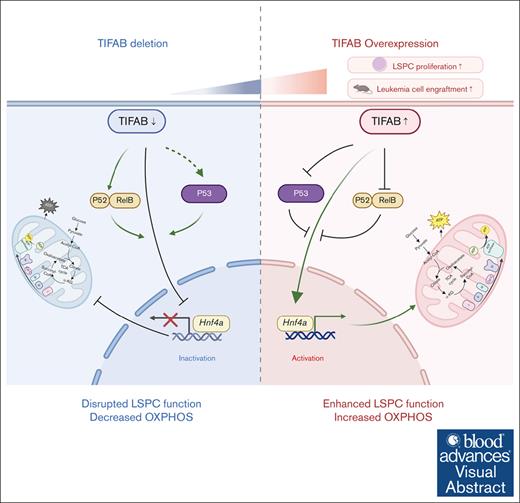
Key Points
TIFAB regulates oxidative phosphorylation in AML stem/progenitor cells.
HNF4A, a newly identified downstream target of TIFAB, rescues the metabolic defects caused by Tifab deletion in AML stem/progenitor cells.
Visual Abstract
Tumor necrosis factor (TNF) receptor–associated factor (TRAF)–interacting protein with forkhead-associated domain B (TIFAB), an inhibitor of NF-κB signaling, plays critical roles in hematopoiesis, myelodysplastic neoplasms, and leukemia. We previously demonstrated that Tifab enhances KMT2A::MLLT3–driven acute myeloid leukemia (AML) by either upregulating Hoxa9 or through ubiquitin-specific peptidase 15–mediated downregulation of p53 signaling. In this study, we show that Tifab deletion in KMT2A::MLLT3–induced AML impairs leukemia stem/progenitor cell (LSPC) engraftment, glucose uptake, and mitochondrial function. Gene set enrichment analysis reveals that Tifab deletion downregulates MYC, HOXA9/MEIS1, mTORC1 signaling, and genes involved in glycolysis and oxidative phosphorylation. By comparing genes upregulated in TIFAB-overexpressing LSPCs with those downregulated upon Tifab deletion, we identify hepatocyte nuclear factor 4 alpha (Hnf4a) as a key TIFAB target, regulated through the inhibition of NF-κB component RelB, which suppresses Hnf4a in leukemia cells. HNF4A, a nuclear receptor involved in organ development, metabolism, and tumorigenesis, rescues the metabolic defects caused by Tifab deletion and enhances leukemia cell engraftment. Conversely, Hnf4a knockdown attenuates TIFAB-mediated enhancement of LSPC function. These findings highlight the critical role of the TIFAB-HNF4A axis in KMT2A::MLLT3–induced AML and uncover a novel regulator in leukemia biology.
Introduction
Acute myeloid leukemia (AML) is a highly aggressive hematologic malignancy characterized by uncontrolled proliferation and impaired differentiation of hematopoietic stem and/or progenitor cells (HSPCs).1-6 As the most common form of acute leukemia in adults, AML has seen advancements in the development of small molecules targeting mutated signaling pathways and epigenetic regulators. However, conventional chemotherapy has largely remained unchanged for >3 decades, leading to high mortality rates, frequent relapses, and substantial recurrences within 3 years for many patients.7-11 Unlike other cancers, leukemia stem/progenitor cells (LSPCs) primarily rely on mitochondrial oxidative phosphorylation (OXPHOS) to generate adenosine triphosphate (ATP) for survival.12-14 Consequently, targeting OXPHOS has emerged as a selectively cytotoxic strategy against AML stem cells.13-19
TNF receptor–associated factor 6 (TRAF6) and TRAF-interacting protein with a forkhead-associated domain (TIFA) are key signaling molecules that mediate NF-κB activation. TIFA’s homolog, TIFA B (TIFAB), lacks a TRAF6-binding motif and acts as a negative regulator of TIFA-TRAF6 signaling.20 Recent structural, biochemical, and cell-based analyses have revealed that TIFAB forms a stable heterodimer with TIFA, inhibiting TIFA dimer formation, which is crucial for NF-κB activation, and suppressing TIFA-TRAF6 signaling.21,Tifab is localized within the commonly deleted region on chromosome 5q in both myelodysplastic syndrome and AML.22 Recent studies suggest that the loss of Tifab disrupts normal hematopoiesis by amplifying TLR4-TRAF6 signaling,23 whereas its deletion impairs leukemic cell function and development by enhancing p53 signaling in KMT2A::MLLT3−driven AML. Conversely, increased TIFAB suppresses p53, promoting leukemia cell function through enhanced activity of the ubiquitin-specific peptidase 15.24 Our recent findings demonstrate that enforced TIFAB expression accelerates KMT2A::MLLT3−induced AML by upregulating HOXA9 and enhancing LSPC OXPHOS.25 However, the mechanisms through which TIFAB regulates OXPHOS and LSPC metabolism remains unclear.
Hepatocyte nuclear factor 4 alpha (HNF4A), a transcription factor within the nuclear receptor superfamily, plays critical roles in organ development, metabolism, and tumorigenesis by regulating genes involved in glucose metabolism, OXPHOS, and fatty acid oxidization.26-29 Despite its importance in these processes, HNF4A’s role in AML has been scarcely studied, with only 1 report suggesting its potential involvement in glucose metabolism regulation in AML by suppressing HNF4A expression.30 In this study, we investigate TIFAB’s role in regulating LSPC metabolism and identify Hnf4a as a novel downstream target. Notably, HNF4A overexpression rescues the metabolic deficiencies caused by Tifab deletion, underscoring the significance of TIFAB in AML pathogenesis.
Methods
Mice
All experiments were conducted in accordance with the guidelines set forth by the Institutional Animal Care and Use Committee, under approved protocol 2020-0031. Tifab knockout (KO) mice on a C57BL6 background have been previously described.23 Recipient mice (CD45.1; stock 002014) were purchased from The Jackson Laboratory. All mouse strains used were aged between 8 and 12 weeks, with both male and female mice included.
Mouse leukemia cells
All experiments were performed using leukemia cells derived from mouse KMT2A::MLLT3−induced leukemia, with or without additional retroviral transduction. For the experiments described in supplemental Figure 2, vector- or TIFAB-transduced mouse RUNX1::RUNX1T1−induced AML cells were used.
