases with TP53 VAFs <10%. There were 6 cases with <10% blasts and 7 cases with >10% blasts that were not classified as TP53-mutated disease by ICC because the TP53 VAF failed to meet the required level of >10%. However, these cases showed equal or worse OS compared with cases with TP53 VAFs of >10%. EB, excess blasts.
OS in cases with TP53 VAFs <10%. There were 6 cases with <10% blasts and 7 cases with >10% blasts that were not classified as TP53-mutated disease by ICC because the TP53 VAF failed to meet the required level of >10%. However, these cases showed equal or worse OS compared with cases with TP53 VAFs of >10%. EB, excess blasts.
Discussion
The ICC and WHO5 classifications both introduced new categories of myeloid disease defined by the presence of TP53 abnormalities. In both classifications, determination of TP53 mutant status requires accounting for the number of TP53 mutations; VAF; TP53 copy number; and, per the ICC, the number of karyotypic abnormalities. Furthermore, the significance of monoallelic vs biallelic TP53 abnormalities depends on blast percentage. The divergent diagnostic criteria between the classifications further increases diagnostic complexity. Here, we evaluated the impact of the new classifications on the diagnosis of TP53-mutated myeloid disease. Overall, 120 of 188 (64%) cases were classified differently by the ICC and WHO5. This included 24 of 80 (30%) cases with <20% blasts; nearly all cases of AML (96/102 [95%]) were classified discrepantly, because of the introduction of the AML-TP53 disease category only in the ICC.
In total, 51 of 188 (27%) cases occurred in the setting of prior cytotoxic chemotherapy. Although the WHO5 retains a specific category of myeloid neoplasms after cytotoxic therapy, this diagnosis requires first that diseases fulfill criteria for a specific myeloid neoplasm, with the “post cytotoxic therapy” designation then added as an appendix. This is similar to the strategy adopted by the ICC, which recommends adding the diagnostic qualifier of “therapy related” after classification as a specific myeloid neoplasm. The initial disease classification into a specific category of MDS or AML is thus not affected by a history of prior therapy in either the WHO5 or ICC. Therefore, the mechanism of specifying prior chemotherapy exposure is overall similar between the 2 classifications, despite the different language used. Nevertheless, we found that a history of prior cytotoxic chemotherapy exposure had no significant impact on OS in patients with TP53-mutated myeloid neoplasms, which is consistent with prior work.11,20
Discrepant diagnoses for cases with <10% blasts were predominantly due to the fact that, in cases with a single TP53 mutation, complex karyotype is considered evidence of biallelic TP53 inactivation only by the ICC. However, when comparing cases meeting WHO5 (and ICC) criteria for biallelic TP53 abnormalities to cases with a single TP53 mutation and complex karyotype, we found no differences in clinicopathologic features, distribution or type of TP53 mutations, or OS. Therefore, our findings support the inclusion of complex karyotype as evidence of a second TP53 abnormality, as defined by the ICC. Similar results were seen for cases with 10% to 20% blasts. Diagnosis in these cases was further complicated by the presence of the MDS/AML category in ICC, which requires only a monoallelic TP53 abnormality. Conversely, WHO5 maintains the requirement for biallelic TP53 inactivation in cases with 10% to 20% blasts. However, all cases in this group showed a complex karyotype. Therefore, if complex karyotype were considered evidence of a second TP53 abnormality, our data suggest that TP53-mutated cases with 10% to 20% blasts would only very rarely show monoallelic TP53 inactivation, and that most of these cases would therefore be classified as TP53-mutated disease by both the ICC and WHO5, despite the differing diagnostic criteria.
Previous studies have shown particularly poor outcomes in patients with AML harboring TP53 mutations, even among subsets of adverse-risk disease.11,15 In line with this, the ICC created a new category for AML-TP53, distinct from AML-MR. In contrast, the WHO5 does not delineate a category of AML defined by TP53 mutation, and these cases are most commonly classified as AML-MR based on cytogenetic or other molecular abnormalities. However, we find that AML-TP53 differs significantly from wild-type–TP53 AML-MR with respect to multiple clinicopathologic features. Indeed, AML-TP53 shows a distinct genetic profile, characterized by fewer somatic gene mutations and increased chromosomal complexity. Importantly, these biological differences were associated with significantly worse OS in patients with AML-TP53. In our cohort, patients with AML-TP53 were less likely to undergo stem cell transplant, the only potentially curative therapy for poor-risk MDS/AML, which we have previously shown to be, at least in part, because of poor response to therapy and an increased risk of infection, among other factors.31 Overall, our findings support the inclusion of AML-TP53 as a distinct disease category associated with particularly poor outcomes in the context of current therapeutic options.
The significance of the TP53 VAF in TP53-mutated myeloid disease remains uncertain, with prior studies showing conflicting results. We evaluated whether various TP53 VAF thresholds correlated with survival in patients with AML-TP53 and found no impact on survival with increasing VAF cutoffs in our cohort. At the other end of the spectrum, it remains unclear how best to interpret cases with TP53 mutations at low VAFs. The WHO5 includes no VAF criteria with respect to TP53 mutations, whereas the ICC requires a TP53 VAF of >10% to diagnose TP53-mutated disease. However, in our cohort, TP53 VAFs of <10% were not correlated with improved survival, with patients with MDS or AML with TP53 VAFs of <10% showing equal or worse outcomes compared with patients with VAFs of >10%. Although the number of patients in these groups is small, our data suggest that a 10% VAF cutoff may not, in isolation, sufficiently discriminate between different categories of disease. However, the absence of VAF criteria in the WHO5 may also be insufficient. Ultimately, the significance of TP53 mutations relates to the loss of normal TP53 protein function, which is often evidenced by karyotypic complexity. Thus, incorporation of cytogenetic data may help delineate the significance of low VAF TP53 mutations, as has been suggested previously.5 Indeed, none of the patients with CCUS in our cohort showed complex karyotypes, despite 3 of 6 patients having TP53 VAFs of >10%. The functional status of the TP53 protein in these cases is thus likely distinct from that in patients with a relatively low VAF TP53 mutation but with a highly complex karyotype. Additional studies with larger numbers of patients are therefore needed to determine which criteria, including VAF cutoffs and/or cytogenetic findings, among others, are necessary to best interpret these cases. With the designation of new categories of TP53-mutated myeloid disease in both the ICC and WHO5 classifications, our understanding of these disease entities will undoubtedly continue to grow. Ultimately, the findings presented here may help guide work on future classifications systems.
Authorship
Contribution: S.A.H. and E.F.M. conceived of, and designed, the study, collected and analyzed the data, and wrote the manuscript; and L.A.L and A.C.S. assisted with data collection and manuscript preparation.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Emily F. Mason, Department of Pathology, Microbiology, and Immunology, Vanderbilt University Medical Center, 445 Great Circle Rd, Nashville, TN 37228; email: emily.f.mason@vumc.org.
References
Author notes
Original data are available on request from the corresponding author, Emily F. Mason (emily.f.mason@vumc.org).
The full-text version of this article contains a data supplement.