HSPC and LSPC isolation, proliferation, apoptosis, glucose uptake, and mitochondrial analysis
We used antibodies from Thermo Fisher Scientific and BD Biosciences, including CD3, CD4, CD8, B220, CD11b, Gr-1, Ter119, Sca1, and CD117. For HSPC isolation, defined as lineage−cKit+Sca-1+ cells in this study, whole bone marrow (BM) cells were incubated with a cocktail of biotinylated anti-mouse antibodies targeting CD3, CD45R/B220, CD11b, Gr-1, and Ter119 from BD Biosciences. Lineage depletion was then performed using BD IMag streptavidin particles Plus-DM. Afterward, cells were stained with Sca-1 phycoerythrin (PE), ckit BV421, and streptavidin PE-CF594.
For LSPC proliferation (Ki67+) and apoptosis (caspase-3 positive) analysis, BM cells were first stained with surface markers to define the progenitor population, followed by fixation and permeabilization with Cytofix/Cytoperm (BD Biosciences). Intracellular staining was then conducted using Ki67 PE or caspase-3 PE. In this study, LSPCs were defined as lineage (CD3, CD4, CD8, B220, and Ter119)–negative, Sca1–, Mac-1/CD11b+, and cKit(bright)+ leukemia cells.
To measure glucose incorporation and cellular reactive oxygen species (ROS), 5 × 105 leukemia cells were stained with Mac1-PE or -allophycocyanin (APC) and cKit-BV421 or -A700 to define the progenitor population. Cells were subsequently labeled with 2-NBDG (2-(N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino)-2-deoxyglucose, 10 μM; N13195; Thermo Fisher Scientific), or for green fluorescent protein (GFP)-expressing leukemia cells, using the Glucose Uptake Assay Kit-Red (Dojindo Laboratories). CellROX (0.75 μM; C10422) was used for 30 minutes at 37°C to measure ROS levels. For mitochondrial mass assessment, leukemia cells were incubated with MitoTracker Green or Red (30 nM for 30 minutes; M7514 or M7512; Thermo Fisher Scientific) at 37°C, then stained with Mac1-PE or -APC and cKit-BV421 or -A700 to define the progenitor population. Mitochondrial membrane potential was assessed by first staining leukemia cells with Mac1-APC and cKit-BV421 to define the progenitor population, followed by staining with tetramethylrhodamine ethyl ester (TMRE) (20 nM for 30 minutes; T669; Thermo Fisher Scientific).
All analyses were performed using the LSRII flow cytometer, and cell sorting was conducted with the FACSAria. Data were analyzed using FlowJo software (BD Life Sciences).
DNA constructs and virus production
The plasmid murine stem cell virus (pMSCV)-KMT2A::MLLT3-internal ribosome entry site (IRES)-GFP has been previously described.31,32 The murine Hnf4a plasmid was purchased from Addgene (catalog no. 33002) and subcloned into the BamH1/EcoRI sites of pMSCV-IRES-mCherry using In-Fusion cloning, following the manufacturer’s instructions (Takara Bio). pLKO-mCherry was used for Hnf4a knockdown.31 The sequences for Hnf4a knockdown are as follows: (1) forward: 5′–CACCG AACCCTTGCCGGCATGGATA–3′; reverse: 5′–AAAC TATCCATGCCGGCAAGGGTT C–3′; (2) forward: 5′–CACCG GTAGTCGGCCATATCCATGC–3′; reverse: 5′–AAAC GCATGGATATGGCCGACTAC C – 3′; and (3) forward: 5′–CACCG GGTCGCCACAGATGGCGCAC–3′; reverse: 5′–AAAC GTGCGCCATCTGTGGCGACC C–3′. Transient transfections of 293T cells were performed using Lipofectamine 3000 to cotransfect the MSCV vectors with pCL-Eco or the pLKO vectors with psPAX2 and pMD2.G for retrovirus or lentivirus production.
Western blotting
Leukemia cells (CD11b+cKit+) were lysed in 1× lysis buffer (10Χ RIPA Buffer; Cell Signaling Technologies; catalog no. 9806S) with protease inhibitors. A total of 40 μg of cellular lysates were loaded onto 4% to 15% precast polyacrylamide gels (Bio-Rad; catalog no. 4568084), transferred to polyvinylidene fluoride membranes, and immunoblotted with antibodies against HNF4A (ab41898, Abcam), TIFAB (PA5-24376, Invitrogen), NDUFA9 (ab14713), SDHA (ab14715), SDHB (ab14714), SDHC (ab15999), SDHD (ab189945), UQCRC2 (ab14745), MTCO1 (ab14705), ATP5B (ab14730) (all from Abcam), or Beta-ACTIN (Santa Cruz). Membranes were imaged using the ChemiDoc Touch Imaging system (Bio-Rad) and quantified with ImageJ software. All experiments were replicated 3 times.
Seahorse assays
Seahorse Mito Stress Test, Seahorse Glycolysis Stress Test, and Seahorse Mito Fuel Flex Test were conducted using a 96-well Seahorse Bioanalyzer XF 96 according to the manufacturer's instructions (Agilent Technologies). Data analysis was performed using Wave Desktop software (Agilent Technologies). For all assays, CD11b+cKit+ leukemia cells (1 × 105 cells per well; 5 replicates) were seeded in poly-D-lysine (Sigma, P6407)–coated 96-well XF96 microplates (Agilent Technologies, 102417-100). Cells were incubated in 180 μL of the corresponding assay medium in a 37°C non-CO2 incubator for 1 hour before the assay.
For the Mito Stress Test, assay medium was prepared by supplementing Seahorse XF RPMI medium with 1-mM pyruvate, 2-mM glutamine, and 10-mM glucose. Mitochondrial function was analyzed by measuring the oxygen consumption rate (OCR) under basal conditions and after sequential injection of 1.5-μM oligomycin (Sigma-Aldrich; 871744), 1-μM carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP) (Sigma-Aldrich; C2920), 1-μM antimycin A (Sigma-Aldrich; catalog no. A8774), and 1-μM rotenone (Sigma-Aldrich; R8875).
For the glycolysis stress test, assay medium was prepared by supplementing Seahorse XF RPMI with 2-mM glutamine. Glycolytic function was analyzed by measuring the extracellular acidification rate (ECAR) under basal conditions and after sequential injection of 10-mM glucose (Agilent; 103577-100), 1.5-μM oligomycin (Sigma-Aldrich; catalog no. 871744), and 50-mM 2-deoxy-D-glucose (Sigma-Aldrich; catalog no. D6134).
For the Mito Fuel Flex Test, assay medium was prepared by supplementing Seahorse XF RPMI medium with 1-mM sodium pyruvate, 2-mM L-glutamine, and 10-mM glucose. Mitochondrial fuel use was assessed by measuring OCR in the presence or absence of specific fuel pathway inhibitors. The 3 major metabolic substrates (pyruvate, fatty acids, and glutamine) were targeted using UK5099 (2 μM; mitochondrial pyruvate carrier inhibitor), etomoxir (4 μM; carnitine palmitoyltransferase 1A inhibitor), and Bis-2-(5-phenylacetamido-1,3,4-thiadiazol-2-yl)ethyl sulfide (BPTES) (3 μM; glutaminase inhibitor). Fuel dependency was assessed by sequentially inhibiting the pathway of interest, followed by the 2 alternative pathways. Fuel capacity was determined by inhibiting the 2 alternative pathways, followed by the pathway of interest. Fuel Flexibility was calculated by subtracting the fuel dependency from the fuel capacity for each pathway of interest.
13C6-glucose tracing analysis
CD11b+cKit+ leukemia cells (2 × 106 cells per sample, in triplicates) were cultured in glucose-free RPMI-1640 medium supplemented with 2000 mg/L 13C6-glucose for 24 hours. After incubation, the cells were washed 3 times with ice-cold phosphate-buffered saline. The cells were then lysed with ice-cold methanol, water and 1-mM tricarballylic acid in a 40:20:1 ratio and then centrifuged at 14,000 g for 10 min at 4 °C. The supernatant was mixed with 300 μL of chloroform, vigorously vortexed for 30 seconds, and centrifuged at 3000g for 3 minutes at 4°C. The upper polar phase was collected and dried using a SpeedVac. Samples were derivatized with 20 μL of 4% methoxyamine-hydrochloride in pyridine for 30 minutes at 45°C, followed by 25 μL of N-tert-butyldimethylsilyl-N-methyltrifluoroacetamide (mtBSTFA) + 1% trimethylchlorosilane (t-BDMCS) for 60 minutes at 45°C. After centrifugation at 14 000g for 10 minutes at 4°C, the supernatant was collected for analysis by gas chromatography mass spectrometry (GC-MS) using an Agilent 5977B system (Agilent Technologies, Santa Clara, CA) equipped with an HP-5 ms column (Agilent Technologies). The injector temperature was set to 300°C, and 1 μL of each sample was injected. GC temperature ramped from 60 to 325°C at a rate of 6.5°C/min. Helium was used as the carrier gas. Metabolite analysis was performed using electron impact ionization and selected ion monitoring mode. Data were processed with MassHunter software (Agilent Technologies) for metabolite annotation and chromatographic peak integration, whereas IsoCorrectoR was used to correct for natural isotope abundance and calculate the isotopomer distribution for each metabolite.33
In vitro colony-forming assays
Fluorescence-activated cell sorting (FACS)–sorted LSPCs (GFP+Lin–CD11b+cKit+; 100 cells per 24-well plate) from KMT2A::MLLT3 AML mice were plated in methylcellulose media (Methocult M3434; STEMCELL Technologies). Colonies were counted after 5 to 7 days.
In vivo leukemogenesis
Lineage-negative cKit+Sca-1+ BM cells, sorted via flow cytometry from wild-type (WT) or Tifab KO mice, were cultured overnight in StemSpan Medium (StemCell Technologies), supplemented with 50 ng/mL stem cell factor (SCF), 50 ng/mL thrombopoietin (TPO), 50 ng/mL FMS-like tyrosine kinase 3 (FLT3) ligand, and 10 ng/mL interleukin-3 (IL-3) (all from Peprotech, Rocky Hill, NJ). The following day, the cells were plated on retrovirus-loaded, retronectin-coated 6-well plates and spin-infected at 1000g for 90 minutes at room temperature. Two days later, the cells were transplanted retro-orbitally into lethally irradiated (950 cGy; single dose) CD45.1 recipient mice (100 000 cells per recipient), along with 2 × 105 to 3 × 105 radioprotective CD45.1+ BM cells. For secondary transplantation, LSPCs from the spleen or BM of primary recipient mice were sorted and transplanted at a dose of 20 000 cells per recipient into sublethally irradiated (6.5 Gy) CD45.1 mice.
RNA-sequencing (RNA-seq) and transcriptome analysis
Total RNA was extracted from sorted leukemic stem cells (LSCs) (lin−ckit+Sca1−CD16+CD34+) obtained from mice receiving KMT2A::MLLT3 transduced WT or TIFAB KO HSPCs. Complementary DNA synthesis and amplification were performed using the SMARTer Ultra Low Input RNA Kit (Clontech), starting with 20 ng of total RNA per sample, following the manufacturer’s instructions. Libraries were subsequently sequenced on the Illumina NovaSeq 6000 platform in paired-end mode, with a read length of 150 base pairs. Differential expression analysis was performed using the DESeq2 v1.30.1 R package.34 Genes were considered differentially expressed between 2 groups of samples when the DESeq2 analysis yielded an adjusted P value of <.01, and the log2-fold change in gene expression was ≥1.5 or less than or equal to –1.5. Gene set enrichment analysis (GSEA) was conducted according to a previous report using the software downloaded from the GSEA website (http://software.broadinstitute.org).35
Statistical analysis
For data that passed the normality test in Prism software, comparisons between 2 groups were conducted using Student t test and 1-way analysis of variance, followed by Tukey post hoc test, which was used for comparisons among multiple groups. For nonnormally distributed data, 2-group comparisons used the Mann-Whitney U test, whereas multiple group comparisons were assessed with the Kruskal-Wallis test, followed by Dunn post hoc test. Survival analyses were conducted using Kaplan-Meier curves with log-rank tests. Results are presented as mean ± standard error of mean, with statistical significance defined as P value <.05 (∗P < .05; ∗∗P < .01).
Results
Enforced expression of TIFAB heightens LSPC OXPHOS
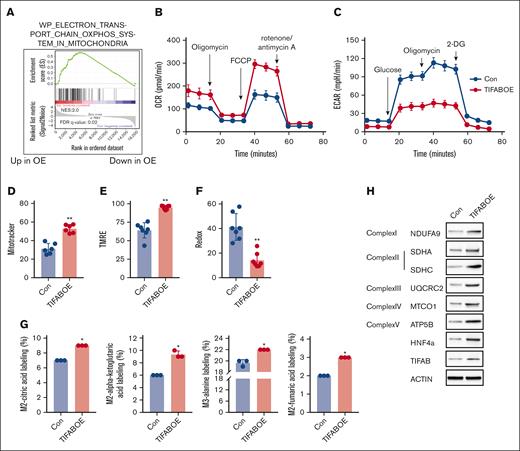
AML stem and progenitor cells rely primarily on mitochondrial OXPHOS for ATP production to support survival.14,36,37 In our recent studies, we demonstrated that enforced TIFAB expression enhances AML cell repopulation by upregulating HOXA9.25 GSEA on TIFAB-overexpressing LSPCs identified the “electron transport chain OXPHOS system in mitochondria” as the most significantly upregulated pathway (Figure 1A). Consistent with this gene expression profile, functional analysis using a Seahorse Bioscience XF96 extracellular flux analyzer showed that, in the murine KMT2A::MLLT3 AML model, TIFAB-overexpressing LSPCs (CD11b+cKit+) exhibited higher OCRs and lower ECARs than control LSPCs expressing an empty vector (Figure 1B-C; supplemental Figure 1A-B).
TIFAB overexpression enhances LSPC OXPHOS activity. (A) GSEA of RNA-seq data comparing vector control (Con) and TIFAB-overexpressing (TIFAB OE) LSPCs isolated from leukemic mice transplanted with either empty vector or TIFAB OE KMT2A::MLLT3 leukemia cells, indicating upregulation of OXPHOS with TIFAB overexpression. (B-C) OCR (Mito Stress test) (B) and ECAR (Glycolysis Stress test) (C) in Con and TIFAB OE LSPCs (n = 5). (D-F) Mitochondrial mass (n = 6) (D), mitochondrial membrane potential (n = 7) (E), and ROS levels (n = 7) (F) were assessed using MitoTracker, tetramethylrhodamine ethyl ester (TMRE), and CellROX, respectively, in Con and TIFAB OE LSPCs. (G) Fractional enrichment of 13C6-labeled intermediate metabolites in Con and TIFAB OE LSPCs, measured by GC/MS (n = 3). ∗P < .05; ∗∗P < .01. Tests used in panels D-G, Mann-Whitney U test. (H) Western blot analysis of key components of the ETC complexes I, II, III, IV, and V, as well as HNF4A and TIFAB in Con and TIFAB OE LSPCs (n = 3). ACTIN served as a loading control. FDR, false discovery rate; NES, normalized enrichment score.
TIFAB overexpression enhances LSPC OXPHOS activity. (A) GSEA of RNA-seq data comparing vector control (Con) and TIFAB-overexpressing (TIFAB OE) LSPCs isolated from leukemic mice transplanted with either empty vector or TIFAB OE KMT2A::MLLT3 leukemia cells, indicating upregulation of OXPHOS with TIFAB overexpression. (B-C) OCR (Mito Stress test) (B) and ECAR (Glycolysis Stress test) (C) in Con and TIFAB OE LSPCs (n = 5). (D-F) Mitochondrial mass (n = 6) (D), mitochondrial membrane potential (n = 7) (E), and ROS levels (n = 7) (F) were assessed using MitoTracker, tetramethylrhodamine ethyl ester (TMRE), and CellROX, respectively, in Con and TIFAB OE LSPCs. (G) Fractional enrichment of 13C6-labeled intermediate metabolites in Con and TIFAB OE LSPCs, measured by GC/MS (n = 3). ∗P < .05; ∗∗P < .01. Tests used in panels D-G, Mann-Whitney U test. (H) Western blot analysis of key components of the ETC complexes I, II, III, IV, and V, as well as HNF4A and TIFAB in Con and TIFAB OE LSPCs (n = 3). ACTIN served as a loading control. FDR, false discovery rate; NES, normalized enrichment score.
Further investigation into mitochondrial function showed an increase in mitochondrial mass (MitoTracker; Figure 1D), heightened mitochondrial membrane potential (tetramethylrhodamine ethyl ester; Figure 1E), and reduced levels of ROS (CellROX; Figure 1F) in TIFAB-expressing LSPCs compared with controls. To explore the mechanism by which TIFAB overexpression promotes OXPHOS, we performed an isotope tracing experiment using 13C6-glucose as a tracer and monitored tricarboxylic acid (TCA) cycle metabolites using GC/MS in LSPCs overexpressing TIFAB or control vector. TIFAB overexpression led to an increase in levels of glucose-derived carbon utilization in the TCA cycle, as demonstrated by the increased fractional enrichment of Mass plus 2 (M2)-citric acid, M2-alpha ketoglutaric acid, M2-fumaric acid, and M3-alanine, compared with the control cells (Figure 1G). The TIFAB-overexpressing leukemia cells were generated by transducing KMT2A::MLLT3 LSPCs with MSCV-TIFAB-IRES-mCherry. To assess metabolic differences between high and low TIFAB-expressing leukemia cells, we sorted TIFAB-expressing LSPCs into mCherry-high (top 25%) and mCherry-low (bottom 25%) populations, because mCherry expression correlates with TIFAB levels, following a previously described strategy38 (supplemental Figure 1C). Our results demonstrated that TIFAB-high leukemia cells had higher OXPHOS activity/OCR and lower glycolysis/ECAR than TIFAB-low cells (supplemental Figure 1D-G). Additionally, 13C6-glucose labeling experiments revealed that TIFAB-high leukemia cells exhibited increased fractional enrichment of M3-pyruvic acid, M3-lactic acid, and M3-alanine compared with TIFAB-low cells, indicating greater glucose use for OXPHOS than TIFAB-low cells (supplemental Figure 1H).
To elucidate TIFAB’s influence on the TCA cycle, we assessed the expression of key components of the electron transport chain (ETC) complexes I to V using western blot. Our data indicate that TIFAB overexpression significantly elevated multiple ETC complex components (Figure 1H; supplemental Figure 1I). These findings demonstrate that TIFAB enhances OXPHOS at multiple levels, contributing to increased LSPC functionality.
We further examined whether TIFAB upregulation exerts similar metabolic effects across different AML subtypes. Using a murine model of AML driven by RUNX1::RUNX1T1 (AE9a isoform),38,39 we found that forced expression of TIFAB increased the frequency of LSPCs (defined as lin–sca1–cKit+ in this model) and enhanced OXPHOS while reducing glycolysis (supplemental Figure 2). This indicates that TIFAB broadly influences LSPC metabolism across AML subtypes.
Tifab deletion disrupts the multifaceted cellular functions of LSPCs
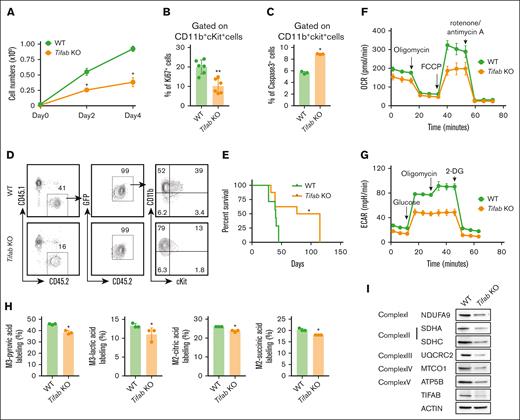
To determine whether TIFAB serves as a critical regulator of LSPC OXPHOS and its associated cellular and molecular mechanisms, we generated KMT2A::MLLT3−driven AML using Tifab KO lineage-negative Sca1+cKit+ cells, as previously described.24,31 Consistent with prior findings,24,Tifab deletion significantly inhibited AML cell proliferation, as demonstrated by cell counting and Ki67 staining (Figure 2A-B), and increased LSPC apoptosis (Figure 2C). Notably, Tifab deletion delayed LSPC engraftment and extended the survival of mice that receiving transplant (Figure 2D-E). In contrast to the elevated OCR and reduced glycolysis/ECAR observed in TIFAB-overexpressing LSPCs, Tifab-ablated LSPCs displayed reduced OCR and ECAR (Figure 2F-G; supplemental Figure 3A-B). We also performed the Seahorse fuel flex test to assess the mitochondrial capacity to oxidize glucose, amino acids, and fatty acids. Our data suggest a trend, although not statistically significant, indicating that Tifab-deleted LSPCs increase their dependence on fatty acids to compensate for reduced reliance on glucose and amino acids40,41 (supplemental Figure 3C). In contrast, TIFAB-overexpressing LSPCs appeared to depend more on glucose and amino acids for OXPHOS (supplemental Figure 3D). Furthermore, 13C6-glucose labeling indicated that Tifab deletion led to decreased levels of glucose-derived carbon utilization, demonstrated by the reduced M3-pyruvic acid, M3-lactic acid, M2-citric acid, and M2-succinic acid compared with controls (Figure 2H). The absence of Tifab also resulted in reduced expression of key ETC components, spanning complex I to V (Figure 2I; supplemental Figure 3E). These findings align with those seen in TIFAB-overexpressing LSPCs (Figure 1), further supporting the role of TIFAB as a positive regulator of LSPC OXPHOS.
Tifab deletion impairs cellular functions of LSPCs. (A) Cell counting of WT and Tifab KO LSPCs. Ten thousand CD11b+cKit+ leukemia cells were plated and counted every other day for 4 days. (B) Percentage of Ki67+ LSPCs in mice that received transplant with WT or Tifab KO KMT2A::MLLT3 leukemia cells. (C) Percentage of caspase-3–positive LSPCs in WT and Tifab KO LSPCs. (D) Representative flow cytometry profiles of mice engrafted with WT or Tifab KO LSPCs, assessed 8 weeks after transplantation. (E) Survival curve of sublethally irradiated recipient mice receiving 20 000 sorted LSPCs from WT or Tifab KO KMT2A::MLLT3 leukemic mice (WT, n = 7; Tifab KO, n = 8). (F-G) OCR (F) and ECAR (G) in WT and Tifab KO LSPCs (n = 5). (H) Fractional enrichment of 13C6-labeled intermediate metabolites in Con and Tifab KO LSPCs, measured by GC/MS (n = 3). (I) Western blot analysis of key components of ETC complexes I, II, III, IV, and V, along with TIFAB, in WT and Tifab KO LSPCs (n = 3). ACTIN was used as a loading control. ∗P < .05; ∗∗P < .01. Statistical test used in panels A-C and H was Mann-Whitney U test; for panel E, log-rank test.
Tifab deletion impairs cellular functions of LSPCs. (A) Cell counting of WT and Tifab KO LSPCs. Ten thousand CD11b+cKit+ leukemia cells were plated and counted every other day for 4 days. (B) Percentage of Ki67+ LSPCs in mice that received transplant with WT or Tifab KO KMT2A::MLLT3 leukemia cells. (C) Percentage of caspase-3–positive LSPCs in WT and Tifab KO LSPCs. (D) Representative flow cytometry profiles of mice engrafted with WT or Tifab KO LSPCs, assessed 8 weeks after transplantation. (E) Survival curve of sublethally irradiated recipient mice receiving 20 000 sorted LSPCs from WT or Tifab KO KMT2A::MLLT3 leukemic mice (WT, n = 7; Tifab KO, n = 8). (F-G) OCR (F) and ECAR (G) in WT and Tifab KO LSPCs (n = 5). (H) Fractional enrichment of 13C6-labeled intermediate metabolites in Con and Tifab KO LSPCs, measured by GC/MS (n = 3). (I) Western blot analysis of key components of ETC complexes I, II, III, IV, and V, along with TIFAB, in WT and Tifab KO LSPCs (n = 3). ACTIN was used as a loading control. ∗P < .05; ∗∗P < .01. Statistical test used in panels A-C and H was Mann-Whitney U test; for panel E, log-rank test.
Tifab deletion disrupts multiple signaling pathways regulating LSPC function
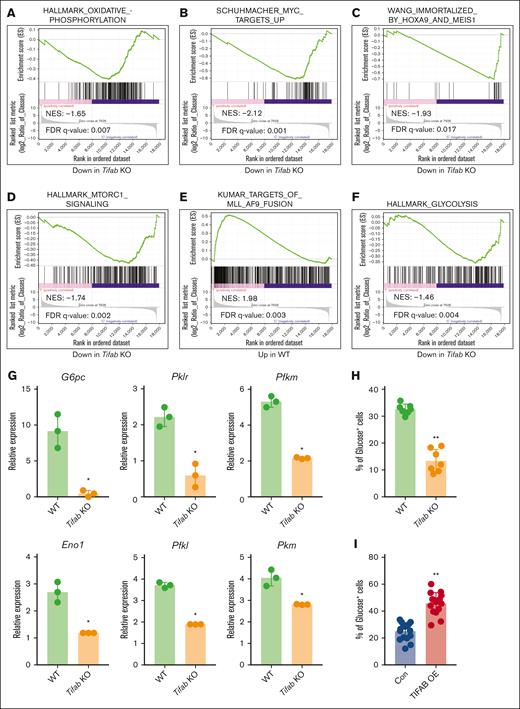
To investigate the molecular mechanisms by which Tifab deletion impairs leukemogenesis, we performed RNA-seq on FACS-sorted control and Tifab-deleted LSPCs from mice with fully developed leukemia. GSEA revealed that Tifab deletion downregulated OXPHOS, MYC targets, HOXA9/MEIS signaling, mTORC1 signaling, and KMT2A::MLLT3 targets (Figure 3A-E), all of which are crucial for AML development and LSPC function. These findings align with the observed reductions in OCRs/ECARs and intermediate metabolites, as shown in Figure 2. Additionally, Tifab ablation downregulated genes related to glycolysis (Figure 3F), which are essential for KMT2A::MLLT3−induced AML, in which LSPCs use glucose via OXPHOS.42,43 We validated the downregulation of glycolysis-related gene expression in Tifab-deleted LSPCs compared with controls (Figure 3G). Moreover, Tifab deletion impaired LSPC glucose uptake, as demonstrated by reduced glucose incorporation (Figure 3H). Although Tifab deletion would be expected to shift metabolism from OXPHOS to glycolysis, the data suggest that glycolytic compensation is impaired due to the downregulation of glycolysis-related genes. Conversely, TIFAB overexpression enhanced glucose uptake (Figure 3I). Collectively, these findings suggest that Tifab deletion impairs multiple genes and pathways involved in LPSC maintenance and metabolism.
Tifab deletion disrupts signaling pathways regulating LSPC function. (A-F) GSEA of RNA-seq data comparing WT and Tifab KO LSPCs. Tifab KO LSPCs exhibit downregulation of gene sets associated with OXPHOS (A), Myc targets (B), HOXA9 and MEIS1 signaling (C), MTORC1 signaling (D), KMT2A::MLLT3 fusion targets (E), and glycolysis (F), compared with WT LSPCs. (G) Quantitative polymerase chain reaction analysis of glycolysis-related enzyme expression in WT and Tifab KO LSPCs. Representative results from 3 independent experiments, each performed with 3 replicates. (H-I) Glucose uptake in WT and Tifab KO (n = 7) (H) or Con and TIFAB OE (n = 16) LSPCs (I). ∗P < .05; ∗∗P < .01. In panel G, Mann-Whitney U test was used; in panels H-I, Student t test was used. FDR, false discovery rate; NES, normalized enrichment score.
Tifab deletion disrupts signaling pathways regulating LSPC function. (A-F) GSEA of RNA-seq data comparing WT and Tifab KO LSPCs. Tifab KO LSPCs exhibit downregulation of gene sets associated with OXPHOS (A), Myc targets (B), HOXA9 and MEIS1 signaling (C), MTORC1 signaling (D), KMT2A::MLLT3 fusion targets (E), and glycolysis (F), compared with WT LSPCs. (G) Quantitative polymerase chain reaction analysis of glycolysis-related enzyme expression in WT and Tifab KO LSPCs. Representative results from 3 independent experiments, each performed with 3 replicates. (H-I) Glucose uptake in WT and Tifab KO (n = 7) (H) or Con and TIFAB OE (n = 16) LSPCs (I). ∗P < .05; ∗∗P < .01. In panel G, Mann-Whitney U test was used; in panels H-I, Student t test was used. FDR, false discovery rate; NES, normalized enrichment score.
Hnf4a is a downstream target of TIFAB, negatively regulated by NF-κB
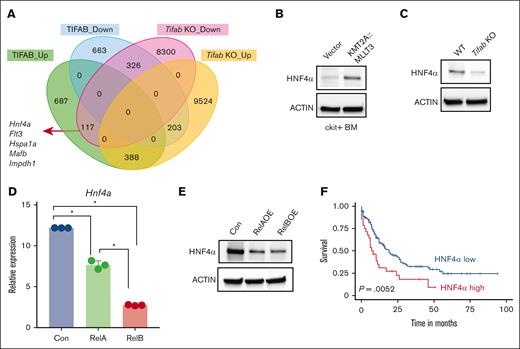
To identify genes directly influenced by TIFAB that are crucial in LSPC maintenance and metabolism, we analyzed RNA-seq data comparing genes upregulated by TIFAB overexpression and downregulated by Tifab deletion. Using a significance threshold of P value <.01 and log2 fold change less than –1.5 or >1.5, we found that most gene expression changes attributed to TIFAB were indirect, with only 117 genes meeting these criteria (Figure 4A; supplemental Table 1). Among these, Hnf4α emerged as a relevant candidate. Hnf4a encodes HNF4A, a nuclear receptor known to regulate genes involved in glucose metabolism, OXPHOS, organ development, and cell differentiation, particularly in liver and intestinal epithelial cells.26,27
Hnf4a is a downstream target of TIFAB, negatively regulated by NF-κB. (A) Venn Diagram illustrating genes upregulated or downregulated in TIFAB OE KMT2A::MLLT3 LSPCs (compared with vector-expressed, GSE178853) and Tifab KO LSPCs (compared with WT). Among these genes, 117 were upregulated in TIFAB OE LSPCs while downregulated in Tifab KO LSPCs, including Hnf4a. (B-C) Expression of HNF4A protein in vector- and KMT2A::MLLT3−transduced cKit+ WT BM cells (B) or WT and Tifab KO LSPCs (n = 3). ACTIN served as a loading control. (D-E) Hnf4a mRNA (D) or protein (E) levels in KMT2A::MLLT3 LSPCs expressing either vector, RelA, or RelB. ∗P < .05, Mann-Whitney U test. (F) Higher expression of HNF4A mRNA in patients with AML (TCGA database) correlates with worse survival. P = .0052, log-rank test.
Hnf4a is a downstream target of TIFAB, negatively regulated by NF-κB. (A) Venn Diagram illustrating genes upregulated or downregulated in TIFAB OE KMT2A::MLLT3 LSPCs (compared with vector-expressed, GSE178853) and Tifab KO LSPCs (compared with WT). Among these genes, 117 were upregulated in TIFAB OE LSPCs while downregulated in Tifab KO LSPCs, including Hnf4a. (B-C) Expression of HNF4A protein in vector- and KMT2A::MLLT3−transduced cKit+ WT BM cells (B) or WT and Tifab KO LSPCs (n = 3). ACTIN served as a loading control. (D-E) Hnf4a mRNA (D) or protein (E) levels in KMT2A::MLLT3 LSPCs expressing either vector, RelA, or RelB. ∗P < .05, Mann-Whitney U test. (F) Higher expression of HNF4A mRNA in patients with AML (TCGA database) correlates with worse survival. P = .0052, log-rank test.
We observed that KMT2A::MLLT3 transduction increased HNF4A expression in cKit+ BM cells compared with those transduced with the empty vector (Figure 4B; supplemental Figure 4A). Deletion of Tifab resulted in decreased HNF4A protein levels (Figure 4C; supplemental Figure 4B), whereas TIFAB overexpression led to increased HNF4A protein levels (Figure 1H). Because TIFAB acts as an inhibitor of the NF-κB pathway,20 we explored whether NF-κB signaling mediates TIFAB’s regulation of HNF4A. We measured Hnf4a messenger RNA (mRNA) levels in KMT2A::MLLT3 LSPCs expressing either vector, RelA (V-Rel Avian Reticuloendotheliosis Viral Oncogene Homolog A, NF-kB subunit), or RelB (V-Rel Avian Reticuloendotheliosis Viral Oncogene Homolog B, NF-kB subunit). RelB expression more profoundly inhibited Hnf4a mRNA than RelA (Figure 4D), a finding confirmed at the protein levels (Figure 4E; supplemental Figure 4C). These results suggest that TIFAB regulates HNF4A primarily through inhibition of RelB.
Given the unclear role of HNF4A in AML, we analyzed the The Cancer Genome Atlas (TCGA) AML data set and found a correlation between elevated HNF4A expression and poor prognosis (Figure 4F). Analysis of TIFAB and HNF4A expression across AML subtypes revealed variability, with a trend toward correlation in certain subtypes, such as AML with complex karyotype, normal karyotype, and KMT2A::MLLT3 (supplemental Figure 4D). However, the low number of cases limited our ability to draw definitive conclusions. Collectively, these data indicate that TIFAB regulates HNF4A through NF-κB inhibition, and HNF4A levels are associated with AML prognosis.
HNF4A overexpression restores Tifab deletion−induced LSPC dysfunction
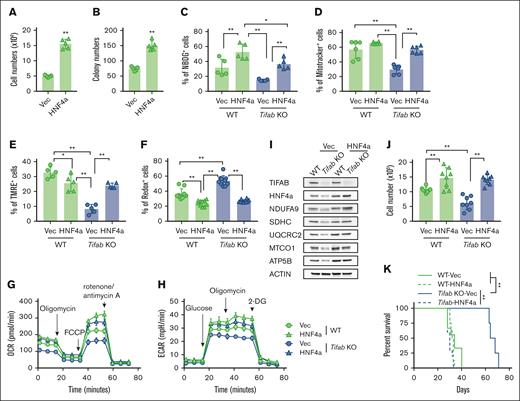
To assess HNF4A’s functional role, we overexpressed HNF4A in KMT2A::MLLT3 LSPCs, which enhanced leukemia growth and colony formation in vitro (Figure 5A-B). We further examined whether HNF4A could compensate for the metabolic defects caused by Tifab deletion. HNF4A overexpression significantly enhanced glucose uptake (Figure 5C) and restored mitochondrial mass (Figure 5D), membrane potential (Figure 5E), ROS levels (Figure 5F), as well as the OCR and ECAR in Tifab-deleted LSPCs (Figure 5G-H; supplemental Figure 5A-B). HNF4A also mitigated the reduction in ETC component expression in TIFAB-deleted LSPCs, although it had minimal effects or only mildly increased expression in WT LSPCs (Figure 5I; supplemental Figure 5C). HNF4A overexpression enhanced cell proliferation and rescued the proliferation defect caused by Tifab deletion (Figure 5J). Additionally, enforced HNF4A expression improved leukemia development, restoring the defect caused by Tifab deletion (Figure 5K). In contrast, Hnf4a knockdown impaired engraftment in a competitive in vivo assay, with a more pronounced effect observed in control LSPCs than in TIFAB-overexpressing cells (supplemental Figure 5D-E). These findings suggest KMT2A:MLLT3 leukemia cells depend on HNF4a, and TIFAB’s leukemia-promoting effects are at least partially mediated by HNF4A. Collectively, our data establish HNF4A as a key downstream target of TIFAB and a critical regulator of LSPC metabolism.
HNF4A overexpression rescues Tifab deletion–induced LSPC dysfunction. (A-B) HNF4A overexpression increases LSPC proliferation (n = 5) (A) and enhances colony formation (n = 5) (B). (C) Glucose uptake assessed by NBDG incorporation in WT or Tifab KO LSPCs transfected with either vector- (Vec) or HNF4A-expressing viruses (n = 5). (D-F) Mitochondrial mass (n = 5) (D), mitochondrial membrane potential (n = 5) (E), and ROS levels (n = 10) (F) in WT or Tifab KO LSPCs transfected with either Vec- or HNF4A-expressing viruses, measured using MitoTracker, TMRE, and CellROX, respectively. (G-H) OCR (G) and ECAR (H) in WT or Tifab KO LSPCs transfected with either Vec- or HNF4A-expressing viruses (n = 5). (I) Western blot analysis of key components of ETC complexes I, II, III, IV, and V, as well as TIFAB and HNF4A, in WT and Tifab KO LSPCs transfected with either Vec- or HNF4A-expressing viruses (n = 3). ACTIN served as a loading control. (J) Proliferation of WT or Tifab KO LSPCs transfected with either Vec- or HNF4A-expressing viruses (n = 8). (K) Survival curve of sublethally irradiated recipient mice transplanted with Vec- or HNF4A-transduced WT or Tifab KO KMT3A::MLLT3 LSPCs (n = 8 in each group). ∗P < .05; ∗∗P < .01. Statistical tests used in panels A-B, Mann-Whitney U test; in panels C-F and J, 1-way analysis of variance and Tukey multiple comparisons test; in panel K, log-rank test.
HNF4A overexpression rescues Tifab deletion–induced LSPC dysfunction. (A-B) HNF4A overexpression increases LSPC proliferation (n = 5) (A) and enhances colony formation (n = 5) (B). (C) Glucose uptake assessed by NBDG incorporation in WT or Tifab KO LSPCs transfected with either vector- (Vec) or HNF4A-expressing viruses (n = 5). (D-F) Mitochondrial mass (n = 5) (D), mitochondrial membrane potential (n = 5) (E), and ROS levels (n = 10) (F) in WT or Tifab KO LSPCs transfected with either Vec- or HNF4A-expressing viruses, measured using MitoTracker, TMRE, and CellROX, respectively. (G-H) OCR (G) and ECAR (H) in WT or Tifab KO LSPCs transfected with either Vec- or HNF4A-expressing viruses (n = 5). (I) Western blot analysis of key components of ETC complexes I, II, III, IV, and V, as well as TIFAB and HNF4A, in WT and Tifab KO LSPCs transfected with either Vec- or HNF4A-expressing viruses (n = 3). ACTIN served as a loading control. (J) Proliferation of WT or Tifab KO LSPCs transfected with either Vec- or HNF4A-expressing viruses (n = 8). (K) Survival curve of sublethally irradiated recipient mice transplanted with Vec- or HNF4A-transduced WT or Tifab KO KMT3A::MLLT3 LSPCs (n = 8 in each group). ∗P < .05; ∗∗P < .01. Statistical tests used in panels A-B, Mann-Whitney U test; in panels C-F and J, 1-way analysis of variance and Tukey multiple comparisons test; in panel K, log-rank test.
Discussion
TIFAB was initially identified as an inhibitor of NF-κB signaling.20 More recently, it has been shown to form a stable heterodimer with TIFA, inhibiting TIFA dimer formation and suppressing TIFA-TRAF6 signaling, ultimately leading to the inhibition of NF-κB activation.21 Consistent with this, Tifab deletion disrupts normal hematopoiesis by intensifying TLR4-TRAF6 signaling.23 TIFAB’s role in AML has also become evident more recently. For instance, its deletion impairs leukemia development and leukemia cell function by augmenting p53 signaling,24 whereas forced expression of TIFAB upregulates HOXA9 in KMT2A::MLLT3−induced AML.25
In this study, we reveal a new role for TIFAB in regulating LSPC metabolism. GSEA shows that deletion of TIFAB downregulates several signaling pathways critical for LSPC function, including MYC signaling, HOXA9/MEIS signaling, mTORC1 signaling, and notably, glycolysis and OXPHOS.13,42-51 Our findings are consistent with this analysis, demonstrating that Tifab deletion decreases glucose uptake, glycolysis, OXPHOS, and the expression of ETC complex proteins, whereas forced TIFAB expression enhances metabolic processes in LSPCs. Collectively, these data suggest that TIFAB plays multifaceted roles in LSPCs, with significant implications for AML metabolism.
Previous studies by Starczynowski et al show that patients with AML with the highest levels of TIFAB expression were significantly enriched for mixed-lineage leukemia 1 (MLL)–rearranged leukemia. Its expression is highest in MLL-rearranged leukemic cell lines MV4;11 and RS4;11, compared with other human AML cell lines.24 Our current work expands on this, demonstrating that TIFAB’s role in AML extends beyond AML with KMT2A::MLLT3 fusion, also affecting AML with RUNX1::RUNX1T1. However, whether TIFAB has a broader role in AML stem/progenitor cells warrants further investigation.
Mechanistically, we identify HNF4A as a key effector regulated by TIFAB in LSPCs. HNF4A is known to control the basal expression of many genes involved in glucose transport, glycolysis, and OXPHOS.26,27,52 However, its role in AML remains largely unexplored. A recent report suggests that downregulation of HNF4A may inhibit the leukemia cell glucose metabolism.30 Examination of HNF4A RNA expression in the TCGA data set revealed that a subset of patients with AML expresses relatively high levels of HNF4A, with higher expression correlating with poor prognosis. Furthermore, the expression of HNF4A and TIFAB appears to correlate, although a definitive conclusion is precluded by the low number of cases in the data set. Importantly, our data show that HNF4A promotes LSPC proliferation, increases glucose uptake and OXPHOS, and mitigates the metabolic deficiencies induced by TIFAB deletion. Conversely, Hnf4a knockdown reduces AML cell engraftment, demonstrating that HNF4A is a bone fide downstream target of TIFAB.
We recently demonstrated that TIFAB regulates HOXA9 in KMT2A::MLLT3−induced AML.25 Interesting, jumonji domain containing 1C (JMJD1C), a JmjC-containing H3K9 demethylase, has been shown to cooperate with HOXA9 to enhance cell proliferation and leukemogenesis by upregulating glycolysis and OXPHOS in HOXA9-dependent AML.53 Whether HNF4A influences AML by regulating HOXA9 remains an open question for future studies.
TIFAB is not a transcription factor, so how does it regulate HNF4A expression? TIFAB has been identified as an NF-κB pathway inhibitor, and NF-κB signaling can either positively or negatively regulate HNF4A expression in a context-dependent manner.28 We previously showed that TIFAB overexpression suppresses the NF-κB component RelB.25 Our current data demonstrate that RelB more effectively suppresses HNF4A expression than RelA, suggesting that TIFAB positively regulates HNF4A expression primarily through the suppression of RelB. Because TIFAB also regulates ubiquitin specific peptidase (USP)-mediated p53 signaling,24 and mutant p53 has been shown to enhance HNF4A activity,54 whereas WT p53 inhibits the transactivation function of HNF4A,55 the role of p53 in regulating HNF4a expression in leukemia warrants future investigation. Nevertheless, our data underscore a new role of TIFAB in modulating LSPC metabolism and identify HNF4A as a novel regulator of LSPC metabolism. Further characterization of HNF4A’s role in AML stem/progenitor cells, particularly its regulation of LSPC metabolism, will provide valuable insights into targeting LSPC metabolism.
Acknowledgments
The authors thank Karina Inacio Carvalho (Case Western Reserve University) for assisting with cell sorting.
This research was partially supported by the Translational Research Shared Resource of the Case Comprehensive Cancer Center (P30 CA043703). J.Z. was supported by the Department of Pathology startup funds (Case Western Reserve University and University Hospitals) and the Science and Technology Foundation of Liaoning Province-PhD startup fund (number 2023-BS-109). This work was further supported by the National Cancer Institute of National Institutes of Health (R01CA237006) and National Heart Lung and Blood Institute of National Institutes of Health (R01HL176819), as well as the Department of Pathology startup funds (C.Z.). D.T.S. was supported in part by the National Cancer Institute (R01CA271455 and R01CA275007), National Heart Lung and Blood Institute (R35HL166430), and National Institute of Diabetes and Digestive and Kindney diseases (U54DK126108) of National Institutes of Health, Cincinnati Children’s Hospital Research Foundation, Cancer Free Kids, and Blood Cancer Discoveries Grant program through the Leukemia & Lymphoma Society, The Mark Foundation for Cancer Research, and The Paul G. Allen Frontiers Group.
Authorship
Contribution: Y.W., Y.X., Q.D., J.Z., K.N., M.M., L.F., H.D.C., and N.P. conducted the experiments and analyzed the data; D.T.S. generated the Tifab knockout mice; Y.W. and N.B. conducted the RNA-sequencing and The Cancer Genome Atlas (TCGA) data analyses; D.T.S. contributed to data interpretation and manuscript revision; C.Z. conceived and supervised the study; and Y.W. and C.Z. wrote the paper.
Conflict-of-interest disclosure: D.T.S. serves on the scientific advisory board at Kurome Therapeutics; is a consultant for and/or received funding from Kurome Therapeutics, Captor Therapeutics, Treeline Biosciences, and Tolero Therapeutics; and has equity in Kurome Therapeutics. The remaining authors declare no competing financial interests.
Correspondence: Chen Zhao, Case Western Reserve University, Wolstein Research Building, Room 6523, 2103 Cornell Rd, Cleveland, OH 44106-7288; email: cxz545@case.edu.
References
Author notes
The data were deposited in the National Center for Biotechnology Information’s Sequence Read Archive database (BioProject ID PRJNA1094963).
The full-text version of this article contains a data supplement.